Summary
Cilia/flagella are highly conserved organelles that play diverse roles in cell motility and sensing extracellular signals. Motility defects in cilia/flagella often result in primary ciliary dyskinesia (PCD). However, the mechanisms underlying cilia formation and function, and in particular the cytoplasmic assembly of dyneins that power ciliary motility, are only poorly understood. Here we report a novel gene, kintoun (ktu), involved in this cytoplasmic process. This gene was first identified in a medaka mutant, and found to be mutated in PCD patients from two affected families as well as in the pf13 mutant of Chlamydomonas. In the absence of Ktu/PF13, both outer and inner dynein arms are missing or defective in the axoneme, leading to a loss of motility. Biochemical and immunohistochemical studies show that Ktu/PF13 is one of the long-sought proteins involved in pre-assembly of dynein arm complexes in the cytoplasm before intraflagellar transport loads them for the ciliary compartment.
Cilia project from the surface of most eukaryotic cells and consist of a microtubule-based axoneme surrounded by a specialized ciliary membrane. Cilia that contain dynein arms are motile and play diverse roles in cell motility, fluid transport, and even in patterning the body of vertebrate embryos. A well-known example is left-right (LR) specification determined by cilia-generated fluid flow in the embryonic mouse node1. Because of their critical and diverse functions, cilia have been linked to a variety of diseases2 such as polycystic kidney disease (PKD; MIM173900)3 and primary ciliary dyskinesia (PCD; MIM242650)4.
PCD, a heterogeneous disorder with an incidence of 1 in 20,000 to 60,000, is characterized by recurrent respiratory tract infections, male infertility and randomization of LR body asymmetry4. The combination of PCD and situs inversus is also referred to as Kartagener syndrome (MIM 244400). The widespread distributions and functions of motile cilia and flagella well explain the multiple phenotypes of PCD. Recent linkage and mutational analyses with PCD patients have so far identified six genes (DNAH5, DNAI1, DNAH11, OFD, RPGR and TXND3) responsible for PCD5,6,7,8,9,10. Four of these six genes encode subunits of axonemal dyneins, highlighting the importance of dynein motors to ciliary motility.
Medaka fish, Oryzias latipes, is an emerging model vertebrate system11 with a high quality draft genome12and a number of unique mutants13,14. Our recent mutagenesis screen in medaka identified kintoun (ktu, formerly named knt) as a mutant showing organ laterality defects due to altered ciliary motility15. Here we show that the ktu gene encodes a novel cytoplasmic protein conserved from ciliated unicellular organisms to higher mammals. Furthermore, we have identified mutations in the homologous gene of two human PCD families and in pf1316, a Chlamydomonas mutant with paralyzed flagella. In all affected organisms the cilia develop normally but exhibit complete or partial loss of outer and inner dynein arms (ODAs, IDAs), leading to loss of motility. Biochemical data demonstrate that Ktu is not a dynein subunit, but is instead required in the cytoplasm for pre-assembly of dynein arm complexes before they become transported to their destined functional sites within the axoneme17.
Results
The medaka ktu mutant is defective in axonemal dynein arms
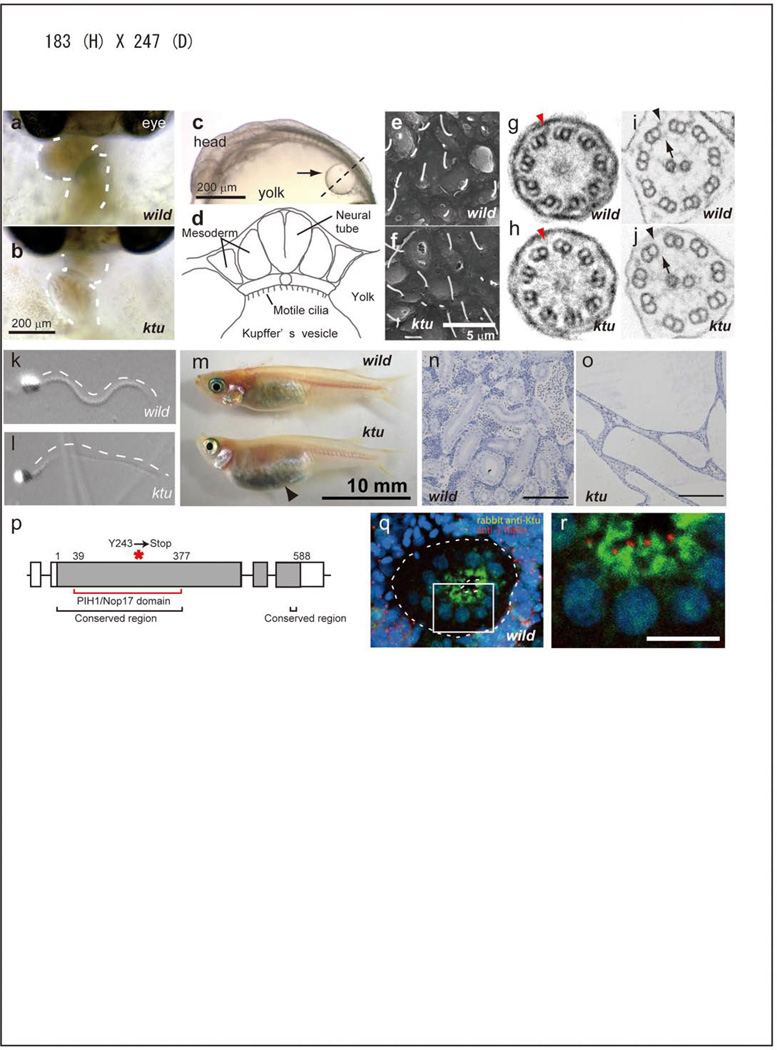
ktu is a typical randomized LR mutant leading to situs inversus (Fig. 1a,b). Mutant fish lack directional liquid flow in Kupffer’s vesicle (KV) (Fig. 1c, d)15, an organ functionally equivalent to the mouse node in terms of LR specification18,19. Although the number and length of cilia in KV appear normal (Fig. 1e, f), their motility is completely lost (Supplementary movies S1, S2). ktu homozygous fish are viable but develop PKD (Fig. 1m–o, Supplementary Fig. S2a). Furthermore, male mutant fish exhibit impaired sperm motility, leading to reduced fertility (Fig. 1k, l; Supplementary movies S3, S4). Ultrastructurally, affected cilia/flagella exhibit partial or complete loss of ODAs and IDAs (Fig. 1g–j). This loss of dynein arms is more severe in KV cilia than in sperm flagella (see Supplementary Table S1).
Figure 1. Medaka ktu mutant.
a, b, Heart looping (dashed line) at 3 days postfertilization (dpf). c, d, Medaka Kupffer’s vesicle (KV, arrow in e) at 1.5 dpf and its schematic structure. e, f, Scanning EM of cilia in KV. g–j, Transmission EM of KV cilia (g, h; 9+0) and sperm flagella (i, j; 9+2), showing a partial or complete loss of ODA (arrowheads) and IDA (arrows) in ktu. k, i, Swimming sperm. m, The belly of the mutant adult (3-month-old) becomes expanded (arrowhead). n, o, PKD in ktu mutant kidney. p, The exon-intron structure of ktu, encoding a protein (solid box) with the conserved N-terminal domain (black bracket) and a domain weekly homologous to yeast PIH1/NOP17 (red bracket). A premature stop codon introduced in ktu is indicated by an asterisk. q, r, Overlay and high-power images of a nephric tubule (dotted lines) double-stained with anti-Ktu (green) and γ-tubulin (red) antibodies. Nuclei are in blue.
Positional cloning identified a candidate gene encoding 588 amino acids, with a single nucleotide change between homozygous ktu and wild-type embryos, creating a premature stop codon (c.841C>A [p.Y243X]) (Fig. 1p; Supplementary Fig. S2d). Knockdown experiments with morpholino anti-sense oligonucleotides and rescue experiments with wild-type mRNA confirmed that we correctly identified the responsible gene (Supplementary Table S2 and S3). Ktu shows homology to the human hypothetical protein encoded by c14orf104 (NM_018139.2). Database searches also identified homologous genes in other vertebrates, insects, Chlamydomonas, and Tetrahymena but not in C. elegans, A. thaliana, S. cerevisiae or D. discoideum (Supplementary Fig. S2e). Their N-terminal half as well as a short stretch of the C-terminal end are relatively conserved among all Ktu homologs (Fig. 1p, Supplementary Fig. S1). The conserved N-terminal region shows a weak similarity to NOP17, a yeast nucleolar protein involved in assembly of pre-rRNA processing complexes20,21,22.
RT-PCR analysis detected ktu expression in medaka embryos and nearly all organs of adult mice, with higher expression in mouse tissues known to have motile cilia/flagella, such as brain and testis (data not shown). The subcellular localization of Ktu was then examined using an antibody specific for medaka Ktu. In the stained sperm, only the cytoplasm but not the flagellum is positive for Ktu (Supplementary Fig. S2g). In renal tubules, which are lined by a single layer of tall columnar cells with either single or multiple motile cilia at the apical surface (Supplementary Fig. S2b, c)23. Ktu is localized in the apical cytoplasm around the γ-tubulin-positive pericentriolar region, not in the cilia (Fig. 1q, r). In most cases, Ktu and γ-tubulin signals did not co-localize. We conclude that Ktu is mainly concentrated in the apical cytoplasm.
Mutations in KTU cause PCD in humans
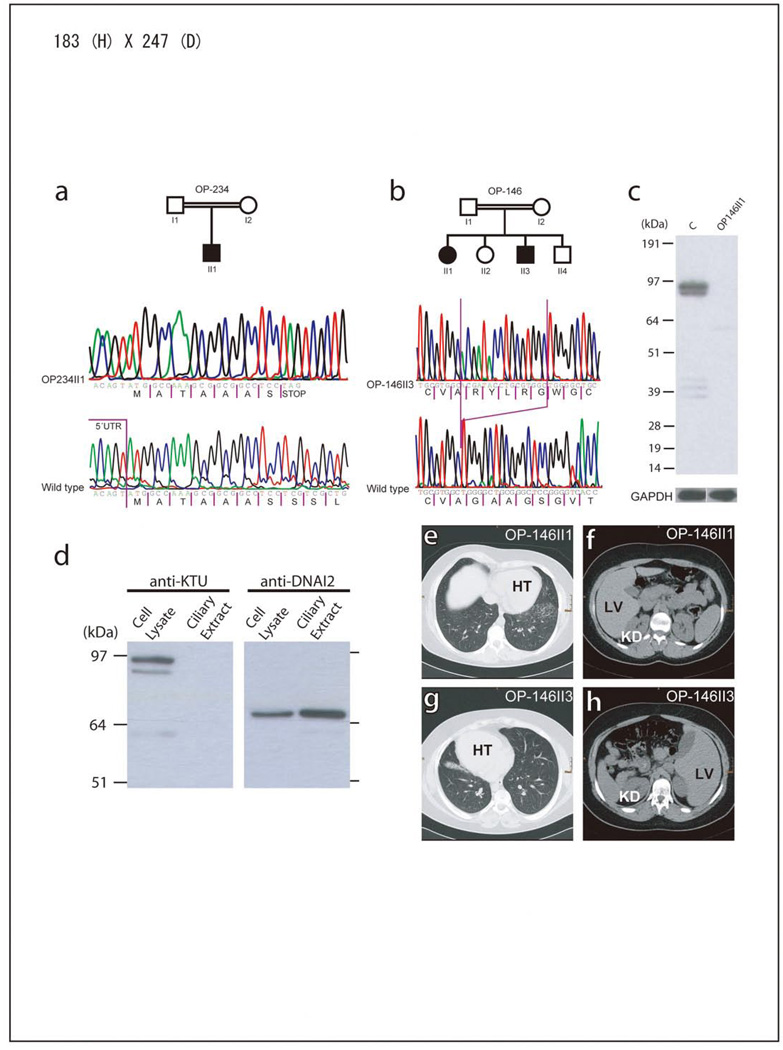
The observed ktu phenotype in medaka led us to consider the human orthologue KTU as a promising candidate gene for human PCD. KTU (previously C14orf104) is located on chromosome 14q21.3 and consists of three exons encoding cDNAs of 2,511 bp or 2,367 bp (the latter an in-frame splice variant lacking exon2) (Fig. 2c). We have screened affected individuals originating from 112 PCD families for the presence of a mutation in their KTU gene. In two consanguineous families we identified homozygous loss-of-function mutations (OP-146: c.1214^1215insACGATACCTGCGTGGC [p.G406Rfs89X]; OP-234: c.23C>A [p.S8X]) which is consistent with homozygosity by descent (Fig. 2a, b). All parents were heterozygous for the detected mutations and, in family OP-146, the mutation clearly co-segregated with the disease status. In Western blots of cell lysates from control respiratory cells, an anti-KTU antibody detects a specific band at ~95kDa, compatible with the size of the predicted 837 amino acid KTU protein encoded by all three exons (Fig. 2c). In contrast, no specific band was detected for the affected children of family OP-146, confirming a loss-of-function mutation in the KTU gene (Fig. 2c). Consistent with the immunohistochemistry in medaka, human KTU was detectable in the cytoplasm but not in ciliary fractions of respiratory cells (Fig. 2d).
Figure 2. Recessive loss-of-function KTU mutations.
a, b. Homozygous mutations (c.C23A [pS8X]; c.1214^1215insACGATACCTGCGTGGC [p.G406Rfs89X]) in two PCD patients. c, KTU is detectable in control respiratory cells but not in KTU mutant cells. The double band is probably explained by distinct alternative splicing products. GAPDH (loading control). d, In control respiratory ciliary extracts KTU is absent, whereas DNAI2 is abundant. KTU is only present in the cytoplasm. (e–h), Consistent with randomization of left/right body asymmetry OP-146-II-1 exhibits situs solitus (e, f) and her brother (OP-146-II-3) situs inversus totalis (g, h). Both have chronic lung disease. HT; heart, LV; liver, KD; kidney.
All three affected patients have suffered from chronic otitis media and sinusitis, and recurrent pneumonia since birth. Two of the affected (OP-146 II3; OP-234 II1) had developed bronchiectasis. In OP-234 II1 bronchiectasis was so severe that the destroyed middle lobe had to be removed because of persistent pneumonia. Complete situs inversus was present in two of the three affected individuals (OP-146 II1; OP-234 II1) with KTU mutations, which is consistent with randomization of LR body asymmetry (Fig. 2e–h). Interestingly, unlike in medaka ktu, their kidneys were of normal size and devoid of cysts (Fig. 2e–h). This probably reflects a difference in the origin of kidneys, mesonephric for fish and metanephric for mammals. High-speed video microscopy demonstrated complete immotility of respiratory cilia in all three affected patients and of sperm in both affected males (Supplementary movies S5–8 and S9, 10).
Transmission electron microscopy (EM) of respiratory cilia and sperm tails identified abnormal axonemal dynein arms (Supplementary Fig. S3a). To further corroborate ultrastructural findings, we performed high-resolution immunofluorescence microscopy in control and KTU mutant respiratory cells using specific monoclonal antibodies against dynein arm components (See Supplementary Fig. S4 for antibody specificity and Fig. S3 for staining data). We observed the absence of ODA components DNAH5 (gamma heavy chain) and DNAI2 (intermediate chain) from the distal ciliary axonemes, but some residual staining was present in proximal proteins (Supplementary Fig. S3b, d), indicating that loss of KTU more strongly affects the assembly of distally located type 2 ODA complexes that specifically contain dynein β-HC (heavy chain) orthologue DNAH9 (Supplementary Fig. S3c)24. In sperm, we observed complete loss of DNAI2 (Supplementary Fig. S5). We then analyzed localization of DNALI1, the orthologue of Chlamydomonas p28, which is a component of several IDA subtypes25,26. Consistent with IDA defects, the antibody to DNALI1 failed to show any positive signal in ciliary axonemes of these patients (Supplementary Fig. S3e). Hence we conclude that in man, like in medaka, KTU mutations cause PCD with combined ODA and IDA defects. Interestingly, combined ODA/IDA defects were only present in a small proportion of analyzed PCD patients (17 of 112), of which approximately 12% (2 of 17) are caused by mutations in the KTU gene. In contrast, mutations in DNAH5, a major causative gene for PCD27, only affect ODA components without mis-localization of the IDA component, DNALI1 (data not shown).
Chlamydomonas KTU homolog PF13 is required for dynein pre-assembly
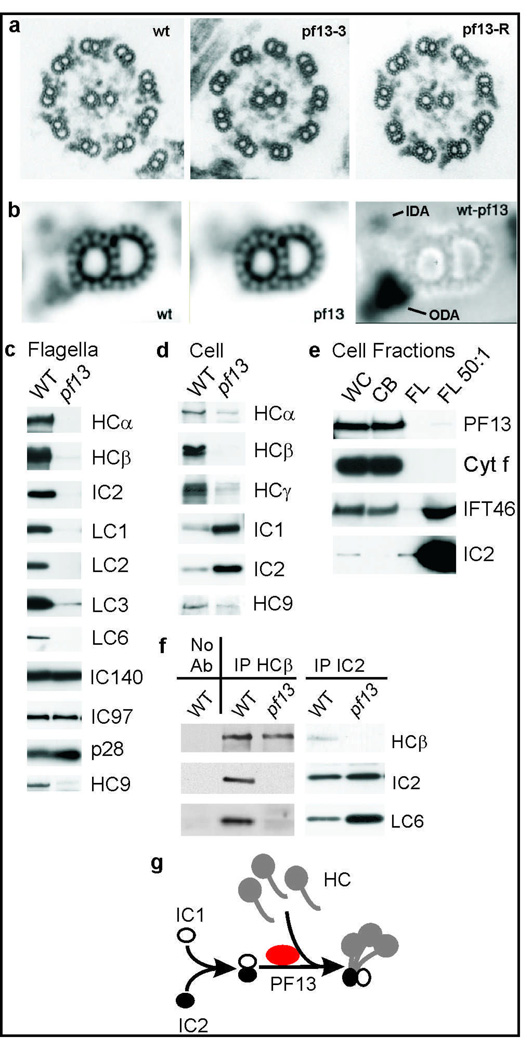
During this study, we found that the KTU homolog in Chlamydomonas, a single-celled alga used extensively for analysis of cilia/flagella, is PF13, a locus required for ODA assembly16. The Chlamydomonas pf13 gene was identified by cloning DNA from the insertion site of transformation-generated mutant allele pf13-3 (Supplementary Fig. S6)28. Gene identity was confirmed by rescuing the mutant phenotype with wild-type genomic clones (Supplementary Table S5), and by the absence of the gene product in mutant cells (Supplementary Fig. S4f). Sequence of genomic and cDNA clones revealed that PF13 encodes a protein homologous to vertebrate Ktu (Supplementary Fig. S1). As expected, Western blots show that PF13 is exclusively localized in the cytoplasm but not in flagella (Fig. 3e), a pattern distinct from that of intraflagellar transport (IFT) components such as IFT46, which are present in both cell body and flagellar fractions (Fig. 3e).
Figure 3. PF13 is the Chlamydomonas homolog of Ktu.
a, micrographs of axonemes from wild type (wt), pf13-3, and pf13 transformed with λ1A (pf13-R). b, averages of 108 wild type and 118 pf13 doublets, and a difference image (wt-13). c–f, Western blots of C. reinhardtii proteins. ODA proteins (HCα, HCβ, IC2, LC1, LC2 and LC3), IDA proteins (IC140, IC97, and p28) and dynein c heavy chain HC9 in pf13 in flagella (c) and cytoplasm (d). e, PF13 occurs in whole cells (WC) and cell bodies (CB) but not in flagella. Absence of cytochrome (Cyt) f demonstrates lack of cell body contamination in flagellar fractions. FL 50:1, a 50-fold excess of flagella. f, IPs of ODA dyneins from wild-type and pf13 cell extracts probed with antibodies to ODA subunits HCβ, IC2 and LC6. g, A hypothesized role of PF13 in ODA assembly.
Unlike most other Chlamydomonas ODA assembly mutants, which swim slowly with reduced flagellar beat frequencies25, pf13 mutant cells are paralyzed16. EM image averages show that a density is missing from the IDA region in pf13 axonemes (Fig. 3a, b), consistent with IDA depletion in vertebrate mutant cilia (Fig. 1j and Supplementary Fig. S3e), which likely accounts for the paralyzed phenotype25. Western blots show that ODA subunits are greatly depleted from pf13 flagella, but IDA subunits IC97 and IC140 (needed for assembly of heterodimeric I1 dynein) remain at normal levels (Fig. 3c). Unlike in human patients, light chain p28 (homolog of DNALI1) also remains at wild-type levels, perhaps related to the ability of p28 to assemble independently of dynein heavy chains in Chlamydomonas29. Consistently, at least one p28-associated IDA heavy chain, HC9 (the motor subunit of monomeric dynein c) is greatly reduced in pf13 flagella (Fig. 3c).
Chlamydomonas cells disassemble and reassemble their flagella once each cell cycle, and maintain a sizable cytoplasmic pool of flagellar subunits. Some Chlamydomonas dynein assembly mutations prevent pre-assembly of a dynein complex in this cytoplasmic pool17, whereas others appear to block trafficking of complexes into flagella by IFT30. Western blots show that IDA HC9 and all three ODA heavy chains are depleted from pf13 cytoplasm, whereas ODA intermediate chains accumulate above normal levels (Fig. 3d), indicating that pf13 blocks an early step in dynein assembly. To further test the effects of pf13 on dynein assembly, HCβ and IC2 subunits were immunoprecipitated from wild-type and pf13 cytoplasmic extracts. Western blots (Fig. 3f) confirm that intermediate chains (IC2) and light chains (LC6) co-precipitate with heavy chains (HCβ) from wild-type extracts17, but show that the small remaining amount of HCβ present in pf13 extracts fails to co-precipitate IC2 or LC6 and thus HCβ has not assembled with other subunits. Furthermore, anti-IC2 co-precipitates IC2 and LC6 (as well as IC1 and LC2; data not shown), demonstrating that intermediate and light chains can still pre-assemble in pf13 cytoplasm. We conclude that PF13/Ktu is needed for ODA heavy chain stability, possibly by aiding assembly of heavy chains with other subunits, as diagrammed in Figure 3g.
Mouse Ktu interacts with Hsp70 and dynein intermediate chain, DNAI2
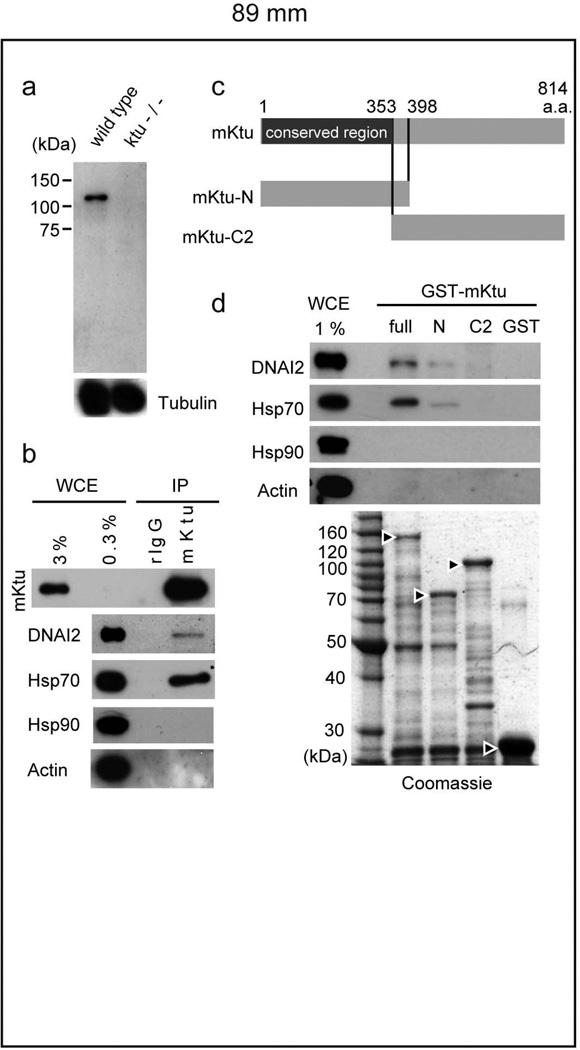
The above results implicate Ktu in the cytoplasmic assembly of dynein complexes in algae. To explore this possibility in vertebrates, we performed immunoprecipitations (IP) using rabbit anti-mouse Ktu (mKtu) antibody (Fig. 4a). mKtu is expected to have a similar function to medaka and human counterparts, because mKtu rescued the phenotype of medaka ktu mutants (Supplementary Table S4) and ktu-knockout mice exhibit PCD-like phenotypes (unpublished data). As shown in Fig. 4b, our IP experiment demonstrated that mKtu co-precipitates with at least one of the intermediate chains, DNAI2 (IC2 in Chlamydomonas), which shows defective assembly in human (Supplementary Fig. S3d) and Chlamydomonas (Fig. 3c) mutant cilia. This was further supported by a glutathione S-transferase (GST) pull-down assay using three mKtu constructs (Fig. 4c): both full length and (more weakly) the conserved N-terminal half have the ability to pull down DNAI2, while the C-terminal region fails to show such interaction (Fig. 4d).
Figure 4. Ktu binds to dyneins and Hsp70.
a, The anti-mouse (m) Ktu antibody detects a single band of 110 –120 kDa, slightly larger size than expected (814 amino acids, 88.3 kDa), which is undetectable in mutant mice. b, Immunoprecipitates with anti-mKtu or rabbit (r) IgG (control) from wild-type mouse testis extracts probed with anti-DNAI2, anti-Hsp70, anti-Hsp90 and anti-Actin. c, Full and truncated constructs of mKtu used in d. d, GST pull-down assay. Immunoblots with the indicated antibodies after incubation of GST-recombinant proteins with testis extracts. WCE, whole-cell extracts used. The amount of GST-recombinant proteins was confirmed by Coomassie staining (Arrowheads indicate GST-proteins used for the assay).
To obtain mechanistic insight into mKtu function, we globally looked for interacting proteins by applying MALDI-TOF mass/mass spectrometry to immunopreciptates from mouse testis extracts. This analysis identified Hsp70 as one of the major interacting proteins (Supplementary Fig. S7). Among molecular chaperones examined, Hsp70 and Hsp90 are the most highly expressed in mouse testis (data not shown), but only Hsp70 co-immunoprecipitates with mKtu (Fig. 4b). This was further confirmed by GST-pull-down assay (Fig. 4d).
Discussion
In the present study, forward genetics of medaka and Chlamydomonas has been successfully applied to isolate a novel gene required in the cytoplasm for ciliary function, which has unique properties compared with other PCD genes. The isolated gene, called ktu/pf13, is conserved among animals and ciliated unicellular organisms. As expected, this gene is not found in organisms that lack cilia, such as yeast and Arabidopsis, or that have only non-motile cilia (C. elegans). Consistently, a previous genomic comparison categorized a partial Chlamydomonas PF13 sequence (MOT45), as a gene only found in organisms with motile cilia31. Thus Ktu is an ancient protein essential for the motility of all eukaryotic cilia/flagella. The phenotypic similarity of the mutants in three organisms examined suggests that the function of Ktu/PF13 for dynein arm pre-assembly has been evolutionarily conserved from algae to humans.
Our biochemical analyses provided important insight into the proposed function of Ktu/PF13 as a facilitator of dynein pre-assembly, by revealing specific defects in the interaction of intermediate and heavy chains in the cytoplasm (Fig. 3g). Indeed, it has been shown that assembled dynein arms form a cytoplasmic precursor pool and are transferred to a loading zone around the basal body where IFT comes into play17,32. The finding of reduced heavy chain and increased intermediate chain abundance in pf13 cytoplasm (Fig. 3d) is consistent with an altered pre-assembly process. Furthermore, IP and GST-pull-down assays demonstrated interaction between mKtu and one of the intermediate chains, DNAI2. However, only a small fraction of the dynein co-precipitated with Ktu (Fig. 4), suggesting transient and/or indirect interaction during complex formation. Since Hsp70 was found to be a major interacting protein, Ktu/Pf13 could work as a co-chaperone to localize general functions of molecular chaperones (holding, assembling and protecting of proteins from degradation) to dynein arm formation33. Although Hsp70 is also found in cilia, axonemal Hsp70 is exclusively associated with central pair microtubules, not dyneins34. We therefore believe Hsp70 in these IP fractions is not a subunit of an axonemal dynein, but rather facilitates dynein pre-assembly in conjunction with Ktu. Such a role for Ktu/Pf13 would be consistent with the proposed role of its distant homologue, NOP17, as a co-chaperone to Hsp90 for assembly of yeast pre-mRNA processing complexes21,22. Although much progress has been made in understanding the mechanisms of IFT35,36, very little is known about the cytoplasmic process for pre-assembly of dynein complexes. Ktu/PF13 will thus be a good starting point to understand this cytoplasmic process.
METHODS SUMMARY
The medaka ktu mutant
ktu was isolated in our recent ENU-induced mutagenesis screening14 as a recessive mutant showing LR randomization in heart looping.
Antibodies to Ktu/Pf13
We generated polyclonal antibodies to medaka Ktu by immunizing rabbit with two peptides (GSPNPTSDPQNENQTRV, GKKIQKRNERPEHEVKN), to mouse Ktu with His-tagged full-length Ktu and to Chlamydomonas Pf13 with GST-fusion protein containing amino acids 3–460. The antibody to human KTU is commercially available, as targeted against anonymous uncharacterized protein C14orf104 (Atlas Antibodies). This antibody did not work well in immunohistochemistry in respiratory cells.
Immunohistochemistry and immunoblotting
For medaka tissues, paraffin-embedded sections (5 µm) were deparaffined and boiled in 1 mM EDTA for 17 min. using microwave and blocked in 5% skim milk, before incubation with primary antibodies. For high-resolution immunofluorescence microcopy and immunoblotting human respiratory epithelial cells were analyzed as previously described24. Monoclonal mouse anti-DNAH5 (aa42–aa325) and anti-DNALI1 (aa3–aa280) antibodies were raised by immunization of mice with bacterial expressed His-tagged protein truncations. Polyclonal rabbit anti-DNAH9 (aa75–aa220) antibodies were raised by immunization of rabbit with bacterial expressed maltose-tagged protein truncations.
Chlamydomonas pf13 mutant
The PF13 gene was cloned from insertional allele pf13-328 (Supplementary Fig. S6) which was previously created by transformation of a nit2 strain with plasmid pMN60, and was provided by Dr. Paul Lefebvre (Univ Minn). The pf13-3 allele was used for all work presented.
Mouse testis cytosolic extract
Seminiferous tubules were taken from the testis of adult male mice (B6). Homogenized samples were filtered through cell strainer and centrifuged. The supernatant was then ultracentrifuged and used as testis cytosolic extracts for immunoprecipitation and pull-down assay.
Patients and mutational analysis
Signed and informed consent was obtained from patients fulfilling diagnostic criteria of PCD4. The three KTU exons were amplified by PCR and sequenced bidirectionally. In specimen from patients with KTU mutations ciliary and sperm flagella beat was assessed using a high-speed video microscopy system24.
Methods
Immunohistochemistry
For immunohistochemistry of renal tubules were fixed in 4% paraformaldehyde (PFA) in PBST for 1 hr. After dehydration, they were paraffin-embedded and sectioned at 5 µm. The sections were deparaffined and boiled in 1 mM EDTA for 17 min. using microwave and blocked in 5% skim milk in PBST. The Ktu signal was amplified with VECTASTAIN ABC elite kit (Vector Laboratory, UK) and TSA-Alexa647 (Molecular Probes).
Antibodies used include mouse monoclonal antibodies to α-tubulin (DM-1A, SIGMA, MO), γ-tubulin (GTU-88, SIGMA, MO) and acetylated- α-tubulin (6-11B-1, SIGMA, MO). Secondary antibodies (Alexa Fluor 488 and Alexa Fluor 555) were from Molecular Probes/Invitrogen (Eugene, OR). DNA was stained with propidium iodide (SIGMA, MO).
Video microscopy of cilia
Medaka sperm motility was captured using 40 × lens on Olympus BX61 equipped with ARTCAM-130MI-BW (ARTRAY, Japan). Movies were acquired about 100 frames per second and played back at 30 frames per second. KV cilia movement was captured using a 60 × water immersion lens on Olympus high-speed microscope system (Olympus, Japan) equipped with FASTCAM-MAX (Photron, Japan).
Electron microscopy for medaka
Medaka embryos were dechorionated, using hatching enzyme in 1 × Yamamoto ringer, before fixation. Dechorionated embryos were incubated in 1 × Yamamoto ringer until the 8-somite stage. Embryos were fixed and processed as previously described37. For electron microscopy of the sperm flagella, collected sperm were processed as described38.
GST pull-down assay
pGEX 4T-2 plasmids (GE Healthcare, PA) containing mKtu full length/N/C2 cDNA were used. Transformed and IPTG-induced E.coli (BL21 codon+) were sonicated in solubilization buffer (40 mM Tris-HCl (pH 7.5), 0.5 mM EDTA, 0.5% Triton-X100). After centrifugation, supernatants were mixed with glutathione sepharose 4B (GE Healthcare, PA) at 4 °C. Glutathione beads were washed with solubilization buffer for 3 times and resuspended with 100 µl of binding buffer (50mM Tris-HCl pH 7.5, 150 mM KCl, 5mM MgCl2, 0.8% NP-40).
GST and GST-mKtu full length/ N/ C2 proteins were conjugated with glutathione beads. After each bead was resuspended with 100 µl of binding buffer, 200 µl of mouse testis cytosolic extracts were added and mixtures were rotated at 4 °C for 40 min. Beads were washed with binding buffer for 3 times, proteins were separated by SDS-PAGE and analyzed by western blotting with DNAI2 monoclonal antibody (M01, clone 1C8, Abnova, Taiwan), and alpha-Tubulin (DM 1A, SIGMA, MO). Additional antibodies used include Hsp70 (BD Biosciences, 610607), Hsp90 (BD Biosciences, 610418), and Actin (polyclonal anti-goat IgG, , Santa Cruz).
Immunoprecipitation from mouse testis extracts
Anti-mKtu antibody and rabbit control IgG were chemically cross-linked to protein A beads using DMP (Dimethyl pimelimidate, SIGMA). Prepared mouse testis extracts were mixed with 1/4 volume of IP buffer (50mM Tris-Hcl (pH7.5), 150mM KCl, 5mM MgCl2, 0.8% NP-40), then ~15µl of anti-mKtu/rIgG beads (1µg/µl) was added. Mixture was rotated for 3 hours at 4 °C, and then washed with ~20-fold reaction-volume of IP buffer. Immunoprecipitates were eluted from antibody-coupled beads by Elution buffer (100mM Glycine (pH2.0), 150mM NaCl) and neutralized by 1/10 eluted volume of 1.5M Tris-HCl (pH8.8).
For mass-spectrometric analysis, see Supplementary Fig. S7.
Immunoblotting
Protein extracts were prepared from human respiratory epithelial cells. Proteins were separated on a NuPAGE 4–12% bis-tris gel (Invitrogen, Karlsruhe, Germany) and blotted onto a PVDF membrane (Amersham). The blot was processed for ECL plus (Amersham) detection using anti-KTU (Atlas antibodies, Sweden)/anti-GAPDH (1:1000) and anti-rabbit/anti-mouse HRP (1:5000) antibodies (Santa Cruz, Heidelberg, Germany).
Immunofluorescence analysis for human respiratory cells
Respiratory epithelial cells were obtained by nasal brush biopsy (cytobrush plus, Medscand Malmö, Sweden) and suspended in cell culture medium. Sperm cells were washed with PBS. Samples were spread onto glass slides, air dried and stored at −80°C until use. Staining was performed as described previously24. Antibodies used: mouse anti-acetylated-α-tubulin antibodies (SIGMA, Germany), monoclonal mouse anti-DNAI2 antibodies (Abnova Corporation ,Taiwan), polyclonal rabbit anti-α/β-tubulin (Cell Signaling Technology Inc., MA). Highly cross-adsorbed secondary antibodies (Alexa Fluor 488, Alexa Fluor 546) were obtained from Molecular Probes (Invitrogen). DNA was stained with Hoechst 33342 (Sigma).
Patients and families
Signed and informed consent was obtained from patients and family members using protocols approved by the Institutional Ethics Review Board at the University of Freiburg. We studied DNA from a total of 112 PCD patients originating from unrelated families. Clinical information of the three patients carrying homozygous KTU mutations was obtained by review of the medical records.
Mutational Analysis
Genomic DNA was isolated by standard methods directly from blood samples or from lymphocyte cultures after Epstein-Barr virus transformation. The 3 exons of KTU (C14ORF104) were amplified by PCR of 3 genomic fragments and sequenced bidirectionally by using BigDye Terminator v3.1 Cycle Sequencing Kit (Perkin Elmer). Samples were analyzed on an Applied Biosystems 3730xl DNA Analyzer. Sequence data were evaluated using the Codoncode software (CodonCode Corporation, MA). Segregation analyses were performed by sequencing.
High-speed video analysis of ciliary beat in human cells
Ciliary and sperm flagella beat was assessed with the SAVA system (Sisson-Ammons Video Analysis of ciliary beat frequency). Trans-nasal brush biopsies and sperm ejaculate were rinsed in cell culture medium and immediately viewed with an Olympus IMT-2 inverted phase-contrast microscope equipped with a Redlake ES-310 Turbo monochrome high-speed video camera (Redlake, AZ) and a 40× objective. Digital image sampling was performed at 125 frames per second and 640×480 pixel resolution.
Electron microscopy of Chlamydomonas
Axonemes were isolated, fixed and processed for thin section electron microscopy as previously described39. Averages were generated using digital images from scanned negatives and were normalized to a region on the outside wall of the doublet microtubules. Normalization, averaging, and creation of subtracted images followed previously described procedures40.
Cloning the PF13 gene
See Supplementary Fig. S6.
Antibodies to Chlamydomonas proteins
A portion of the PF13 cDNA spanning amino acids 3–460 was cloned into pGEX-2T, and a bacterially expressed GST fusion protein was gel-purified and used to raise two rabbit polyclonal antisera, R1087 and R1088.
Additional antibodies used include polyclonal anti-IFT4641, polyclonal anti-HCα17, monoclonal anti-HCβ, anti-IC1, and anti-IC242, monoclonal anti-HCγ 12γB43 and polyclonal anti-LC1, -LC2, -LC3 and -LC644. Antibodies against inner row dyneins included polyclonal anti-p2826, polyclonal anti-IC97 (W. Sale, Emory Univ), polyclonal anti-IC14045, and polyclonal anti-HC9. The HC9 antibody was generated by cloning nt 859–1421 of the HC9 cDNA coding region, spanning aa 287–47346, into pGEX2T and using the resulting GST fusion protein to immunize rabbits (see Supplementary Fig. S4e for antibody specificity). As a marker for cytoplasmic proteins, we used a polyclonal antibody against chloroplast cytochrome f, the product of the chloroplast PetA gene47. Immunoprecipitation was done as previously described17.
Supplementary Material
Acknowledgments
We thank Dr. Cecilia Lo (National Heart Lung and Blood Institute, NIH) and Dr. Deborah Morris-Rosendahl (Institute for Human Genetics, Freiburg) for critical reading of this manuscript. We are grateful to Ms. Sugimoto M., Ito-Igarashi A., Nakaguchi K., Minami S., Park Y.H., Mochizuki Y., Ozawa Y., Ohki K., Obata T., Heer A. and Reinhardt C. for excellent fish care and/or experimental assistance. We also thank Dr. A. Shimada and Mr. D. Nihei for their help in medaka experiments, J. Freshour and M. Nakatsugawa for help with Chlamydomonas, and S. King, H. Qin, W. Sale and D. Stern for antibodies. Our mutant screening was carried out mainly at the National Institute of Genetics (NIG), supported by NIG Cooperative Research Program (2002–2006). This work was supported in part by Grants-in-Aid for Scientific Research Priority Area Genome Science and Scientific Research (A and B), Global COE Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, Yamada Science Foundation, and a Bio-Design Project of the Ministry of Agriculture, Forestry and Fisheries of Japan. DK was a research fellow supported by the 21th century COE program of the University of Tokyo, MEXT, Japan. This work was supported by grants to H.O. from the “Deutsche Forschungsgemeinschaft” DFG Om 6/4, GRK1104, BIOSS and the SFB592, and to DRM from the NIH, GM44228. We would like to acknowledge the sequencing activities by Katja Borzym and the Seq-Team at MPI-MG, which was supported by the German Ministry of Science and Education (BMBF) by grant: NGFN-2: "01GR0414-PDN-S02T17" to RR. We are grateful for the support by the "Primare Ciliaere Dyskinesie and Kartagener Syndrom e.V."
Footnotes
Author Contributions Research planning and supervision by H.Omran, D.R.M. and H.T.; medaka genetics and phenotypic analyses by D.K., T.T. and H.T.; biochemical experiments using mouse testis by T.T., S.K. and Y.W.; high-speed video microscopy of medaka KV cilia by C.H., H.M., H.K., D.K. and A.M.; EM of medaka cilia/flagella by H.H. and R.K.; experiments on human PCD by H. Omran, H. Olbrich, N.T.L, M.F., H.Z., H.S. and R.R.; Chlamydomonas experiments by D.R.M, Q.Z., G.L., E.O., T.Y. and R.K.; and Paper writing by H.Omran, D.R.M. and H.T.
References
- 1.Okada Y, et al. Mechanism of nodal flow: A conserved symmetry breaking event in left-right axis determination. Cell. 2005;121(4):633. doi: 10.1016/j.cell.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8(11):880. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 3.Wilson PD. Polycystic kidney disease. N Engl J Med. 2004;350(2):151. doi: 10.1056/NEJMra022161. [DOI] [PubMed] [Google Scholar]
- 4.Zariwala MA, Knowles MR, Omran H. Genetic defects in ciliary structure and function. Annu Rev Physiol. 2007;69:423. doi: 10.1146/annurev.physiol.69.040705.141301. [DOI] [PubMed] [Google Scholar]
- 5.Olbrich H, et al. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left-right asymmetry. Nat Genet. 2002;30(2):143. doi: 10.1038/ng817. [DOI] [PubMed] [Google Scholar]
- 6.Pennarun G, et al. Loss-of-function mutations in a human gene related to Chlamydomonas reinhardtii dynein IC78 result in primary ciliary dyskinesia. Am J Hum Genet. 1999;65(6):1508. doi: 10.1086/302683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartoloni L, et al. Mutations in the DNAH11 (axonemal heavy chain dynein type 11) gene cause one form of situs inversus totalis and most likely primary ciliary dyskinesia. PNAS. 2002;99(16):10282. doi: 10.1073/pnas.152337699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Budny B, et al. A novel X-linked recessive mental retardation syndrome comprising macrocephaly and ciliary dysfunction is allelic to oral-facial-digital type I syndrome. Hum Genet. 2006;120(2):171. doi: 10.1007/s00439-006-0210-5. [DOI] [PubMed] [Google Scholar]
- 9.van Dorp DB, Wright AF, Carothers AD, Bleeker-Wagemakers EM. A family with RP3 type of X-linked retinitis pigmentosa: an association with ciliary abnormalities. Hum Genet. 1992;88(3):331. doi: 10.1007/BF00197269. [DOI] [PubMed] [Google Scholar]
- 10.Duriez B, et al. A common variant in combination with a nonsense mutation in a member of the thioredoxin family causes primary ciliary dyskinesia. PNAS. 104(9):3336. doi: 10.1073/pnas.0611405104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wittbrodt J, Shima A, Schartl M. Medaka--a model organism from the far East. Nat Rev Genet. 2002;3(1):53. doi: 10.1038/nrg704. [DOI] [PubMed] [Google Scholar]
- 12.Kasahara M, et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447(7145):714. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- 13.Furutani-Seiki M, et al. A systematic genome-wide screen for mutations affecting organogenesis in Medaka, Oryzias latipes. Mechanisms of Development. 2004;121(7–8):647. doi: 10.1016/j.mod.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 14.Yokoi H, et al. Mutant analyses reveal different functions of fgfr1 in medaka and zebrafish despite conserved ligand-receptor relationships. Developmental Biology. 2007;304(1):326. doi: 10.1016/j.ydbio.2006.12.043. [DOI] [PubMed] [Google Scholar]
- 15.Hojo M, et al. Right-elevated expression of charon is regulated by fluid flow in medaka Kupffer's vesicle. Dev Growth Differ. 2007;49(5):395. doi: 10.1111/j.1440-169X.2007.00937.x. [DOI] [PubMed] [Google Scholar]
- 16.Huang B, Piperno G, Luck DJ. Paralyzed flagella mutants of Chlamydomonas reinhardtii. Defective for axonemal doublet microtubule arms. J Biol Chem. 1979;254(8):3091. [PubMed] [Google Scholar]
- 17.Fowkes ME, Mitchell DR. The Role of Preassembled Cytoplasmic Complexes in Assembly of Flagellar Dynein Subunits. Mol Biol Cell. 1998;9(9):2337. doi: 10.1091/mbc.9.9.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Essner JJ, et al. Kupffer's vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 2005;132(6):1247. doi: 10.1242/dev.01663. [DOI] [PubMed] [Google Scholar]
- 19.Kramer-Zucker, Albrecht G, et al. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer's vesicle is required for normal organogenesis. Development. 2005;132(8):1907. doi: 10.1242/dev.01772. [DOI] [PubMed] [Google Scholar]
- 20.Gonzales FA, Zanchin NI, Luz JS, Oliveira CC. Characterization of Saccharomyces cerevisiae Nop17p, a novel Nop58p-interacting protein that is involved in Pre-rRNA processing. J Mol Biol. 2005;346(2):437. doi: 10.1016/j.jmb.2004.11.071. [DOI] [PubMed] [Google Scholar]
- 21.Zhao R, et al. Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J Cell Biol. 2008;180(3):563. doi: 10.1083/jcb.200709061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boulon S, et al. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J Cell Biol. 2008;180(3):579. doi: 10.1083/jcb.200708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mochizuki E, et al. Fish mesonephric model of polycystic kidney disease in medaka (Oryzias latipes) pc mutant. Kidney Int. 2005;68(1):23. doi: 10.1111/j.1523-1755.2005.00378.x. [DOI] [PubMed] [Google Scholar]
- 24.Fliegauf M, et al. Mislocalization of DNAH5 and DNAH9 in Respiratory Cells from Patients with Primary Ciliary Dyskinesia. Am. J. Respir. Crit. Care Med. 2005;171(12):1343. doi: 10.1164/rccm.200411-1583OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamiya R. Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int Rev Cytol. 2002;219:115. doi: 10.1016/s0074-7696(02)19012-7. [DOI] [PubMed] [Google Scholar]
- 26.LeDizet M, Piperno G. The light chain p28 associates with a subset of inner dynein arm heavy chains in Chlamydomonas axonemes. Mol Biol Cell. 1995;6(6):697. doi: 10.1091/mbc.6.6.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hornef N, et al. DNAH5 mutations are a common cause of primary ciliary dyskinesia with outer dynein arm defects. Am J Respir Crit Care Med. 2006;174(2):120. doi: 10.1164/rccm.200601-084OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tam LW, Lefebvre PA. Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics. 1993;135(2):375. doi: 10.1093/genetics/135.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto R, Yanagisawa HA, Yagi T, Kamiya R. A novel subunit of axonemal dynein conserved among lower and higher eukaryotes. FEBS Lett. 2006;580(27):6357. doi: 10.1016/j.febslet.2006.10.047. [DOI] [PubMed] [Google Scholar]
- 30.Ahmed NT, Mitchell DR. ODA16p, a Chlamydomonas flagellar protein needed for dynein assembly. Mol Biol Cell. 2005;16(10):5004. doi: 10.1091/mbc.E05-07-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318(5848):245. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmed TN, et al. ODA16 aids axonemal outer row dynein assembly through an interaction with the intraflagellar transport machinery. J Cell Biol. 2008 doi: 10.1083/jcb.200802025. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young JC, Barral JM, Ulrich Hartl F. More than folding: localized functions of cytosolic chaperones. Trends Biochem Sci. 2003;28(10):541. doi: 10.1016/j.tibs.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell BF, et al. ATP production in Chlamydomonas reinhardtii flagella by glycolytic enzymes. Mol Biol Cell. 2005;16(10):4509. doi: 10.1091/mbc.E05-04-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirokawa N, Takemura R. Molecular motors and mechanisms of directional transport in neurons. Nat Rev Neurosci. 2005;6(3):201. doi: 10.1038/nrn1624. [DOI] [PubMed] [Google Scholar]
- 36.Rosenbaum JL, Witman GB. Intraflagellar transport. Nature Reviews Molecular Cell Biology. 2002;3(11):813. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 37.Hagiwara H, Shibasaki S, Ohwada N. Abnormal cilia in human uterine tube epithelium. Journal of Clinical Electron Microscopy. 1990;23:493. [Google Scholar]
- 38.Kamiya R. Mutations at twelve independent loci result in absence of outer dynein arms in Chylamydomonas reinhardtii. J Cell Biol. 1988;107(6 Pt 1):2253. doi: 10.1083/jcb.107.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell DR, Sale WS. Characterization of a Chlamydomonas insertional mutant that disrupts flagellar central pair microtubule-associated structures. J Cell Biol. 1999;144(2):293. doi: 10.1083/jcb.144.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mastronarde DN, et al. Arrangement of inner dynein arms in wild-type and mutant flagella of Chlamydomonas. J Cell Biol. 1992;118(5):1145. doi: 10.1083/jcb.118.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hou Y, et al. Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J Cell Biol. 2007;176(5):653. doi: 10.1083/jcb.200608041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell DR, Rosenbaum JL. Protein-protein interactions in the 18S ATPase of Chlamydomonas outer dynein arms. Cell Motil Cytoskeleton. 1986;6(5):1986. doi: 10.1002/cm.970060510. [DOI] [PubMed] [Google Scholar]
- 43.King SM, Otter T, Witman GB. Characterization of monoclonal antibodies against Chlamydomonas flagellar dyneins by high-resolution protein blotting. Proc Natl Acad Sci U S A. 1985;82(14):4717. doi: 10.1073/pnas.82.14.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DiBella LM, et al. Differential light chain assembly influences outer arm dynein motor function. Mol Biol Cell. 2005;16(12):5661. doi: 10.1091/mbc.E05-08-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang P, Sale WS. The Mr 140,000 intermediate chain of Chlamydomonas flagellar inner arm dynein is a WD-repeat protein implicated in dynein arm anchoring. Mol Biol Cell. 1998;9(12):3335. doi: 10.1091/mbc.9.12.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yagi T, et al. An axonemal dynein particularly important for flagellar movement at high viscosity. Implications from a new Chlamydomonas mutant deficient in the dynein heavy chain gene DHC9. J Biol Chem. 2005;280(50):41412. doi: 10.1074/jbc.M509072200. [DOI] [PubMed] [Google Scholar]
- 47.Chen X, Kindle KL, Stern DB. The initiation codon determines the efficiency but not the site of translation initiation in Chlamydomonas chloroplasts. Plant Cell. 1995;7(8):1295. doi: 10.1105/tpc.7.8.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.