Abstract
Background and Objectives
Shiga toxin-producing Escherichia coli (STEC) strains are human pathogens linked to hemorrhagic colitis and hemolytic uremic syndrome. Shiga toxins (Stx1 and Stx2) are the major virulence factors of these strains. The aim of this work was to study the prevalence and distribution of stx 1 and stx 2 gene in E. coli O157:H7 and non-O157:H7 strains isolated from cattle in Shiraz, Iran.
Materials and Methods
Four hundred and twenty samples consisted of recto-anal mucosal swabs were collected from cattle. They were checked for the presence of the stx1 and stx2 gene using multiplex-PCR every 1 week over a 1-year period (2007-2008).
Results
A total of 146 strains carrying the stx1 and stx2 gene were isolated from 51 (12.14%) cattle. Overall, 15 (3.57%) were identified as O157:H7 and 131 (31.19%) revealed to be non-O157:H7. Both stx2 and stx1 genes were detected in 51 (34.93%) STEC isolates. Genotypes stx1 and stx2 were detected in 15 (10.27%) and 78 (53.42%) respectively. Seasonal distribution of stx genes revealed high percentage of positive animals in warm seasons. The gene sequence similarity ranged from 94 to 100%.
Conclusion
Frequency of stx1 and stx2 in animals and its relation to human disease is not well understood in Iran. The high prevalence of STEC in cattle seems to parallel that which is usually observed in warm seasons and it also parallels occurrence of human STEC. The higher prevalence of the stx2 gene than stx1 in strain populations isolated from cattle indicates a risk alert of E. coli O157:H7 being shed by cattle in these populations. Appropriate measures are now needed to prevent the spread of this life-threatening foodborne disease in our country.
Keywords: STEC, stx1, stx2, cattle, Iran
INTRODUCTION
The broad group of E. coli are known as Enterohemorrhagic Escherichia coli (EHEC), including E. coli O157:H7 and non-O157 (1). EHEC refers to a subset of Shiga toxin-producing Escherichia coli (STEC) strains found to cause human and sometimes animal disease (2). They are linked to development of hemorrhagic colitis (HC), hemolytic uremic syndrome (HUS) (3) and thrombotic hospitalization and intensive care (4). STEC O157:H7 strains were first isolated from cattle in Argentina in 1977, although the strains were identified as such 10 years later (5).
Cattle are considered the primary reservoir of both O157:H7 and non-O157 STEC bacteria (2). Cattle frequently excrete the bacteria in their feces (6). The illness is often linked to the consumption of contaminated and undercooked ground beef. Although other means of transmission (7, 8) have been reported (8).
Several virulence factors have gained importance for the pathogenesis of the STEC infections (9). However, the complete list of bacterial virulence determinants necessary for STEC to cause EHEC-related disease is not known. Shiga toxin is the key factor in STEC pathogenesis (2, 10). Shiga toxin is toxic to human colonic, ileal epithelial (11) and endothelial cells (12). Two main groups of Shiga toxins are harbored in STEC (13–15). Shiga toxin 1 (16) is 98% homologous to the Stx produced by Shigella dysenteriae type 1, while Stx2 is about 60% homologous with Stx1 and is antigenically different (9, 17).
E. coli O157:H7 is a common cause of bloody diarrhea in developed countries, but its incidence in developing countries including Iran is not clear. The limited prevalence data in foods and animals in Iran has made the assessment of risks difficult, and also the options for management and control are unclear. In Iran, only a few studies have reported the isolation and characterization of STEC in humans (18–20). E. coli O157:H7/ H- was sporadic reported to be present in up to 11.5% of cattle in Iran (21). Unfortunately, there is no conclusive data from Middle-East particularly from Iran. Therefore, the objective of this study was to determine the frequency of bacterial virulence profiles of stx in STEC O157 and non-157 in cattle in a one-year period.
MATERIALS AND METHODS
Sample collection. In one year, from September 2007 to August 2008, 420 samples consisting of recto-anal mucosal swabs from cattle were collected in Shiraz-Iran. All samples were collected aseptically, placed in coolers, transported to the laboratory, and kept at 4°C. The samples were immediately processed upon arrival to the laboratory. A questionnaire was carried out for risk factor of E. coli O157:H7 determination that included season of the year. The prevalence ratio (PR) was determined using Win Episcope 2.0 at 95.0% level of confidence for association between risk factors.
Seasonal variations in the presence of the stx genes in bacterial populations from rectum were studied by sampling weekly over a 1-year period. For groups of cattle derived from one herd, a random selection of 10% of the total number of cattle was sampled, with a maximum of 10 cattle being sampled. The enumeration of stx gene-carrying bacteria was performed by the MPN-PCR method (Most Probable Number).
Bacterial strains and media. E. coli O157:H7 strain EDL933 (kindly from Prof. David Gally, University of Edinburgh, UK), which produces Stx1 and Stx2, was used as positive control. E. coli O157:H7 strain SHT87 (isolated from cattle in Shiraz-Iran, unpublished) which do not possess the genes stx1 and stx2, was used as negative control. The bacteria were grown in modified tryptic soy broth and on sorbitol MacConkey agar (supplementation with ceffexime and potassium tellorite) (mTSB, SMAC -CT) (Difco, Le Pont de Claix, France) and incubated at 37°C overnight.
Isolation and identification of E. coli. All samples were placed in 15 ml of mTSB and incubated overnight at 37°C. The suspension was thoroughly mixed and allowed to stand for a short period before plating. One loop full of enriched broth was streaked onto SMAC-CT. All agar plates were incubated at 37°C for 24 h. Five to six sorbitol negative colonies per sample were collected and streaked on SMAC again. Finally, the bacteria were streaked onto eosin methylene blue (EMB) agar plates and were incubated same as above. The typical E. coli metallic shine on EMB were characterized by biochemical tests, including conventional indol, methyl red, voges proskauer, citrate and lysine decarboxylase tests. The identity of E. coli O157:H7 was confirmed using an anti-E. coli O157 and H7 antisera agglutination kit (Oxoid DR620).
Nucleic acid isolation. One ml of overnight mTSB culture from all the bacterial strains was employed as template for PCR. Cells were pelleted from the cultures at 3,000 rpm for 5 min (Hermle Z23o MA centrifuge) and then continued by DNPTM Sina-gene kit (Cat No.: 8115C). All isolates were examined for verotoxin virulence genes determinants by PCR.
PCR assay. DNA samples (1µg nucleic acids) were amplified in a 25µl volumes reaction mixture of the following constitution: 2 mM magnesium chloride, 2.5mM for each of dATP, dCTP, dGTP, and dTTP, 2 pM for each of the STX-specific oligonucleotide primers (Oligonucleotide Synthesis Laboratory, Roche, Germany) described in table 1, 1.25 U of Taq polymerase (Fermantas, Sylvius, Lithuania) and the final volume was adjusted with sterile double-distilled water.
Table 1.
Specific oligonucleotide primers used for amplification of stx1 and stx2 gene.
| Oligonucleotides | Size | Reference |
|---|---|---|
| Stx1 F CTT CGG TAT CCT ATT CCC GG | 484 | (35) |
| Stx1 R GGA TGC ATC TCT GGT CAT TG | ||
| Stx2 F CCA TGA CAA CGG ACA GCA GTT | 779 | |
| Stx2 R CCT GTC AAC TGA GCA GCA CTT TG |
The samples were overlaid with 100 µl of mineral oil, denatured at 94°C for 5 min, and subjected to 30 cycles of amplification in a DNA Thermal Cycler (Ependorf Mastercycler Gradient). Parameters for the amplification cycles were: denaturation for 30 s at 94°C, annealing of primers for 30 s at 56°C, and primer extension for 30 s at 72°C with auto- extension. After the last cycle, the PCR tubes were incubated for 10 min at 72°C. Six microliters of the reaction mixture was then analyzed by standard submarine gel electrophoresis (1.5% agarose; 5 V/cm), and the reaction products were visualized by being stained with ethidium bromide (0.5, µg/ml in the running buffer).
Bromocresol broth tubes containing 1% D-sorbitol (Sigma, St. Louis, Mo.) were used for the sorbitol fermentation test. All susceptible strains were separately incubated in 250 µl of phosphate-buffered saline with β -glucuronidase tablet (Diatabs, Rosko, Denmark).The incubation time and temperature was 24h at 37°C.
Sequencing: The strains harboring shiga toxin were sequenced both in reverse and forward with the same primers used for stx genes. The obtained sequences were balsted in NCBI databases. The data was analyzed with SPSS (Statistical Package for Social Sciences) for Windows version 11.5 software with assumed confidence level of 95%.
RESULTS
The majorities of the isolated strains were not able to ferment sorbitol within 24 h and had β-D-glucuronidase activity. Only ten strains were able to ferment sorbitol within 24 h, of which only one belonged to the E. coli O157: H_ serotype. This serotype was also negative for the β-D-glucuronidase test. The data point to the high prevalence of stx2 in our study both in O157 and non-O157.
All animals came to the Shiraz slaughterhouse from farms located in different regions of the state. No significant difference in STEC isolation rate was observed when the cattle were grouped according to their geographical origin.
Isolation and characterization of STEC in cattle. A total of 146/420 (34.76%, 95% CI) STEC strains were isolated from 51 (12.14%) out of the 420 cattle, that posses stx 1 and/or stx 2. Fifteen (3.57%) isolates were classified as E. coli O157:H7 and 131(31.19%) non-O157 (Table 2).
Table 2.
Distribution of E. coli O157, Non-O157 stx1, stx2 Genes in Cattle. Shiraz-2007-8.
| Strains (No.) | Stx1 | Stx2 | Stx1+stx2 | -stx | Total% |
|---|---|---|---|---|---|
| O157: H7 | 6 | 8 | 6 | 6 | 15(3.57) |
| Non- O157 | 60 | 121 | 45 | 15 | 131(31.19) |
| Total | 66 | 129 | 51 | 21 | 146(34.76) |
Isolation and characterization of stx gene-carrying STEC bacteria. Any E. coli isolated harboring at least one shiga toxin gene was considered positive for STEC. Both stx 2 and stx 1 genes were detected in 51(34.93%) isolates, but stx 1 was detected in 15 (10.27%) and stx 2 was detected in 78 (53.42%). One or more cattle from each Shiraz farm was positive for stx. The ratio of stx 2 to stx 1 gene-carrying bacteria was 5.2:1. Except for the six strains that apparently lost the genes, the presence of the stx gene was confirmed by specific PCR for all of these isolates.
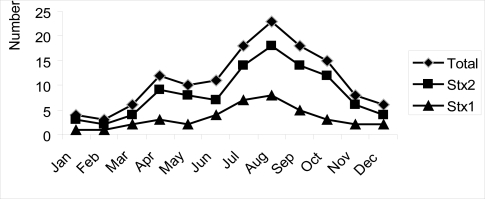
Seasonal distribution of the stx genes. The proportion of each of these bacterial populations that carried stx 2 and/or stx 1 were not similar (Fig. 1). There was a significance seasonal difference for any of the measured parameters, as indicated by an analysis of variance test (P< 0.05). The percentage of positive animals range from 24.28 to 40.9% in warm seasons of May to August (in Iran) compared to winter seasons with the average frequency dropping (8.96 to 11.11%).
Fig. 1.
Seasonality STXs from STEC isolates from cattle Shiraz-Iran.2007–8
Sequencing. Some of the 484 and 779 bp (Table 1) amplimers were sequenced. These phages were isolated from strains summarized in Table 3.
Table 3.
Sequence comparison of stx2 genes in different strains.
| Identity to stx1,2 gene of EDL933 (AE005174.2)% | |||
|---|---|---|---|
| Strains ( Stx1) | Identity to stx1 | Identity to stx2 | Strains ( Stx2) |
| DEC10J, EC108 | 100 | 98 | c466-01B |
| EC127, EC120 | 99 | 98.2 | EC130 |
| BCN26 | 94 | 98.1 | EC176, EC125 |
| EC152 | 99 | 98.6 | EC169, EC131 |
| EC176 | 99 | 97.9 | H2687 serotype O157:NM |
| O157:H7 Str. Sakai | 99 | 99 | I8257, 933W slt-II |
| AB8SF, ECLR2 | 91 | 99.1 | O157:H7 str. Sakai |
DISCUSSION
This is the first study which describes the detection and frequency of major virulence genes of STEC isolated from cattle in Shiraz, Iran. Our data revealed high levels of stx2 gene-carrying bacteria in fecal samples from different cattle. STEC harboring stx 2 was isolated significantly more (53.42%) than STEC stx 1 (10.27%) (P <0. 01). Most human epidemiological studies in Iran have revealed that the prevalence of STEC infection ranges between 0.7 to 15%, but none of them belonged to the O157:H7 serotype (18, 20). Isolation of STEC from bovine reservoirs from other parts of the country has already been documented (8, 21–24). Zahraee Salehi and his colleagues identified STEC O157 among 7 isolates (11.5%), from cattle, whereas non-O157 strains that are frequently associated with sporadic cases of HUS (25, 26), were isolated from 4 (6%) of animals. They showed 5 (8.2%) isolates carried stx genes (21). This finding was in parallel with presence of stx 1 in 35.5 and stx 2 in 49.1% of human isolates (19). This is in contrast with Askari et al. finding with a report of stx 1 and stx 2, among 5% and 1.9% of calves respectively (23). Recently Ludwig Kerstin et al. reported that 71% of children with HUS were due to Stx2-producing E. coli strains (16). In a study in the USA, stx 1 gene was not detected in any strains tested while 93.1% of the isolates were found to carry the stx 2 gene (27).
The gene belonging to strains detected from animals showed more expression of protein toxin than human samples (28), hence the strain within animal origin maintain the characteristic and are more cytotoxic than the gene from human origin (16). This supports the suggestion that cattle may have been the source of the organism for the HUS patients.
The seasonal shedding of STEC and distribution of stx ranged from 8.69 to 40.9%. There are significant seasonal differences in the levels of shedding of stx gene-carrying bacteria. The results revealed decrease in the number of stx gene carrying bacteria during the winter. These results are in agreement with those studies indicating that STEC shedding has seasonal variations by cattle in different countries (5). High isolation rate was observed in late spring and summer in the UK (29), Sweden, Washington State in the USA (30) and Italy (5). In this study, we observed a marked decrease in the prevalence of STEC from October, which in Iran is the beginning of fall, to the end of spring (June). This trend seems to parallel what is usually observed in the summer for the occurrence of human STEC, with very few episodes notified during the winter season (31).
Sequence variations in stx 1 and stx 2 genes of fifteen E. coli O157 isolates in this study were investigated. The similarity ranged from 94 to 100%. As demonstrated in the present investigation, the genetic diversity of organisms causing disease is considerable. (GenBank accession number DQ235775).
In conclusion, there is no data available about the frequency of Stx2 and Stx1 in animal and people in close contact to HUS patients in Iran. The greater observation of the stx2 gene relative to the stx 1 gene in strain populations indicates a risk alert of this gene between these populations (32). Some studies have revealed that strains possessing only stx 2 are potentially more virulent than strains harboring stx 1 or even strains carrying both stx 1 and stx 2 (17, 33). It is of note that most HUS-associated clinically relevant STEC isolates produce Stx2, but at least in Europe, Stx1 is rarely highly relevant (32). Stx2 has been found to be approximately 400 times more toxic (as quantified by LD50 in mice) than Stx1(4).
These findings are important for public health and preventive veterinary medicine. Therefore, emergency cautions are necessary to decrease the incidence of STEC infections in animals and people. In order to achieve this, good hygienic practice and HACCP systems are necessary from the farm to the family table especially in the abattoirs to prevent contamination of meat and abattoir environment with intestinal content.
ACKNOWLEDGEMENTS
This work was supported by grants from the Razi Vaccine and Serum Research Institute.
REFERENCES
- 1.Louie M, Azavedo J, Handelsman M, Clark C, Ally B, Dytoc M. Expression and characterization of the eaeA gene product of Escherichia coli serotype O157:H7. Infect Immun. 1993;61:4085–4092. doi: 10.1128/iai.61.10.4085-4092.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Terrance MA, Genevieve ABG, Mildred RB, Koohmaraie M. Prevalence and Characterization of Non-O157 Shiga Toxin-Producing Escherichia coli on Carcasses in Commercial Beef Cattle Processing Plants. App Environ Microbiol. 2002;68:4847–4852. doi: 10.1128/AEM.68.10.4847-4852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hussein SH, Brandolyn HT, Hudson AG. Verotoxin producing Escherichia coli in sheep grazing an irrigated pasture or arid rangeland forages. VTEC in Sheep. 2003;4:358–364. doi: 10.1177/153537020322800405. [DOI] [PubMed] [Google Scholar]
- 4.Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR, et al. Hemorrhagic colitis associated with a rare Escherichia coli serotype. New Eng J Med. 1983;308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 5.Bonardi S, Maggi E, Bottarelli A, et al. Isolation of verocytotoxin-producing Escherichia coli O157:H7 from cattle at slaughter in Italy. Vet Microbiol. 1999;67:203–211. doi: 10.1016/s0378-1135(99)00039-5. [DOI] [PubMed] [Google Scholar]
- 6.Molina PM, Parma AE, Sanz M. Survival in acidic and alcoholic medium of shiga toxin-producing Escherichia coli O157:H7 and non-O157:H7 isolates in Argentina. BMC Microbiol. 2003;13:3–17. doi: 10.1186/1471-2180-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonough PL, Rossiter CA, Rebhun RB, Stehman SM, Lein DH, Shin SJ. Prevalence of Escherichia coli O157:H7 from cull dairy cows in New York state and comparison of culture methods used during preharvest food safety investigations. J Clin Microbiol. 2000;38:318–322. doi: 10.1128/jcm.38.1.318-322.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jamshidi A, Bassami MR, Rasooli M. Isolation of Escherichia coli O157:H7 from ground beef samples collected from beef markets, using conventional culture and polymerase chain reaction in Mashhad, northeastern Iran. Iran J Vet Res, Shiraz Univers. 2008;9:72–76. [Google Scholar]
- 9.Paton JC, Paton AW. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin Microbiol Rev. 1998;11:450–479. doi: 10.1128/cmr.11.3.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Acheson DW. How does Escherichia coli O157:H7 testing in meat compare with what we are seeing clinically? J Food Protect. 2000;63:819–821. doi: 10.4315/0362-028x-63.6.819. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt H, Scheeff J, Huppertz HI, Frosch M, Karch H. scherichia E.coli O157:H7 and O157:H2 strains that do not produce Shiga toxin: phenotypic and genetic characterization of isolates associated with diarrhea and hemolytic-uremic syndrome. J Clin Microbiol. 1999;37:3491–3496. doi: 10.1128/jcm.37.11.3491-3496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Obrig TG. Washington, D.C.: 1998. Interaction of Shiga toxins with endothelial cells. [Google Scholar]
- 13.Lindgren SW, Samuel JE, Schmitt CK, O'Brien AD. The specific activities of Shiga-like toxin type II (SLT-II) and SLT-II-related toxins of enterohemorrhagic Escherichia coli differ when measured by Vero cell cytotoxicity but not by mouse lethality. Infect Immun. 1994;62:623–631. doi: 10.1128/iai.62.2.623-631.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt H, Scheef J, Morabito S, Caprioli A, Wieler LH, Karch H.A. new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. Appl Environ Microbiol. 2000;66:1205–1208. doi: 10.1128/aem.66.3.1205-1208.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinstein DL, Jackson MP, Samuel JE, Holmes RK, OBA D. Cloning and sequencing of a Shiga-like toxin type II variant from an Escherichia coli strain responsible for edema disease of swine. J Bacteriol. 1988;170:4223–4230. doi: 10.1128/jb.170.9.4223-4230.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ludwig K, Karmali MA, Sarkim V, Bobrowski C, Petric M, Karch H, et al. Antibody response to Shiga toxins Stx2 and Stx1 in children with enteropathic hemolytic-uremic syndrome. J Clin Microbiol. 2001;39:2272–2279. doi: 10.1128/JCM.39.6.2272-2279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli . Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aslani MM, Bouzari S. An epidemiological study on Verotoxin-producing Escherichia coli (VTEC) infection among population of northern region of Iran Mazandaran and Golestan provinces. Europ J Epidemiol. 2003;18:345–349. doi: 10.1023/a:1023602416726. [DOI] [PubMed] [Google Scholar]
- 19.Jomezadeh N, Farajzadeh Sheikh A, Khosravi AD, Amin M. Detection of Shiga Toxin Producing E. coli Strains Isolated from Stool Samples of Patients with Diarrhea in Abadan Hospitals. Iran J Biolog Sci. 2009;9:820–824. [Google Scholar]
- 20.Salmanzadeh-Ahrabi S, Habibi E, Jaafari F, Zali MR. Molecular epidemiology of Escherichia coli diarrhoea in children in Tehran. Ann Trop Paediat. 2005;25:35–39. doi: 10.1179/146532805X23335. [DOI] [PubMed] [Google Scholar]
- 21.Zahraei Salehi T, Mahzounieh M, Asadian F, Khosravi M. Virulence genes in Escherichia coli isolates from calves in shahrekord area, Iran; 16th European Congress of Clinical Microbiology and Infectious Disease; Nice, France: 2006. [Google Scholar]
- 22.Shekarforoosh S, Tahamtan Y, Pourbakhsh A. Detection and frequency of stx2 gene in E.coli O157 and O157 strains isolated from sheep carcasses in Shreaz-Iran. Pakistan J Biomed Sci. 2008;11(8):1085–1092. doi: 10.3923/pjbs.2008.1085.1092. [DOI] [PubMed] [Google Scholar]
- 23.Sepehriseresht S, Zahraei Salehi T, Sattari M, Tadjbakhsh H, Aslani MM. Detection of shigatoxigenic Escherichia coli from fecal samples of calves and cattle by molecular and serological methods. Comparat Clin Pathol. 2008;3:12–17. [Google Scholar]
- 24.Askari Badouei M, Zahraei Salehi T, Rabbani Khorasgani M, Tadjbakhsh H, Nikbakht Brujeni G. Occurrence and characterisation of enterohaemorrhagic Escherichia coli isolates from diarrhoeic calves. Comparat Clin Pathol. 2009;9:201–205. [Google Scholar]
- 25.Rahimi E, Momtaz H, Hemmatzadeh F. The prevalence of Escherichia coli O157:H7, Listeria monocytogenes and Campylobacter spp. on bovine carcasses in Isfahan, Iran. Iran J Vet Res. 2008;9:365–370. [Google Scholar]
- 26.de Sablet T, Bertin Y, Vareille M, Girardeau JP, Garrivier A, Gobert AP, et al. Differential expression of stx2 variants in Shiga toxin-producing Escherichia coli belonging to seropathotypes A and C. Microbiol. 2008;154:176–186. doi: 10.1099/mic.0.2007/009704-0. [DOI] [PubMed] [Google Scholar]
- 27.Karmali MA, Mascarenhas M, Shen S, Ziebell K, Johnson S, Reid-Smith R, et al. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J Clin Microbiol. 2003;41:4930–4940. doi: 10.1128/JCM.41.11.4930-4940.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keen EJ, Elder RO. Isolation of Shiga-toxigenic Escherichia coli O157 from hide surface and the oral cavity of finished beef feedlot cattle. JAMA. 2002;220:756–763. doi: 10.2460/javma.2002.220.756. [DOI] [PubMed] [Google Scholar]
- 29.Aljaro CG, Muniesa M, Jofre J, Blanch AR. Prevalence of the stx2 gene in coliform populations from Aquatic Environments. Appl Environ Microbiol. 2004;70:3535–3540. doi: 10.1128/AEM.70.6.3535-3540.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonardi S, Maggi E, Pizzin G, Morabito S, Caprioli A. Faecal carriage of Verocytotoxin-producing Escherichia coli O157 and carcass contamination in cattle at slaughter in northern Italy. Int J Food Microbiol. 2001;66:47–53. doi: 10.1016/s0168-1605(00)00491-8. [DOI] [PubMed] [Google Scholar]
- 31.Hancock DD, Besser TE, Kinsel ML, Tarr PI, Rice DH, Paros MG. The prevalence of Escherichia coli O157:H7 in dairy and beef cattle in Washington State. Epidemiol Infect. 1994;113:199–207. doi: 10.1017/s0950268800051633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellingsona JLE, Koziczkowskia JJ, Andersona JL, Carlsonb SA, Sharmab VK. Rapid PCR detection of enterohemorrhagic Escherichia coli (EHEC) in bovine food products and feces. Mol Cell Probes. 2005;19:213–217. doi: 10.1016/j.mcp.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 33.Ludwig K, Sarkim V, Bitzan M, Karmali MA, Bobrowski C, Ruder H, et al. Shiga Toxin-Producing Escherichia coli infection and antibodies against Stx2 and Stx1 in household contacts of children with enteropathic hemolytic-uremic syndrome. J Clin Microbiol. 2002;40:12–17. doi: 10.1128/JCM.40.5.1773-1782.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tesh VL, Burris JA, Owens JW, Gordon VM, Wadolkowski EA, O'Brien AD, et al. Comparison of the relative toxicities of Shiga-like toxins type I and type II for mice. Infect Immun. 1993;61:3392–3402. doi: 10.1128/iai.61.8.3392-3402.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fagen PK, Hornitzky MA, Bettelheim KA, Djordjevic SP. Detection of Shiga-Like Toxin (stx1 and stx2), Intimin (eaeA), and Enterohemorrhagic Escherichia coli (EHEC) Hemolysin (EHEC hlyA) Genes in Animal Feces by Multiplex PCR. Appl Environ Microbiol. 1999;65:868–872. doi: 10.1128/aem.65.2.868-872.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]