Abstract
Metabolomics is an emerging technology that allows researchers to characterize hundreds of small molecules that comprise the metabolome. We sought to determine metabolic differences in depressed and non-depressed subjects. The sample consisted of a depressed group of patients with heart failure enrolled in an NIMH-supported clinical trial of sertraline versus placebo in depressed heart failure patients, and a non-depressed comparator group of heart failure patients. Plasma was obtained from blood samples provided by participants at baseline, and samples were profiled on GC-MS and LC-MS metabolomics platforms for biochemical content. A number of biochemicals were significantly different between groups, with depressed subjects showing higher concentrations of several amino acids and dicarboxylic fatty acids. These results are consistent with prior findings where changes in neurotransmitter systems and fatty acid metabolism were shown to associate with the depressed state. It is unclear what role heart failure may have played in these differing concentrations.
Introduction
Metabolomics tools enable us to study the metabolome, the repertoire of small molecules present in cells and tissue. The identities, concentrations, and fluxes of these substances are the final product of interactions between gene expression, protein expression, and the cellular environment 1–4. The metabolome thus defines a metabolic state as regulated by net interactions between gene and environment influences and provides information that can possibly bridge the gap between genotype and phenotype. Metabolic signatures for such disorders could result in the identification of biomarkers for disease, for disease progression or for response to therapy.1, 5
Major depressive disorder is associated with dysregulation of multiple neurotransmitter systems, including serotonergic and noradrenergic systems6 and other neurotransmitters such as gamma-aminobutyric acid (GABA)7, 8 and glutamic acid.9, 10 Previously, we undertook a metabolomic analyses in a small sample of 9 depressed, 11 remitted and 10 never-depressed older adults.11 Metabolites that were altered in currently depressed patients compared with controls included GABA, glycerol, and several fatty acids, including oleate, stearate, and linoleate. Relative to currently depressed patients, remitted patients showed elevated concentrations of the ketone 3-hydroxybutanoic acid. As noted in a recent review, previous studies have found differences in n-3 and n-6 long-chain polyunsaturated fatty acids in depressed patients.12
In the present study, we sought to expand our previous work, here in a more homogeneous sample of depressed patients enrolled in a pharmacological clinical trial and a similar group of non-depressed controls. We hypothesized that there would be differences between depressed and control groups in both lipid metabolism and in metabolic pathways related to neurotransmitter biosynthesis. Further, we expected to see differences in other metabolites, given the large numbers of compounds examined using this powerful technology.
Methods
Participants
Depressed patients were participants in SADHART-CHF, a NIMH-sponsored, prospective, randomized, double-blind, placebo-controlled study that is designed to assess the safety and efficacy of sertraline in the treatment of heart failure patients with major depression (clinicaltrials.gov registry number NCT00078286). Study participants were primarily recruited from an academic center (Duke University Medical Center, the study’s primary center), and two community hospitals and clinics (Durham Regional Hospital and Alamance Hospital), and their affiliated clinics. The study population consisted of patients who were at least 45 years of age with diagnosis of chronic heart failure of any etiology. Eligible patients met the Diagnostic and Statistical Manual of Mental Disorders Fourth Edition (DSM-IV) criteria for major depressive disorder and had a history of chronic systolic heart failure characterized by left ventricular ejection fraction ≤ 45% and New York Heart Association class ≥ II. They were not taking antidepressant medications at the beginning of the study.
Non-depressed control subjects were recruited from Duke University Medical Center during the period when the above depressed patients were recruited. Controls met the same criteria for heart failure as the depressed subjects.
Recruitment
Depressed subjects
After initial, signed consent, patients with heart failure from the primary recruiting site (Duke University Medical Center) were screened for depressive symptoms using the Beck Depression Inventory Scale (BDI).13 Patients whose BDI scores were equal to or greater than 10 were given a second consent form to sign followed by the Diagnostic Interview Schedule (DIS) a structured psychiatric interview to assess MDD. Patients recruited from the satellite sites (Community hospitals and their clinics) were approached directly for participation in the trial prior to formal BDI screening. Depressed patients also consented to provide a blood sample for future laboratory analyses.
Non-depressed control subjects
After initial, signed consent, patients with heart failure from the primary recruiting site (Duke University Medical Center) were screened for depressive symptoms using the BDI). Patients whose BDI scores were less than 10 were given a second consent form to sign followed by the DIS, a structured psychiatric interview to assess MDD. Informed consent was obtained from each patient in order to examine their medical records for evidence of current or past depression diagnosis or current use of antidepressant medication. Individuals were excluded from the control group if they screened positive for any psychiatric disorder, were currently using antidepressant medication, or had current or past history of depression documented on their chart. Subjects who met the control group criteria also provided written informed consent to obtain a blood sample for metabolomic analysis.
Blood samples
Sample collection
We obtained blood samples from 40 currently depressed individuals and 40 non-depressed control subjects. Samples were collected using a Vacutainer system (Ref#367983) containing clot activator and gel serum separator and were centrifuged using a Sorvall Model RT 7 Plus Centrifuge at a g force of 1864 (3000 rpm for 10 minutes at a fixed angle of 34°). The resulting plasma was aliquoted in cryogenic vial tubes (Fisher Scientific microtube 2mL Catalog#: NC9739217) and frozen at −80° C. Of note, 21 of the controls had blood drawn using Vacutainer tubes (Ref# 362788) containing the chelating agent ethylenediaminetetraacetic acid (EDTA). Once all samples were collected, the tubes were marked with a unique number to blind them and were sent to Metabolon, Inc (Durham, NC), where they were inventoried, immediately stored at −80° C and then extracted using a proprietary solvent extraction method. None of the samples were previously frozen or thawed.
Sample preparation
At the time of analysis, samples were thawed and extracts prepared according to Metabolon’s standard protocol,14 which is designed to remove protein and dislodge small molecules bound to protein or physically trapped in the precipitated protein matrix. All aliquots and remaining samples were refrozen until the time of analysis. Extracts were then split for analysis on the gas chromatography (GC) and liquid chromatography (LC) platforms.
Data collection
Data were collected over a 4-day platform run with samples randomly distributed and balanced for depressed and control groups across each platform day. Also included were 24 technical replicates of process standard samples created from a homogeneous pool containing a mixture of human plasma samples (“MTRX”). The MTRX samples were used to monitor process variation and were treated independently throughout the process as if they were study samples. All process samples (MTRX, GROBs – a mixture of organic components used to assess GC column performance, process blanks, etc.) were spaced evenly among the injections for each day and all experimental samples were randomly distributed throughout each day’s run.
Data normalization and imputation
Data for each individual compound were normalized by calculating the median values for each run. This minimizes any inter-day instrument gain drift, but does not interfere with intra-day sample variability. For a given metabolite, missing values were assumed to be below the limit of detection and were imputed with the observed minimum after the normalization step.
Statistical Analysis
Welch’s two sample t-test and an analysis of covariance (ANCOVA) were used to analyze the data. The statistical analyses were performed on (natural) log-transformed data to account for increases in data variance that occur as the level of response is increased. T-test comparisons were calculated using the program ‘R’ from the Free Software Foundation, Inc (available at http://www.r-project.org). ANCOVA was calculated using JMP version 8 (SAS Institute, Inc, Cary, NC). ANCOVA analyses were used to account for additional variable in the data including subject age, statin use and ejection fraction (EF).
As mentioned earlier, this data set consisted of two sets of control samples, 19 collected in tubes with clot activator and gel for plasma separation and 21 with tubes containing EDTA. Conversely, plasma samples isolated from the depressed subjects were collected with only the clot activator and gel for plasma separation and therefore did not contain EDTA. For the statistical analysis of this data, we initially identified biochemicals that were significantly different within the control group based on the presence or absence of EDTA. Then, for biochemicals with a significant EDTA difference within the control group, we proceeded with the 19 non-EDTA samples in subsequent analyses with the depressed group. For those biochemicals without a significant EDTA difference within the control group, we used all 40 control group subjects in comparison with the depressed group. Random forest classification was used to classify subjects.15 Random forests are based on a consensus of a large number of decision trees which split the data into groups based on the biochemicals which provide the best separation between groups. The error rate from the random forest will give an estimate of how well one can predict new data. Random forest classification results were obtained with the use of the randomForest package in R.16
Two parameters were evaluated when considering statistical significance, namely the p-value and the q-value.17 We used a p-value less than 0.05 for significance coupled with a q-value (describing the false discovery rate, FDR)18 of less than 0.10 as an indication of high confidence in a result. The q-values estimate cumulative FDR in the context of multiple tests.
Results
Plasma samples were obtained from 40 depressed and 40 control subjects with heart failure (see Table 1). Depressed subjects were younger than controls (58.5 and 66.4 years of age, respectively) but the two groups had similar ratios based on sex (about one-quarter female) and race (55% Caucasian). Clinically, the presence of ischemic heart disease was similar between depressed and control subjects. Control subjects had lower ejection fractions, while depressed subjects had higher use of statin drugs.
Table 1.
Characteristics of the sample
| Characteristic | Patient (n = 40) |
Control (n = 40) |
Total (n = 80) |
Statistic, df, p |
|---|---|---|---|---|
| Age, mean years, (std dev) | 58.50 (8.15) | 66.4 (10.75) | 62.45 (10.28) | t = 3.70, 78 df, p = 0.0004 |
| Gender, N (%) Female |
27.50 (11) | 22.50 (9) | 25.00 (20) | χ2 = 0.267, 1 df, p = 0.61 |
| Race, N (%) Caucasian |
55.00 (22) | 55.00 (22) | 55.00 (44) | χ2 = 0.00, 1 df, p = 1.00 |
| Beck Depression Inventory Score, mean (std dev) | 21.3 (7.06) | 4.23 (2.11) | 12.87 (10.04) | t = −14.6, 46.1 df, p <.0001 |
| Cardiac variables | ||||
| Ischemic Heart disease, N (%) | 65.00 (26) | 62.50 (25) | 63.75 (51) | χ2 = 0.05, 1 df, p = 0.82 |
| Ejection Fraction, mean (std dev) | 30.48(8.86) | 24.95 (9.76) | 27.71 (10.13) | t = −2.52, 78 df, p = 0.014 |
| Current use of a Statin Drug, N (%) | 75.00 (30) | 55.00 (22) | 65.00(52) | χ2 = 3.52, 1 df, p = 0.06 |
Metabolomic analysis of the plasma samples obtained from the study population identified 205 known and 218 unknown biochemicals. In the comparison between the depressed and control group, 154 biochemicals were identified that showed statistically significant between-group differences (i.e., p <0.05 and q <0.10). Of the 154 statistically significant biochemicals identified, there were, respectively, 68 and 86 biochemicals that showed fold-increased and fold-decreased concentrations within the depressed subjects when compared with the control subjects. However, when we limited our analysis to only those samples that were collected without EDTA, the comparisons identified 63 significantly different biochemicals: 40 with increased concentrations and 23 with decreased concentrations within depressed subjects compared with their control counterparts.
Depressed versus control comparisons
The initial set of comparisons performed for this study was for the purpose of initially identifying defining metabolites and pathways that are differently regulated in depression. With this goal in mind, numerous differences were initially identified between the depressed and control groups. These differences, detailed below, included biochemicals associated with amino acid metabolism and lipid metabolism.
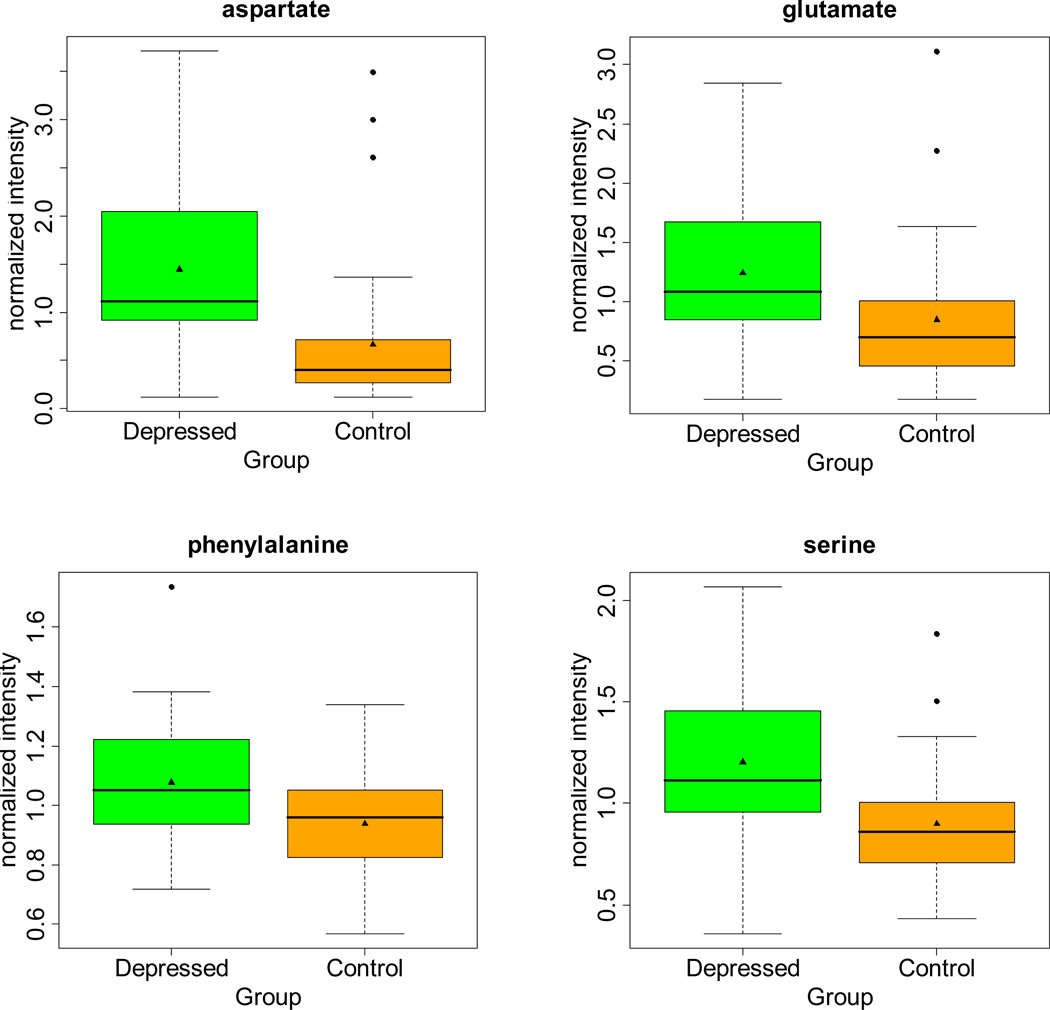
As shown in Table 2 and Figure 1, concentrations of several amino acids were found to differ significantly between depressed and control groups. Within this study, phenylalanine, aspartate acid and serine were among the amino acids that were significantly elevated in depressed subjects compared with controls. In addition, levels of γ-glutamyl amino acids (γ-glutamylleucine and γ-glutamylglutamine) were found to be significantly elevated in depressed subjects in comparison to control subjects. However 3-methyl-2-oxobutyrate, 3-methyl-2-oxovalerate and 4-methyl-2-oxopentanoate, which are associated with valine, leucine and isoleucine metabolism, were not found to be significantly altered in depressed subjects.
Table 2.
Ratio of normalized biochemical concentrations within depressed subjects in comparison with non-depressed controls
| Metabolite | Ratio | P-value | Q-Value |
|---|---|---|---|
| Amino Acids | |||
| Alanine | 1.18 | 0.009 | 0.019 |
| Arginine | 1.56 | <0.001 | 0.001 |
| Aspartate | 2.17 | <0.001 | <0.001 |
| Cystine | 0.69 | 0.009 | 0.019 |
| Dimethylglycine | 0.73 | <0.001 | 0.002 |
| Glutamate | 1.46 | 0.001 | 0.004 |
| Isoleucine | 1.16 | 0.011 | 0.022 |
| Kynurenine | 0.79 | 0.027 | 0.048 |
| Leucine | 1.17 | 0.012 | 0.024 |
| Phenylalanine | 1.15 | 0.004 | 0.010 |
| Pyroglutamine | 0.75 | 0.008 | 0.017 |
| Serine | 1.34 | <0.001 | 0.001 |
| Carbohydrates | |||
| Arabinose | 0.75 | 0.003 | 0.008 |
| Dicarboxylic Fatty Acids | |||
| Azelate (nonanedioate) | 2.28 | <0.001 | <0.001 |
| 3-hydroxysebacate | 1.74 | <0.001 | <0.001 |
| Sebacate | 2.21 | <0.001 | <0.001 |
| Suberate | 1.87 | <0.001 | 0.001 |
| Nucleotide | |||
| Pseudouridine | 0.82 | 0.028 | 0.047 |
| Uridine | 1.18 | 0.043 | 0.061 |
| Peptides | |||
| Aspartylphenylalanine* | 0.191 | 0.003 | 0.086 |
| Gamma-glutamylleucine | 1.23 | 0.024 | 0.042 |
| Glycylvaline | 2.76 | <0.001 | <0.001 |
| Other biochemicals | |||
| Bilirubin | 0.75 | 0.023 | 0.040 |
| Choline | 1.18 | 0.003 | 0.009 |
| Pantothenate | 0.53 | 0.001 | 0.002 |
| Taurocholate | 0.63 | 0.023 | 0.040 |
| Threonate* | 0.54 | 0.001 | 0.066 |
| Urea | 0.72 | 0.001 | 0.003 |
For aspartylphenylalanine and threnoate there was a significant difference in the ratio of concentrations among control samples with and without EDTA. As a result, for these two biochemicals the comparison shown is for samples not containing EDTA.
Figure 1.
Comparisons of amino acid concentrations for patients with depression and non-depressed controls. Concentrations are normalized to mean-scaled values for two laboratory standards.
For analyses related to lipid metabolism we examined concentrations of ketone bodies and dicarboxylic acids (DCAs, see Table 2). There were reduced levels of the ketone body 3-hydroxybutyrate in depressed versus control subjects. A number of DCAs were observed to be elevated in depressed subjects in relation to controls. These include an increase in the levels of azelate, suberate, sebacate and 3-hydroxysebacate. As described below, several DCA were identified through the random forest analysis as some of the most important biochemicals in the discrimination between depressed and non-depressed subjects from this study.
We then performed a series of ANVOVA analyses to examine effects of age, statin use and ejection fraction. Compounds that were identified as having a significant age effect (p<0.05 and q<0.1) were limited to glutamate, hypoxanthine, xanthine and trigonelline. Further analysis indicated while there was a significant age effect with the above mentioned compounds, there were no significant effect detected with either statin use or ejection fraction with any of the compounds detected within this study (p<0.05 and q<0.1).
In other analyses, we examined the effects of severity of heart failure (as measured by New York Heart Association classification) and of severity of depression (as measured by the BDI). After accounting for group differences (depressed versus no depression), there was no significant effect of heart failure severity. We did find a positive association (using Pearson’s correlation) for BDI score and several compounds, obtaining similar results as the previous analysis of depressed versus no depression.
Random Forest Classification
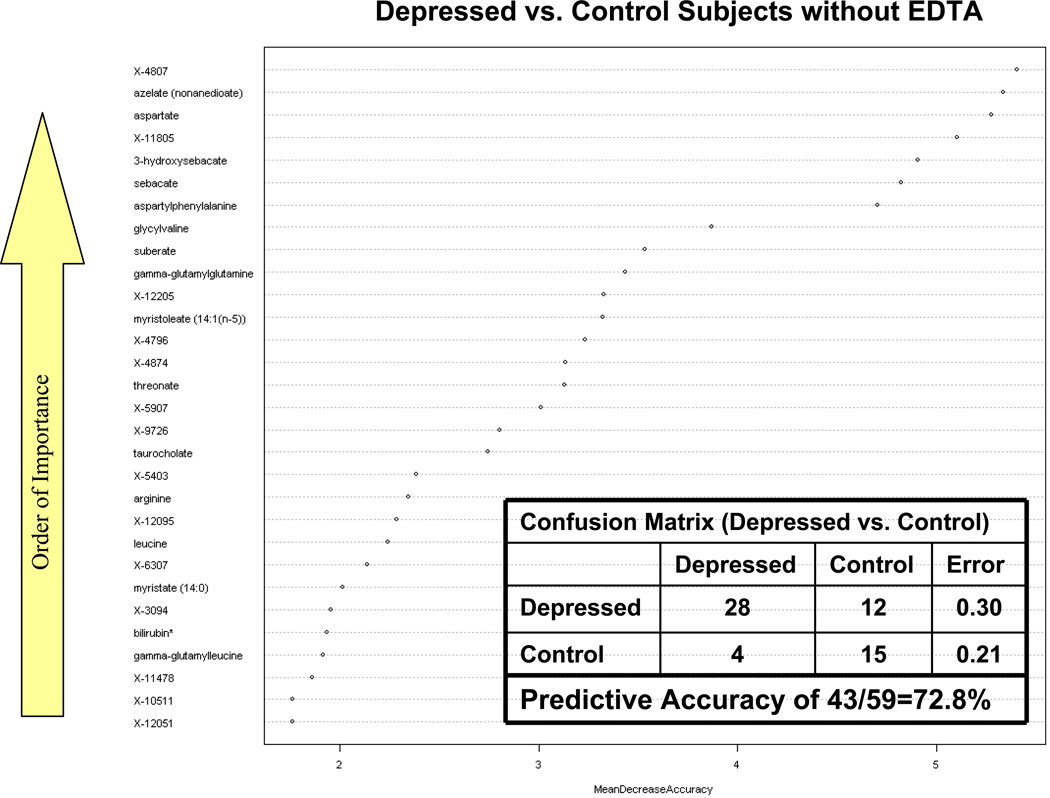
We used the entire metabolite dataset derived from plasma without EDTA for Random Forest analysis to classify depressed and non-depressed subjects in this study. The confusion matrix within Figure 2 shows that 28 of the depressed subjects were correctly classified as depressed, while 12 were misclassified as belonging to the control group. Of the 19 control group subjects without EDTA, 15 were correctly classified as non-depressed subjects while only 4 were incorrectly classified as belonging to the depressed group. From Figure 2, there appear to be seven biochemicals that are particularly important in differentiating the depressed subjects from the control subjects: unnamed biochemical X-4807, azelate, aspartate, X-11805, 3-hydroxysebacate, sebacate and aspartylphenylalanine. Overall, 43 of the 59 plasma samples from groups B and C without EDTA were correctly classified which resulted in a predictive accuracy of 43/59=72.8%. The accuracy rate for discriminating depressed and non-depressed subjects based on plasma samples was much higher than random chance (50%). It is interesting to note that three of the top biochemicals (azelate, 3-hydroxysebacate and sebacate) that contributed to the discrimination of depressed from non-depressed subjects, are DCAs associated with ω-oxidation of fatty acids. These DCAs were found to be elevated in the depressed subjects in relation to controls and are a possible indication of increased ω–oxidation fatty acid metabolism.19 Aspartate, as mentioned earlier, is an important neurotransmitter and was also very effective in discriminating between depressed and control subjects.
Figure 2.
Random forest analysis of depressed and non-depressed control participants. X-axis shows mean decrease in accuracy. Y-axis ranks biochemicals in order of importance from top to bottom in ability to successful differentiate samples between depressed and non-depressed subjects.
Comment
The major findings of this metabolomic study are that we were able to identify differences in concentrations of several metabolites between depressed and non-depressed heart failure patients. Significant differences were found in key metabolic pathways, including amino acid metabolism and lipid metabolism. These findings build on a previous report by our group that identified between-group differences in several fatty acids, glycerol and gamma-aminobutyric acid.11
Several amino acids play an important role as neurotransmitters and studies have shown that major depression may be accompanied by alteration in serum, cerebrospinal fluid or brain concentrations of many excitatory amino acids.20 Glutamate and aspartate, two amino acids detected in this study, are considered to be the main excitatory amino acids in the central nervous system.20, 21
Additionally, glutamate, aspartate and serine play an important role with N-methyl D-aspartate (NMDA) receptors. NMDA receptors are non-specific cation channel receptors which act by mediating fast-synaptic transmission in the brain and are fundamental to memory and behavior.22 Activation of the NMDA receptor can be initiated by the binding of the amino acid glutamate and aspartate. NMDA receptors also require the additional binding of glycine or serine as a co-agonist for functional opening of the ion channel. Within this study, aspartate and serine were significantly elevated in depressed subjects when compared with controls. Glutamate was initially identified as having a significantly elevated level within depressed subjects; however, ANCOVA analysis later indicated that there was a significant age effect associated with glutamate. It should be noted that once accounting for age, the group effect was no longer significant for glutamate. There were other metabolites with a significant age effect, but the group effects remained significant. Several studies have identified elevated levels of glutamate and aspartate in serum samples from depressed subjects in relation to non-depressed controls.20, 21, 23 It has been suggested that brain concentration of these amino acids which exceed those normally found in the synaptic space can cause selective neuronal loss and may be involved in a variety of chronic neurological disorders.24
Higher levels of the branch-chain amino acids (BCAA) leucine and isoleucine were also detected within the depressed subjects in relation to the control group. BCAAs are involved with systemic stress responses, they are regulators of protein synthesis and degradation, and they serve as key precursors for glutamine and alanine synthesis.25 In particular, leucine plays a key role in several key brain metabolic functions, including serving as a precursor of fuel molecules, alpha-ketoisocaproate and ketone bodies, and as a regulator of the activity of some enzymes important for brain energy metabolism.26
Phenylalanine is an essential amino acid and serves as a precursor for the amino acid tyrosine as well as for catecholamines in the body (tyramine, dopamine, epinephrine and norepinephrine). In this study, the levels of phenylalanine were found to be slightly but significantly higher in depressed subjects in relation to controls. It is not clear why this is the case. There may be dietary differences, differential absorption from the gastro-intestinal tract or other factors that would elevate levels in depressed subjects or lower them in controls.
We also examined the data for indicators of oxidative stress in association with depression. Glutathione (GSH) is a tripeptide (L-γ-glutamyl-L-cysteinyl-glycine) that is synthesized in the cytosol from the precursor amino acids glutamate, cysteine and glycine. GSH is the most abundant redox molecule and is critical in maintaining redox status in cells.27 It also plays a key role in the detoxification of reactive oxygen species, reactive nitrogen species and xenobiotic compounds in cells. However, glutathione (reduced or oxidized) was not detected in samples for this study and pyroglutamate (5-oxoproline), which is also an indication of glutathione biosynthesis flux and oxidative stress, failed to show any significant difference between depressed and non-depressed subjects. These results suggest that there was no increase in oxidative stress among the depressed subjects.
While the primary route of fatty acid metabolism is via β-oxidation, other metabolic pathways are known to contribute to fatty acid metabolism. For example, ω-oxidation of fatty acids is known to occur in liver microsomes through a NADPH- and cytochrome P450-dependent process by which the omega carbon of a fatty acid is oxidized to an alcohol then to a carboxylic acid, thereby generating a dicarboxylic acid (DCA). DCAs produced by ω-oxidation can be transported back into the mitochondria or to peroxisomes for β-oxidation. DCAs are produced when there is a defect in fatty acid oxidation or when mitochondrial β-oxidation is overwhelmed.28, 29 A number of DCAs were observed to be elevated in depressed subjects in relation to controls. These include an increase in the levels of azelate, suberate, sebacate and 3-hydroxysebacate. Several DCAs were also identified through the random forest analysis as some of the most important biochemicals in the discrimination between depressed and non-depressed subjects from this study. With regard to other fatty acids, we found depressed heart failure patients had lower concentrations of fatty acids such as docosahexaenoic acid (DHA) compared with non-depressed patients but the differences were not different statistically. This may be due to the relatively small sample size in the study.
In addition to this, we also found decreased level of the ketone body 3-hydroxybutyrate in depressed subjects in comparison with controls. Ketone bodies are generated from acetyl-CoA formed during β-oxidation of fatty acids and can be used as fuel for the brain and central nervous system. In a normal situation, lipids undergo β-oxidation eventually resulting in the accumulation of acetyl-CoA which may then enter the TCA cycle for energy production. However, if the levels of acetyl-CoA reach a point such that they exceed the requirements of the TCA cycle then the excess is directed towards the formation of ketone bodies such as 3-hydroxybutyrate. It is interesting to note from this study that while there was a reduction in the level of 3-hydroxybutyrate that there also was an increase in the levels of several DCAs. This suggests that in the depressed subjects examined for this study, that metabolism of fatty acids may have been a shift away from β-oxidation and redirected towards ω-oxidation. These data are further supported by the findings from the random forest analysis that several of the DCAs were actually the best predictors between the depressed and non depressed subjects. We demonstrated the importance of both amino acids and dicarboxylic acids in differentiating between the depressed and non-depressed groups with a predictive accuracy of 78.2%. However at this time, the relationship of increased DCA formation through ω-oxidation and depression is unclear. As others have noted, in humans, medium-chain dicarboxylic acids, e.g., adipic (C6), suberic (C8), sebacic (C10), and dodecanedioic (C12) acids, derive from the β-oxidation of longer-chain dicarboxylic acids. Long-chain dicarboxylic acids, in turn, are formed from the correspondent fatty acids by ω-oxidation in the microsomial membranes or are consumed with a diet rich in vegetables.30
This metabolomic study advances our understanding of biochemical processes involved with depression. Yet, the study is not without limitations. The sample size, although relatively small, is comparable to other studies that were successful in mapping signatures for neuropsychiatric conditions,31, 32 other medical conditions,33–35 and individuals drugs.5, 36 Absence of a control group without heart failure might also be seen as a limitation. Depressed participants in this study were recruited as part of an antidepressant trial and as such they may differ in some respects from the larger population of depressed patients with systolic dysfunction and class II or higher heart failure. Another potential limitation is lack of anthropomorphic data, e.g., height and weight for determination of Body Mass Index (BMI). Our findings related to fatty acids and depression raise the possibility that metabolic syndrome may be important in depressed heart failure patients, and data related to BMI would have been helpful in this regard.
The fact that about half of the control samples contained EDTA initially posed a problem, but we feel our approach adequately dealt with this issue. Finally, we chose to focus on a more medically homogeneous group of patients with heart failure, so it is not clear whether our findings are generalizable to other depressed adults. Furthermore, future studies will need to examine heart-failure related variables (e.g., ischemic versus non –ischemic heart failure, use of statins and other medications), comorbid medical conditions, and cardiac outcomes.
In summary, in this metabolomic analysis of patients with heart failure, we found that the concentrations of several biochemical compounds differed between depressed and control subjects. Numerous amino acid neurotransmitters were found to be elevated in depressed subjects in comparison to non-depressed control subjects. Depressed subjects had reduced levels of ketone body formation along with increased levels of dicarboxylic acid (DCA) formation which suggested reduced β-oxidation and enhanced ω-oxidation of fatty acids in depressed subjects. Inositol levels were also reduced in the depressed subjects in relation to controls, a finding consistent with previous studies. Finally, we showed that Random forest classification analysis was able to distinguish between depressed and non-depressed samples with a 72.8% success rate. Our study focused on heart failure patients, and it is unclear whether presence of this medical condition in all subjects would have affected the results among depressed and non-depressed groups. Future studies should attempt to replicate these findings in populations with larger sample sizes and with different medical comorbidities. In addition, other markers should be included, such as those related to inflammation or neurohormonal activation, as both have been linked to depression and heart failure.37, 38
Acknowledgements
This study was supported by NIMH Grants P50 MH60451, R01 MH54846, R01 MH63211, R21 MH076178 and K24 MH70027
The authors would like to acknowledge Ms. Mae Burks for her recruitment of study controls interviews and processing of samples for metabolomics analysis
Footnotes
Drs. Steffens, Krishnan and Kaddurah-Daouk have a patent application pending regarding the use of metabolomics in depression. Drs. Karoly and Mitchell are employed by Metabolon, Inc. Drs. Jiang and O’Connor report no conflict of interest related to this manuscript.
Dr. Steffens had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
References
- 1.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annual Review of Pharmacology and Toxicology. 2008;48:653–683. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 2.Kristal BS, Shurubor YI, Kaddurah-Daouk R, Matson WR. High-performance liquid chromatography separations coupled with coulometric electrode array detectors: a unique approach to metabolomics. Methods in Molecular Biology. 2007;358:159–174. doi: 10.1007/978-1-59745-244-1_10. [DOI] [PubMed] [Google Scholar]
- 3.Lindon JC, Holmes E, Nicholson JK. Metabonomics in pharmaceutical R&D. FEBS J. 2007 Mar;274(5):1140–1151. doi: 10.1111/j.1742-4658.2007.05673.x. [DOI] [PubMed] [Google Scholar]
- 4.Harrigan G, Goodacre R. Metabolic Profiling: Its Role in Biomarker Discovery and Gene Function Analysis. Boston: Kluwer Acad Publ; 2003. [Google Scholar]
- 5.Kaddurah-Daouk R, Krishnan KRR. Metabolomics: A global biochemical approach to the study of central nervous system diseases. Neuropsychopharmacology Reviews. 2009;34:173–186. doi: 10.1038/npp.2008.174. 173–186. [DOI] [PubMed] [Google Scholar]
- 6.Delgado PL, Moreno FA. Role of norepinephrine in depression. Journal of Clinical Psychiatry. 2000;61 Suppl 1:5–12. [PubMed] [Google Scholar]
- 7.Brambilla P, Perez J, Barale F, Schettini G, Soares JC. GABAergic dysfunction in mood disorders. Molecular Psychiatry. 2003 Aug;8(8):721–737. doi: 10.1038/sj.mp.4001362. [DOI] [PubMed] [Google Scholar]
- 8.Sanacora G, Saricicek A. GABAergic contributions to the pathophysiology of depression and the mechanism of antidepressant action. CNS & Neurological Disorders - Drug Targets. 2007 Apr;6(2):127–140. doi: 10.2174/187152707780363294. [DOI] [PubMed] [Google Scholar]
- 9.Levine J, Panchalingam K, Rapoport A, Gershon S, McClure RJ, Pettegrew JW. Increased cerebrospinal fluid glutamine levels in depressed patients. Biological Psychiatry. 2000 Apr 1;47(7):586–593. doi: 10.1016/s0006-3223(99)00284-x. [DOI] [PubMed] [Google Scholar]
- 10.Frye MA, Tsai GE, Huggins T, Coyle JT, Post RM. Low cerebrospinal fluid glutamate and glycine in refractory affective disorder. Biological Psychiatry. 2007 Jan 15;61(2):162–166. doi: 10.1016/j.biopsych.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 11.Paige LA, Mitchell MW, Krishnan KR, Kaddurah-Daouk R, Steffens DC. A preliminary metabolomic analysis of older adults with and without depression. International Journal of Geriatric Psychiatry. 2007 May;22(5):418–423. doi: 10.1002/gps.1690. [DOI] [PubMed] [Google Scholar]
- 12.Appleton KM, Rogers PJ, Ness AR. Is there a role for n-3 long-chain polyunsaturated fatty acids in the regulation of mood and behaviour? A review of the evidence to date from epidemiological studies, clinical studies and intervention trials. Nutr Res Rev. 2008 Jun;21(1):13–41. doi: 10.1017/S0954422408998620. [DOI] [PubMed] [Google Scholar]
- 13.Beck AT, Steer RA, Ball R, Ranieri W. Comparison of Beck Depression Inventories -IA and -II in psychiatric outpatients. Journal of Personality Assessment. 1996 Dec;67(3):588–597. doi: 10.1207/s15327752jpa6703_13. [DOI] [PubMed] [Google Scholar]
- 14.Lawton KA, Berger A, Mitchell M, et al. Analysis of the adult human plasma metabolome. Pharmacogenomics. 2008 Apr;9(4):383–397. doi: 10.2217/14622416.9.4.383. [DOI] [PubMed] [Google Scholar]
- 15.Breiman L. Random Forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 16.Liaw A, Wiener M. Classification and regression by Random Forest. R News. 2002;2:18–22. [Google Scholar]
- 17.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B (Methodological) 1995;57:289–300. [Google Scholar]
- 19.Muth A, Jung J, Bilke S, et al. Simultaneous enantioselective analysis of chiral urinary metabolites in patients with Zellweger syndrome. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;792:269–277. doi: 10.1016/s1570-0232(03)00285-x. [DOI] [PubMed] [Google Scholar]
- 20.Maes M, Verkerk R, Vandoolaeghe E, Lin A, Scharpe S. Serum levels of excitatory amino acids, serine, glycine, histidine, threonine, taurine, alanine and arginine in treatment-resistant depression: modulation by treatment with antidepressants and prediction of clinical responsivity. Acta Psychiatrica Scandinavica. 1998;97:302–308. doi: 10.1111/j.1600-0447.1998.tb10004.x. [DOI] [PubMed] [Google Scholar]
- 21.Altamura C, Maes M, Dai J, Meltzer HY. Plasma concentrations of excitatory amino acids, serine, glycine, taurine and histidine in major depression. European Neuropsychopharmacology. 1995;5 Suppl:71–75. doi: 10.1016/0924-977x(95)00033-l. [DOI] [PubMed] [Google Scholar]
- 22.Stephenson FA. Structure and trafficking of NMDA and GABAA receptors. Biochemical Society Transactions. 2006 Nov;34(Pt 5):877–881. doi: 10.1042/BST0340877. [DOI] [PubMed] [Google Scholar]
- 23.Francis PT, Poynton A, Lowe SL, et al. Brain amino acid concentrations and Ca2+-dependent release in intractable depression assessed antemortem. Brain Research. 1989 Aug 14;494(2):315–324. doi: 10.1016/0006-8993(89)90600-8. [DOI] [PubMed] [Google Scholar]
- 24.Meldrum BS. Drugs acting on amino acid neurotransmitters. Advances in Neurology. 1986;43:687–706. [PubMed] [Google Scholar]
- 25.Choudry HA, Pan M, Karinch AM, Souba WW. Branched-chain amino acid-enriched nutritional support in surgical and cancer patients. Journal of Nutrition. 2006;136:314S–318S. doi: 10.1093/jn/136.1.314S. [DOI] [PubMed] [Google Scholar]
- 26.Murin R, Hamprecht B. Metabolic and regulatory roles of leucine in neural cells. Neurochemical Research. 2008;33(2):279–284. doi: 10.1007/s11064-007-9444-4. [DOI] [PubMed] [Google Scholar]
- 27.Forman HJ, Zhang H, Rinna A. Glutathione: Overview of its protective roles, measurement, and biosynthesis. Mol Aspects Med. 2008 Aug 30; doi: 10.1016/j.mam.2008.08.006. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Draye JP, Veitch K, Vamecq J, Van Hoof F. Comparison of the metabolism of dodecanedioic acid in vivo in control, riboflavin-deficient and clofibrate-treated rats. European Journal of Biochemistry. 1988 Dec 1;178(1):183–189. doi: 10.1111/j.1432-1033.1988.tb14442.x. [DOI] [PubMed] [Google Scholar]
- 29.Mortensen PB. C6-C10-dicarboxylic acids in liver and kidney tissue in normal, diabetic ketotic and clofibrate-treated rats. Biochimica et Biophysica Acta. 1986 Aug 14;878(1):14–19. doi: 10.1016/0005-2760(86)90338-3. [DOI] [PubMed] [Google Scholar]
- 30.Mingrone G, Castagneto M. Medium-chain, even-numbered dicarboxylic acids as novel energy substrates: an update. Nutrition Reviews. 2006 Oct;64(10 Pt 1):449–456. doi: 10.1301/nr.2006.oct.449-456. [DOI] [PubMed] [Google Scholar]
- 31.Rozen S, Cudkowicz ME, Bogdanov M, et al. Metabolomic analysis and signatures in motor neuron disease. Metabolomics. 2005;1:101–108. doi: 10.1007/s11306-005-4810-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaddurah-Daouk R. Metabolic profiling of patients with schizophrenia. PLoS Med. 2006 Aug;3(8):e363. doi: 10.1371/journal.pmed.0030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabatine MS, Liu E, Morrow DA, et al. Metabolomic identification of novel biomarkers of myocardial ischemia. Circulation. 2005;112:3868–3875. doi: 10.1161/CIRCULATIONAHA.105.569137. [DOI] [PubMed] [Google Scholar]
- 34.Yang C, Richardson AD, Smith JW, Osterman A. Comparative metabolomics of breast cancer. Pac Symp Biocomput. 2007:181–192. [PubMed] [Google Scholar]
- 35.Yan SK, Wei BJ, Lin ZY, Yang Y, Zhou ZT, Zhang WD. A metabonomic approach to the diagnosis of oral squamous cell carcinoma, oral lichen planus and oral leukoplakia. Oral Oncol. 2008 May;44(5):477–483. doi: 10.1016/j.oraloncology.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Kaddurah-Daouk R, McEvoy J, Baillie RA, et al. Metabolomic mapping of atypical antipsychotic effects in schizophrenia. Molecular Psychiatry. 2007 Oct;12(10):934–945. doi: 10.1038/sj.mp.4002000. [DOI] [PubMed] [Google Scholar]
- 37.Parissis JT, Nikolaou M, Farmakis D, et al. Self-assessment of health status is associated with inflammatory activation and predicts long-term outcomes in chronic heart failure. European Journal of Heart Failure. 2009 Feb;11(2):163–169. doi: 10.1093/eurjhf/hfn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parissis JT, Farmakis D, Nikolaou M, et al. Plasma B-type natriuretic peptide and anti-inflammatory cytokine interleukin-10 levels predict adverse clinical outcome in chronic heart failure patients with depressive symptoms: a 1-year follow-up study. European Journal of Heart Failure. 2009 Oct;11(10):967–972. doi: 10.1093/eurjhf/hfp125. [DOI] [PubMed] [Google Scholar]