Abstract
Background
Surgical site infections (SSIs), the second most common healthcare-associated infections, increase hospital stay and healthcare costs significantly. Traditional surveillance of SSIs is labor-intensive. Mandatory reporting and new non-payment policies for some SSIs increase the need for efficient and standardized surveillance methods. Computer algorithms using administrative, clinical, and laboratory data collected routinely have shown promise for complementing traditional surveillance.
Methods
Two computer algorithms were created to identify SSIs in inpatient admissions to an urban, academic tertiary-care hospital in 2007 using the International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) diagnosis codes (Rule A) and laboratory culture data (Rule B). We calculated the number of SSIs identified by each rule and both rules combined and the percent agreement between the rules. In a subset analysis, the results of the rules were compared with those of traditional surveillance in patients who had undergone coronary artery bypass graft surgery (CABG).
Results
Of the 28,956 index hospital admissions, 5,918 patients (20.4%) had at least one major surgical procedure. Among those and readmissions within 30 days, the ICD-9-CM-only rule identified 235 SSIs, the culture-only rule identified 287 SSIs; combined, the rules identified 426 SSIs, of which 96 were identified by both rules. Positive and negative agreement between the rules was 36.8% and 97.1%, respectively, with a kappa of 0.34 (95% confidence interval [CI] 0.27–0.41). In the subset analysis of patients who underwent CABG, of the 22 SSIs identified by traditional surveillance, Rule A identified 19 (86.4%) and Rule B identified 13 (59.1%) cases. Positive and negative agreement between Rules A and B within these “positive controls” was 81.3% and 50.0% with a kappa of 0.37 (95% CI 0.04–0.70).
Conclusion
Differences in the rates of SSI identified by computer algorithms depend on sources and inherent biases in electronic data. Different algorithms may be appropriate, depending on the purpose of case identification. Further research on the reliability and validity of these algorithms and the impact of changes in reimbursement on clinician practices and electronic reporting is suggested.
Surgical site infections (SSIs) are the second most common healthcare-associated infections (HAI). An estimated 2–5% of inpatient surgical procedures are complicated with an SSI, adding 7–10 days to the hospital stay and $10 billion in direct and indirect health care expenditure [1]. Surveillance of SSIs and feedback of rates to surgeons reduces the incidence of these infections [2].
The Centers for Disease Control and Prevention's (CDC) National Healthcare Safety Network (NHSN) has developed detailed guidelines for the surveillance and definition of HAIs, including SSIs [3]; however, traditional surveillance using these methods tends to be time-consuming and labor-intensive, requiring a large expenditure of time by infection prevention and control department staff [4]. Computer algorithms to identify SSIs using administrative, clinical, laboratory, and pharmacy data collected routinely have shown promise in enhancing and complementing traditional surveillance of SSIs [5–8]; however, these methods have had various degrees of success [9,10].
Certain algorithms and approaches to using electronic data may be more suitable than others, depending on the purpose of case identification. In this paper, we explore the performance characteristics and underlying principles of two computer algorithms for the identification of SSIs using retrospective data that are collected routinely. We also compare the results of these algorithms with the results of traditional surveillance in patients who underwent coronary artery bypass graft (CABG) surgery.
Materials and Methods
The study was conducted at a 745-bed urban tertiary-care teaching hospital. After approval from the Columbia University Medical Center Institutional Review Board (Federalwide Assurance No. 00002636), we extracted data for all inpatient discharges for the year 2007 from three sources. Administrative data, including admission and discharge data; International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) diagnosis; and procedure codes recorded by hospital-based coders, were obtained from the cost accounting and billing system. Microbiology culture results were extracted from the University's Clinical Data Warehouse, which acts as a repository of laboratory, administrative, and clinical data for research and administrative tasks. Finally, a list of all SSIs in patients who underwent CABG in 2007 was obtained from the Department of Infection Prevention & Control, which performs quarterly surveillance for SSIs after CABG procedures using the CDC-NHSN definitions and methods [3]. Infection preventionists review all positive incision cultures and all re-admissions for up to one year for infections at the surgical site, as required by NHSN because of the presence of sternal wires, which are considered implants.
All patient identifiers were replaced by a unique study identification number. Data extraction and analysis were performed with TOAD for DB2 version 3.1.1 (Quest Software, Aliso Viejo, CA) and SAS version 9.1.3 (SAS Institute, Cary, NC).
Administrative data were screened to identify hospital stays that included a major surgical procedure. Surgical procedures from the list of ICD-9-CM codes of procedures from the Patient Safety Protocol of the NHSN Manual were considered major [11]. Re-admissions within 30 days of discharge were linked to the original hospital stay, and the two stays were considered a single hospital episode. Administrative data were screened for ICD-9-CM codes for post-operative infection not elsewhere classified (998.5), infected post-operative seroma (998.51), and other post-operative infection (998.59) after a major procedure and in subsequent qualifying re-admissions.
Microbiology cultures were identified with key words such as “wound,” “abscess,” “surgical,” “drainage,” “body fluid,” or “surgical specimen” in the specimen source, indicative of a culture taken from a relevant site. Because colony-forming units are not reported routinely for cultures taken from these sites, growth of any organism was considered a positive culture. Two of the co-authors (YF, SH) derived a rule dividing organisms implicated commonly in SSIs into commensals or pathogens, which was applied to the culture results to label them potential colonization or potential infection. In cultures that yielded common skin contaminants, those with at least two positive cultures on separate occasions were considered potential infections.
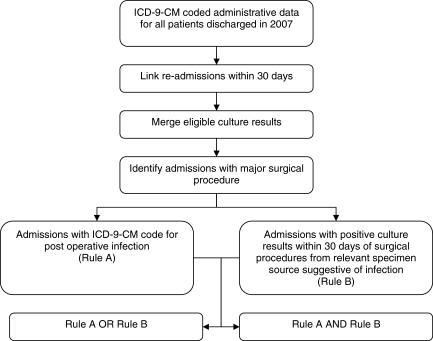
Using these data, two decision rules were formulated and applied independently and together to these hospital stays to identify SSIs. The first rule identified SSIs in linked hospital stays with a major surgical procedure that had ICD-9-CM codes for post-operative infection (Rule A). The second rule identified SSIs in hospital stays with a positive microbiologic culture suggestive of infection from a relevant specimen source within 30 days of a major surgical procedure (Rule B) (Fig. 1).
FIG. 1.
Algorithm to identify surgical site infections using International Classification of Diseases, Ninth Revision, Clinical Modification codes for post-operative infection (Rule A) and culture results (Rule B).
Analysis was conducted in two parts. First, we determined the number of SSIs identified and the percent agreement for the two rules in all patient discharges. Second, we compared the results of these rules with SSIs identified by traditional surveillance in the subset of patients who underwent CABG. For this analysis, discharges were screened for ICD-9-CM codes for CABG only rather than any major surgical procedure. We chose SSIs after CABG to test the validity of the algorithms, as this operation represents a major procedure under surveillance during the study period for which complete data were available. Two of the clinician co-authors conducted a chart review of these data to evaluate the differences between the results of the computer algorithms and surveillance. For discrepant results (SSI identified by traditional surveillance but not by Rule A or B, or vice versa), the electronic medical record was reviewed in detail to identify possible reasons for the discrepancies.
Results
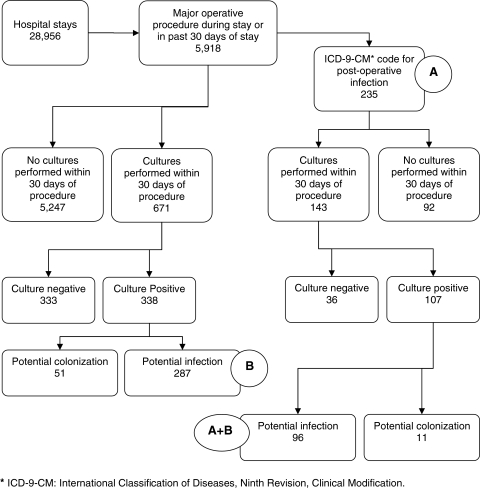
The data set consisted of 33,834 inpatient hospital admissions in 2007 with 28,956 first admissions and 4,878 re-admissions within 30 days. Of these, 5,918 patients (20.4%) had at least one major surgical procedure performed.
In the first part of our analysis, among the 5,918 patients, the ICD-9-CM rule (Rule A) identified 235 SSIs (3.97 per 100 procedures). With regard to Rule B, cultures were sent from a relevant specimen source within 30 days of the procedure in 617 patients (10.43%), 338 (5.71%) of whom had at least one positive culture (Fig. 2). Patients who had undergone heart transplants had the highest rate of cultures obtained within 30 days (40%), followed by those having re-fusion of the spine (20%) or non-transplant cardiac procedures (13.49%). In cultures that grew common skin contaminants, eight patients had at least two separate positive cultures, seven of whom had concurrent growth of pathogenic organisms (data not shown). Of the 338 patients with positive cultures, 287 satisfied the culture-based definition (Rule B) for SSI (4.85 per 100 procedures) (Table 1).
FIG. 2.
Surgical site infections identified using two computer algorithms independently and together.
Table 1.
Surgical Site Infections in Patients Undergoing Major Surgical Procedures Identified Using International Classification of Diseases, Ninth Revision, Clinical Modification Code for Post-Operative Infection (Rule A) or Positive Cultures from Relevant Specimen Sources (Rule B) within 30 Days of Procedure
| |
Culture-based rule (Rule B) |
|
|
|---|---|---|---|
| Positive (% of total) | Negative (% of total) | Total | |
| ICD-9-CM-based rule (Rule A) positive | 96 | 139 | 235 |
| ICD-9-CM-based rule (Rule A) negative | 191 | 5,492 | 5,683 |
| 287 | 5,631 | 5,918 | |
| Kappa=0.34 (95% confidence interval 0.27-0.41) | |||
| Positive agreement 36.8% | |||
| Negative agreement 97.1% | |||
ICD-9-CM=International Classification of Diseases, Ninth Revision, Clinical Modification.
Surgical site infections were identified in 416 patients by the application of either Rule A or Rule B; 96 patients (1.62 per 100 procedures) satisfied both Rule A and Rule B. Positive agreement between Rules A and B was 36.8% and negative agreement was 97.1% with a kappa of 0.34 (95% confidence interval [CI] 0.27–0.41).
In a subset analysis, we compared the results of traditional surveillance with the results of the computer algorithms in patients who underwent CABG surgery in 2007. Of the 22 cases identified by traditional surveillance, Rule A identified 19 (86.4%) (3.29 per 100 procedures), and Rule B identified 13 cases (59.1%), all of which were also Rule A-positive (Table 2). Positive and negative agreement between Rules A and B within these “positive controls” was 81.3% and 50.0% with a kappa of 0.37 (95% CI 0.04–0.70). Rule A and Rule B identified 15 and seven cases, respectively, that were not identified as SSI by traditional surveillance.
Table 2.
Results of Two Computer Algorithms in Patients Identified by Traditional Surveillance as Having Surgical Site Infections after Coronary Artery Bypass Grafting
| |
Culture-based rule (Rule B) |
|
|
|---|---|---|---|
| Positive (% of total) | Negative (% of total) | Total | |
| ICD-9-CM-based rule (Rule A) positive | 13 | 6 | 19 |
| ICD-9-CM-based rule (Rule A) negative | 0 | 3 | 3 |
| 13 | 9 | 22 | |
| Kappa=0.37 (95% confidence interval 0.04–0.70) | |||
| Positive agreement 81.3% | |||
| Negative agreement 50.0% | |||
ICD-9-CM=International Classification of Diseases, Ninth Revision, Clinical Modification.
Three cases were missed by both rules. One patient was found to have an SSI in the physician's private office on follow-up and had no matching admission record in the data set. In a second patient, SSI was diagnosed after the study followup period of 30 days. In the third case, the SSI was diagnosed after the study period of 2007 ended. The culture-based rule missed an additional six cases. Three patients had a single culture with a common skin contaminant, which did not satisfy the positive culture definition of the algorithm, two had no positive cultures, and one patient had positive cultures obtained more than 30 days after the procedure. Of the 22 cases identified by traditional surveillance, only two were diagnosed more than 30 days after the procedure.
Discussion
Identification of SSIs using electronic data collected routinely has several potential uses, including supplementing or aiding traditional surveillance, directing and monitoring quality improvement activities, mandatory reporting, and health services research. Each of these purposes has specific requirements that must be met if SSI identification using electronic data is to prove useful. Use of multiple sources of data in formulating rules to identify infections improves the performance of algorithms [12]. Thus, we augmented ICD-9-CM diagnosis codes with microbiologic culture data and data on surgical procedures in an attempt to increase the utility of these rules for as many purposes as possible.
Short stays and earlier discharges of patients having surgical procedures mean that SSIs are not always diagnosed in the same admission as the procedure. In our dataset, 16/22 SSIs after CABG were diagnosed after discharge, in a subsequent encounter. Most patients with SSIs following CABG at our institution are re-admitted to our hospital (either directly or by transfer from other institutions) for further management. If a patient is admitted to another institution within New York State for SSI management, the institution is required to report the infection to our Department. However, if a patient is managed at a hospital outside the state or as an outpatient, those cases may not be captured, and this is a recognized limitation, as is true at most institutions. Our study was limited further by the fact that we were able to compare rates obtained by electronic surveillance and traditional surveillance by clinicians for only one surgical procedure—CABG. It is possible that there would be variations in the rates of SSI with different procedures. However, even traditional surveillance methods (the gold standard) may yield inaccurate results.
In an earlier version of the rules that did not account for re-admissions, 37% of the admissions with an ICD-9-CM code for post-operative infection did not have a procedure in the same admission, and Rule A and Rule B identified 88 and 41 fewer cases, respectively. Accounting for re-admissions in the final rules allowed us to identify patients with an NHSN operative procedure from a prior admission, aligned our electronic definitions with the NHSN SSI definitions, and prevented over-counting of SSIs in patients with multiple follow-up admissions for post-operative infection. Although ICD-9-CM diagnosis codes are associated with a “present on admission” (POA) indicator [13], this indicator has not been adopted reliably. Improved POA coding is crucial to improve the reliability of electronic data use for the identification of SSIs. Inclusion of microbiologic data, which have precise date–time stamps, made it possible to estimate the time of infection within an encounter, an especially desirable parameter for research into the costs of HAI.
The quality of algorithms to identify SSIs is dependent largely on the availability and accuracy of the data used. The ICD-9-CM codes are assigned for each hospital discharge by expert, trained coders after scrutiny of medical records. In addition, documentation improvement specialists review records to ensure complete and accurate documentation of patients' conditions, including, where indicated, querying physicians for clarification to ensure that the most appropriate ICD-9-CM code is selected. Because ICD-9-CM codes are easily available, they are used widely in health services and health economics research. However, as they are designed primarily for reimbursement purposes, use of ICD-9-CM representations of clinical conditions is prone to misclassification [9,14]. In particular, ICD-9-CM codes for post-operative infection are not always specific to infections at the surgical site. Any algorithms depending solely on ICD-9-CM codes for identification of infection will suffer from these limitations. The newer ICD-10 coding system, scheduled to be rolled out in the U.S. in 2013, has more than five times as many codes as the ICD-9-CM system and thus offers more granularity in describing the etiology and site of infection. However, the single code category for “infection following a procedure” (T81.4) exposes it to some of the same limitations as the ICD-9-CM [15,16].
Microbiologic data are a reliable source for organism identification and date–time stamps. However, specimen sources are not always labeled precisely, leading to the possibility that positive cultures are not related to the surgical site. In creating Rule B, the assumption was made that cultures collected more than 30 days after a procedure were less likely to be related to the surgical site, and hence, these were not included in the algorithm. Nonetheless, we recognize that when an implant is in place, NHSN defines SSIs as occurring as late as one year post-procedure, an event that seemed to be rare according to traditional surveillance of CABG procedures, but which we are missing with our algorithm.
On a more basic level, not all SSIs are identified by cultures; some may be diagnosed on the basis of clinical examination of the surgical site or other signs and symptoms. In institutions in which cultures are obtained more frequently, culture-based Rule B may be the preferable algorithm, but when cultures are obtained less frequently, the ICD-9-CM codes may identify a higher proportion of SSIs. Using either (or both) rules still will result in some misclassification, however.
These advantages and limitations are reflected in our findings. Our ICD-9-CM-based Rule A identified more cases of surveillance-confirmed SSI in CABG patients than did the culture-only Rule B, which did not identify any cases beyond those found by Rule A. However, it also mis-classified more cases as SSI than the more restrictive culture-based rule. To obtain the most accurate SSI rates, prospective in-patient and out-patient surveillance of all surgical patients for the entire at-risk period by expert infection control staff would be required. However, this is neither feasible nor cost effective, nor does it assure that standardized assessments will be used. Hence, the use of electronic data and algorithms such as those we propose here can be helpful and practical for monitoring trends over time and as a supplement to traditional surveillance.
West et al. recently reported similar results; they used the same criteria we did to determine the population at risk; i.e., assignment of ICD-9-CM procedure codes for procedures from the NHSN operative procedures list [17]. However, they used only procedures that were under surveillance by infection control staff during the study period, whereas our list included all NHSN procedures. The secondary ICD-9-CM codes they used were the 140 codes from the Pennsylvania Health Care Cost Containment Councils (PHC4) set for public reporting of HAIs. It is interesting that code 998.59 was important in their analysis. We included this code as the basis for our ICD-9-CM definition of post-operative infection because it was one of the few that were used consistently and reliably. This is similar to the approach of West et al. to include only eight of the 140 most frequently used codes in their models. Hence, our findings are consistent. Their approach using regression models suggested that 998.59 is the best code predictor for SSI, but it did not specifically identify how good. In their previous paper using the same dataset [9] and all PHC4 140 codes, they reported a positive predictive value of 0.42 and a negative predictive value of 0.96 for identifying SSI after CABG surgery. The paper by West et al. also mentioned as a limitation (and suggestion) that post-operative infection codes should be counted only after the procedure. We have done that in this study; that is, our ICD-9-CM rule counts post-operative infection codes only if they were not present on admission in the encounter with the procedure or if they occurred at a subsequent encounter.
Changes in reimbursement policies of the Centers for Medicare and Medicaid Services may provide negative incentives for reporting of untoward events such as SSI. In addition, SSI rates are used as a quality measure, which also provides a disincentive to report all potential SSIs. Unanticipated consequences of these incentives include modifications in ICD-9-CM coding practices or Diagnosis-Related Groups, changes in provider practices regarding the frequency of obtaining microbiologic cultures or prescribing antimicrobial agents, or changes in patterns of surveillance. Monitoring the unintended consequences of payment and reporting incentives has been identified as an important area for future research [18]. Such changes would have unpredictable effects on the reliability and validity of SSI diagnosis, regardless of the surveillance method(s) used.
Because different surveillance strategies are employed and the utility of electronic surveillance differs among institutions, caution is necessary when using SSI rates to compare healthcare quality between institutions. Our results reinforce this caution, because the accuracy of electronic surveillance is highly dependent on the quality of data, the decision rules applied, and local clinical practices. However, as a supplement to traditional surveillance, electronic databases and administrative data show promise for enhancing our ability to identify SSIs accurately.
Conclusions and Recommendations
We found differences in SSI rates employing two algorithms based on electronic data sources. Algorithms using only ICD-9-CM codes may perform better at identifying surveillance-confirmed SSIs in CABG than do culture-only algorithms or a combined algorithm. Despite their limitations, electronic databases and administrative data will be used increasingly to identify adverse events such as SSI. Hence, it is essential to assess how rates of SSI differ depending on the selection of data to be used and the type of procedure and to measure the biases inherent in various datasets. We recommend that each hospital clearly define how SSIs are being identified. We also suggest that further research be conducted on the reliability and validity of various electronic algorithms to identify SSIs and the impact of changes in reimbursement on clinician practices and electronic reporting/recording of SSI data.
Acknowledgments
This study was supported by the National Institute of Nursing Research grant “Distribution of the costs of antimicrobial resistant infections” (R01 NR010822).
Author Disclosure Statement
No conflicting financial interests exist.
References
- 1.Perencevich EN. Sands KE. Cosgrove SE, et al. Health and economic impact of surgical site infections diagnosed after hospital discharge. Emerg Infect Dis. 2003;9:196–203. doi: 10.3201/eid0902.020232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roy MC. Perl TM. Basics of surgical-site infection surveillance. Infect Control Hosp Epidemiol. 1997;18:659–668. doi: 10.1086/647694. [DOI] [PubMed] [Google Scholar]
- 3.Horan TC. Andrus M. Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control. 2008;36:309–332. doi: 10.1016/j.ajic.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Tokars JI. Richards C. Andrus M, et al. The changing face of surveillance for health care-associated infections. Clin Infect Dis. 2004;39:1347–1352. doi: 10.1086/425000. [DOI] [PubMed] [Google Scholar]
- 5.Yokoe DS. Platt R. Surveillance for surgical site infections: The uses of antibiotic exposure. Infect Control Hosp Epidemiol. 1994;15:717–723. doi: 10.1086/646844. [DOI] [PubMed] [Google Scholar]
- 6.Klompas M. Yokoe DS. Automated surveillance of health care-associated infections. Clin Infect Dis. 2009;48:1268–1275. doi: 10.1086/597591. [DOI] [PubMed] [Google Scholar]
- 7.Sands K. Vineyard G. Livingston J, et al. Efficient identification of postdischarge surgical site infections: Use of automated pharmacy dispensing information, administrative data, and medical record information. J Infect Dis. 1999;179:434–441. doi: 10.1086/314586. [DOI] [PubMed] [Google Scholar]
- 8.Spolaore P. Pellizzer G. Fedeli U, et al. Linkage of microbiology reports and hospital discharge diagnoses for surveillance of surgical site infections. J Hosp Infect. 2005;60:317–320. doi: 10.1016/j.jhin.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Stevenson KB. Khan Y. Dickman J, et al. Administrative coding data, compared with CDC/NHSN criteria, are poor indicators of health care-associated infections. Am J Infect Control. 2008;36:155–164. doi: 10.1016/j.ajic.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Leal J. Laupland KB. Validity of electronic surveillance systems: A systematic review. J Hosp Infect. 2008;69:220–229. doi: 10.1016/j.jhin.2008.04.030. [DOI] [PubMed] [Google Scholar]
- 11.National Healthcare Safety Network (NHSN). Atlanta. Centers for Disease Control and Prevention [updated Nov. 1, 2010; cited Dec. 20, 2010] www.cdc.gov/nhsn/library.html www.cdc.gov/nhsn/library.html
- 12.Huang SS. Yokoe DS. Stelling J, et al. Automated detection of infectious disease outbreaks in hospitals: A retrospective cohort study. PLoS Med. 2010;7:e1000238. doi: 10.1371/journal.pmed.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Overview of Hospital-Acquired Conditions (Present on Admission Indicator). Baltimore. Center for Medicare and Medicaid Services [updated Sept. 2, 2010; cited Dec. 20, 2010] www.cms.gov/HospitalAcqCond/ www.cms.gov/HospitalAcqCond/
- 14.Romano PS. Chan BK. Schembri ME. Rainwater JA. Can administrative data be used to compare postoperative complication rates across hospitals? Med Care. 2002;40:856–867. doi: 10.1097/00005650-200210000-00004. [DOI] [PubMed] [Google Scholar]
- 15.Curtis M. Graves N. Birrell F, et al. A comparison of competing methods for the detection of surgical-site infections in patients undergoing total arthroplasty of the knee, partial and total arthroplasty of hip and femoral or similar vascular bypass. J Hosp Infect. 2004;57:189–193. doi: 10.1016/j.jhin.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 16.Gerbier S. Bouzbid S. Pradat E, et al. [Use of the French medico-administrative database (PMSI) to detect nosocomial infections in the University hospital of Lyon] (Fre) Rev Epidemiol Sante Publique. 2011;59:3–14. doi: 10.1016/j.respe.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 17.West J. Khan Y. Murray DM. Stevenson KB. Assessing specific secondary ICD-9-CM codes as potential predictors of surgical site infections. Am J Infect Control. 2010;38:701–705. doi: 10.1016/j.ajic.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 18.Stone PW. Glied SA. McNair PD, et al. CMS changes in reimbursement for HAIs: setting a research agenda. Med Care. 2010;48:433–439. doi: 10.1097/MLR.0b013e3181d5fb3f. [DOI] [PMC free article] [PubMed] [Google Scholar]