Abstract
Background and Objectives
In Iran, anaplasmosis is normally diagnosed with traditional Giemsa staining method. This is not applicable for identification of the carrier animals. The aim of this study was to compare the detection of Anaplasma marginale in two different numbers of microscopic fields (50 and 100) using conventional Giemsa staining method compared with the PCR-RFLP technique.
Materials and Methods
In this study, examinations were performed on 150 blood samples from cattle without clinical signs. Sensitivity and specificity of two microscopic fields (50 and 100 fields) were compared with A. marginale specific PCR-RFLP. The degree of agreement between PCR-RFLP and the two microscopic tests was determined by Kappa (κ) values with 95% confidence intervals.
Results
PCR-RFLP showed that 58 samples were A. marginale, while routine microscopy showed erythrocytes harboring Anaplasma like structures in 16 and 75 blood samples determined in 50 and 100 microscopic fields respectively. Examination of 50 and 100 microscopic fields showed 25.8% and 91.4% sensitivity and 99% and 76.1% specificity compared to 100% sensitivity and specificity by PCR-RFLP. The Kappa coefficient between PCR-RFLP and Microscopy (50 fields) indicated a fair level of agreement (0.29). The Kappa coefficient between PCR-RFLP and Microscopy (100 fields) indicated a good level of agreement (0.64)
Conclusion
Our results showed that the microscopic examination remains the convenient technique for day-to-day diagnosis of clinical cases in the laboratory but for the detection of carrier animal with low bacteremia, microscopy with 100 fields is preferable to Microscopy with 50 fields and molecular methods such as PCR-RFLP can be used as a safe method for identifying cattle persistently infected with A. marginale.
Keywords: Anaplasma marginale, Carrier cattle, Microscopic method, Giemsa staining, PCR-RFLP
INTRODUCTION
Anaplasmosis is an arthropod-born disease of cattle and other ruminants caused by species of the genus Anaplasma (Rickettsiales: Anaplasmataceae) (1). Four species, including Anaplasma marginale, A. centrale, A. bovis and A. phagocytophilum are recognized in blood of Iranian cattle by molecular methods (2–5). Based upon location within the infected erythrocyte, two species of Anaplasma that infect cattle have been described, A. marginale and A. centrale. In addition to having differences in morphology, these species display differences in virulence and geographical distribution. A. centrale causes mild infections in cattle. In contrast, the closely related rickettsia A. marginale is the aetiological agent of acute anaplasmosis, a bovine syndrome characterised by a progressive haemolytic anaemia associated with fever, weight loss, abortion, decreased milk production and in some cases, death of the infected cattle (6).
Anaplasmosis caused by A. marginale is an economically important and widespread disease of cattle in most tropical and subtropical countries, including Iran (7, 8). The infectious agent transmitted either biologically by ticks or mechanically by other arthropod vectors (9). Following transmission, A. marginale invades and multiplies within mature erythrocytes. During acute anaplasmosis, rickettsemia levels exceed 109 infected erythrocytes per ml and the resulting disease is characterized by anemia, weight loss, abortion, and death (10). Recovery from acute anaplasmosis results in persistent infection characterized by repetitive cycles of rickettsemia ranging from approximately 102.5 to 107 infected erythrocytes per ml (10). Persistently infected cattle serve as long-term reservoirs for transmission within herds (7). Detection of persistently infected cattle is important to control the movement of infected cattle into and from disease-free regions.
Conventional method for identification includes examination of blood smears using Giemsa staining, which is accompanied with some critical problems. Diagnosis of A. marginale is performed routinely by morphological identification based on location of inclusion bodies marginally within the erythrocytes (11). Microscopic examination by Giemsa staining of blood smears can only detect levels of >106 infected erythrocytes per ml (12). Giemsa-stained blood smears can be indeed used as a suitable method to detect Anaplasma agents in the animals clinically suspected acute anaplasmosis, but it is not applicable for the determination of pre-symptomatic or carrier animals (13).
Conventional microscopy is time-consuming and tedious. Furthermore, microscopic examination of Giemsa stained blood smears, especially from carrier animals, is accompanied with several problems. First of all, due to the very low amount of infected erythrocytes in carrier animals, the detection of good stained Anaplasma organism is very limited and the microscopy it is not possible to distinguish between A. marginale and A. centrale. Additionally the differentiation between Anaplasma organisms and structures like Heinz bodies, Howell-Jolly bodies or staining artifacts, which often seen in Giemsa stained blood smears need special experiences (14).
Molecular methods, with a high degree of sensitivity and specificity, have been developed to identify A. marginale DNA (13, 15) (16) and polymerase chain reaction (PCR) assay has been considered the “gold standard” for detection of persistently infected cattle (17).
In Iran, anaplasmosis is normally diagnosed with the traditional Giemsa staining method, yet it seems not to be applicable for identifying of the carrier animals (8). The aim of this study was to compare the detection of Anaplasma organisms in two different numbers of microscopic fields (50 and 100) using conventional Giemsa staining method with the PCRRestriction Fragment Length Polymorphism (RFLP) technique.
MATERIALS AND METHODS
Collection of blood Samples. From June 2007 to October 2007, 30 farms in Isfahan province, central part of Iran, were selected for the study based on their history of outbreak of bovine anaplasmosis. Blood samples were collected from jugular vein of 150 Friesian and crossbred cattle ranging between 1 and 9 years. Five hundred micro liters of each collected blood samples was fixed with 1 ml 96% ethanol in 1.5 ml sterile eppendorf tubes. Additionally, two thin blood smears were prepared immediately after each blood collection. The blood smears were air dried, fixed in methanol, stained with Giemsa and analyzed for the presence of A. marginale in the erythrocytes at 100× magnification. In each blood smear both 50 and 100 fields were examined separately by a single observer. All smears carefully examined to estimate the Percent Parasitized Erythrocytes (PPE) as described by Coetzee et al. (2005) (18).
DNA extraction. DNA was extracted using a DNA isolation kit (MBST, Iran) according to the manufacturer's instructions. Briefly, ca. 5 mm3 big pieces of fixed blood samples were first air dried and subsequently lysed in 180 l lysis buffer and the proteins were degraded with 20 l proteinase K for 10 min at 55°C. After addition of 360 l Binding buffer and incubation for 10 min at 70°C, 270 l ethanol (96%) was added to the solution and after vortexing, the complete volume was transferred to the MBST-column. The column was first centrifuged, and then washed twice with 500 l washing-buffer. Finally, DNA was eluted from the carrier using 100 l Elution buffer. The amount of extracted DNA and its purity was measured by OD260 and the ratio of OD260 to OD280 respectively. In addition the extracted DNA was analyzed on agarose gel before use.
PCR. Primers were designed from the published sequence of 16S ribosomal RNA (GenBank accession no. M60313) from A. marginale and were as follows P1 (Forward, 5'AGAGTTTGATCCTGGCTCAG3‘, positions 1 to 20); P2 (Reverse, 5'GTTAAGCCCTGGTATTTCAC3’, positions 558 to 577). Approximately 100 to 500 ng DNA was used for the PCR analysis. The PCR was performed in 100 l total volume including one time PCR buffer, 2.5 U Taq DNA Polymerase (Cinnagen, Iran), 2 l of each primer (P1/P2, 20 M, Cinnagen, Iran), 200 M of each dATP, dTTP, dCTP and dGTP (Fermentas, EU) and 1.5 mM MgCl2 in automated Thermocycler (MWG, Germany) with the following program: 5 min incubation at 95°C to denature double strand DNA, 35-38 cycles of 45 s at 94°C (denaturing step), 45 s at 56°C (annealing step) and 45 s at 72°C (extension step). Finally, PCR was completed with the additional extension step for 10 min. The PCR products were analyzed on 2% agarose gel in 0.5 times Tris-Borate-EDTA (TBE) buffer and visualized using ethidium bromide and UV-illuminator. A molecular mass ladder (100 bp) and positive and negative controls were used for each batch run. Each sample was spiked with positive control A. marginale DNA to detect any inhibition of the PCR that might lead to falsenegative results.
RFLP for A. marginale. One micro liter of the extracted DNA from blood samples was amplified with the primers P1/P2, resulting in a PCR product of 577 bp for all Anaplasma spp. The PCR products were purified from enzyme and salts using PCR-product purification kit (MBST). 10 l of purified PCR product (577 bp) was then cut with 0.1 l restriction endonuclease Bst 1107 I (Roche, 10U/l) in 2.5 l 10 x corresponding buffer and 12.5 l H2O for 1 h by 37°C. As control 10 l PCR products was treated with 2.5 l 10 x corresponding buffer and 12.5 l H2O without adding of enzyme.
Statistical analysis. The degree of agreement between PCR-RFLP and the two microscopical tests was determined by Kappa (κ) values with 95% confidence intervals. We used the PCR-RFLP as the reference test to calculate the relative sensitivity and relative specificity of the microscopical tests.
RESULTS
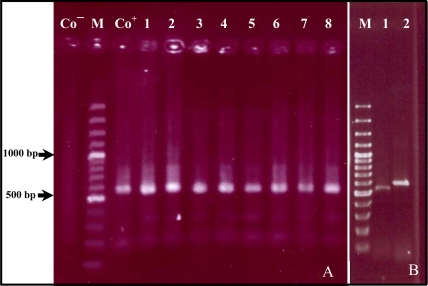
The DNA was extracted from blood samples and analyzed by PCR using primers derived from the 16S rRNA gene. The nucleotide sequence of 16S rRNA gene is highly conserved in Anaplasma spp. and the primers P1/P2 can amplify the corresponding fragments of the gene in all known Anaplasma species. PCR analysis of the DNA isolated from blood samples showed that 58 out of the total 150 blood samples were Anaplasma spp. positive and revealed an expected PCR product of 577 bp in length (Fig. 1A).
Fig. 1.
DNA isolated from blood was analysed by PCR and PCR-RFLP. A: DNA was amplified with primer P1/P2 resulting in PCR product of 577 bp in length (lanes 1-8). B: PCR product of 577 bp (line 2) was cut with restriction endonuclease. BST1107I resulting in DNA fragment of 509 bp (line 1). Co– =Negative control. Co+ =Positive control. M=100 bp molecular marker.
For determination of A. marginale specificity of the PCR products, PCR-RFLP method was used (2, 3). The restriction endonuclease Bst 1107I recognizes the sequence (GTATAC) in corresponding PCR product (577 bp) of A. marginale and makes a cut in position 68, whereas the used restriction enzyme can not cut the corresponding PCR product of other Anaplasma sp. Analysis of all 58 Anaplasma positive PCR products with the restriction endonuclease Bst 1107I showed that all PCR products could be cut in two expected DNA fragments with 509 bp and 68 bp in length respectively (Fig. 1B). Fifty-eight blood samples were A. marginale positive by PCR-RFLP. Ninety-two samples were Anaplasma negative by PCR-RFLP.
Interestingly, Giemsa staining analysis of blood smears showed different results dependent upon the chosen number of examined microscopic fields by 100 x magnification. In 16 out of 150 blood samples, Anaplasma like structures could be identified when the examination was performed in 50 microscopic fields. From these 16 samples, 15 were also PCR-RFLP positive and 1 sample was negative. When the examination was performed in 100 microscopic fields, 75 samples were determined as Anaplasma positive, from which 53 samples were also PCR-RFLP positive. This means that 22 samples were Anaplasma false positive (Table 1).
Table 1.
Comparison of results of PCR-RFLP assay and microscopic examination for A. marginale in 150 cattle blood samples.
| PCR-FLP assay results | Microscopy results | ||||
|---|---|---|---|---|---|
| 50 fields | 100 fields | ||||
| + | − | + | − | ||
| + | 15 | 43 | 53 | 5 | |
| − | 1 | 91 | 22 | 70 | |
The percentage of erythrocytes harboring Anaplasma like structures varied in the positive blood samples from 10-3% to 10-2%. Examination of 50 and 100 microscopic fields showed 25.8% and 91.4% sensitivity and 99% and 76.1% specificity respectively compared to 100% sensitivity and specificity for PCR-RFLP (Table 2).
Table 2.
Sensitivity and specificity of microscopical methods compared to 100% sensitivity and specificity of PCR-RFLP for detection of A. marginale in carrier cattle.
| Method | No. of samples examined | No. of positives detected | Sensitivitya(%) | Specificityb(%) |
|---|---|---|---|---|
| PCR-RFLP | 150 | 58 | 100 | 100 |
| Microscopy(50 fields) | 150 | 16 | 25.8 | 99 |
| Microscopy(100 fields) | 150 | 75 | 91.4 | 76.1 |
Calculated as follows: [number of true positives/(number of true positives + number of false negatives)] × 100.
Calculated as follows: [number of true negatives/(number of true negatives + number of false positives)]× 100.
The Kappa coefficient between PCR-RFLP and Microscopy (50 fields) indicated a fair level of agreement (0.29). The Kappa coefficient between PCR-RFLP and Microscopy (100 fields) indicated a good level of agreement (0.64). The Kappa coefficient between Microscopy with 100 fields and Microscopy with 50 fields indicated a poor level of agreement (0.2).
DISCUSSION
Molecular methods based on DNA with high degree of sensitivity and specificity have been developed (13, 16). PCR methods based on the 16S rRNA gene are already used for differentiation of genus Anaplasma (11) but the sequence strong similarity of this gene in A. marginale and A. centrale do not allow the use of the simple PCR method for discrimination between these two species. The 16S rRNA sequence of A. marginale and A. centrale differed only in two positions within hyper-variable region (V1) and designing of species-specific primers is near impossible (2).
Microscopic examination of Giemsa stained blood smears were used traditionally to diagnose not only the acute anaplasmosis but also to detect the carrier animals in Iran, which is accompanied with serious problems. Serological tests were also developed for the diagnosis of anaplasmosis. But due to the cross reactivity, this method is not suitable for the differential diagnosis of anaplasmosis (19–23).
Our results showed that the traditional microscopic examination of blood smears is not able to detect low bacteremia in carrier cattle. Furthermore, the Anaplasma like structures recognized in erythrocytes are often difficult to differentiate from Heinz bodies, Howell-Jolly bodies or staining artifacts (14). This means, due to the very low amount (10-2% – 10-3%) of infected erythrocyte in the examined carrier cattle, it is very difficult to determine the Anaplasma organisms by simple Giemsa staining, which is performed routinely in the laboratories in Iran. To determine the sensitivity and specificity of the microscopic examination, 50 and 100 microscopical fields were analyzed and compared with the corresponding PCR-RFLP analysis.
Examination of 50 microscopic fields showed 25.8% sensitivity and 99% specificity compared to 100% sensitivity and specificity of PCR-RFLP. With sole use of this method, 16 blood samples were identified as Anaplasma positive, from which 1 sample was false positive. This means that within 150 blood samples, 43 PCR-RFLP positive samples were recognized as false-negative. In Iran, most of the veterinary laboratories examine 50 microscopic fields for detection of parasites in blood smears. Although this microscopic screening is specific, this approach often lacks the desired sensitivity.
Examination of 100 microscopic fields showed 91.4% sensitivity and 76.1% specificity compared to RFLP-PCR results. With this approach, 75 blood samples were recognized as Anaplasma positive from which only 53 samples were Anaplasma PCR-RFLP positive. This means that with this method, 22 blood samples were recognized as false positive samples. Although sensitivity of blood sample examination in 100 microscopic fields is greater than in 50 microscopic fields, but examination 100 microscopic fields yields lower specificity. This means that due to the very low amount (10-2%-10-3%) of infected erythrocytes in carrier cattle, it is very difficult to determine the Anaplasma organisms in the carrier cattle by simple Giemsa staining.
The agreement between PCR-RFLP and Microscopy with 50 fields was fair and the agreement between PCR-RFLP and Microscopy with 100 fields was good. Detection of anaplasma microscopically requires high bacteremia, good smear preparation, proper staining and a well-trained microscopist (in spite of the fact that the technique is cheaper and easier to perform). However, microscopic examination remains the convenient technique for day-to-day diagnosis of clinical cases in the laboratory.
We believe that the microscopic examination can fulfill the desired results for the diagnosis of acute anaplasmosis but for the detection of carrier animal with low bacteremia, Microscopy with 100 fields is preferable to Microscopy with 50 fields. Our results showed that for the detection of cattle infected persistently with A. marginale, the PCR-RFLP can be used as a safe method.
ACKNOWLEDGEMENTS
This work was supported by the Veterinary Department of Isfahan Research Center for Agriculture and Natural Resources and the Veterinary Faculty at University of Tehran.
REFERENCES
- 1.Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, et al. Reorganization of genera in the families Rickettsiaceae and Anaplasma taceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and “HGE agent” as subjective synonyms of Ehrlichia phagocytophila . Int J Syst Evol Microbiol. 2001;51:2145–2165. doi: 10.1099/00207713-51-6-2145. [DOI] [PubMed] [Google Scholar]
- 2.Noaman V, Shayan P. A new PCR-RFLP method for detection of Anaplasma marginale based on 16S rRNA. Vet Res Commun. 2010;34:43–50. doi: 10.1007/s11259-009-9331-3. [DOI] [PubMed] [Google Scholar]
- 3.Noaman V, Shayan P, Amininia N. Molecular diagnostic of Anaplasma marginale in carrier cattle. Iran J Parasitol. 2009;4:31–38. [Google Scholar]
- 4.Noaman V, Shayan P. Molecular detection of Anaplasma phagocytophilum in carrier cattle of Iran-first documented report. Iranian J Microbiol. 2009;1:37–42. [Google Scholar]
- 5.Noaman V, Shayan P. Vet Res Forum 2010. Molecular detection of Anaplasma bovis in cattle from central part of Iran. (in press) [Google Scholar]
- 6.Wannduragala L, Ristic M. Anaplasmosis. In: Woldehiwet Z, Ristic M, editors. Rickettsial and chlamydial diseases of domestic animals. Oxford, U K: Pergamon Press; 1993. pp. 65–87. [Google Scholar]
- 7.Eriks IS, Stiller D, Palmer GH. Impact of persistent Anaplasma marginale rickettsemia on tick infection and transmission. J Clin Microbiol. 1993;31:2091–2096. doi: 10.1128/jcm.31.8.2091-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nazifi S, Razavi SM, Mansourian M, Nikahval B, Moghaddam M. Studies on correlations among parasitaemia and some hemolytic indices in two tropical diseases (theileriosis and anaplasmosis) in Fars province of Iran. Trop Anim Health Prod. 2008;40:47–53. doi: 10.1007/s11250-007-9052-y. [DOI] [PubMed] [Google Scholar]
- 9.Kocan KM, de la Fuente J, Guglielmone AA, Melendez RD. antigens and alternatives for control of Anaplasma marginale Infection in Cattle. Clin Microbiol Rev. 2003;4:698–712. doi: 10.1128/CMR.16.4.698-712.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kieser ST, Eriks IE, Palmer GH. Cyclic rickettsemia during persistent Anaplasma marginale infection in cattle. Infect Immun. 1990;58:1117–1119. doi: 10.1128/iai.58.4.1117-1119.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Z, Luo J, Bai Q, Ma M, Guan G, Yin H. Amplification of 16S rRNA genes of Anaplasma species in China for phylogenetic analysis. Vet Microbiol. 2005;107:145–148. doi: 10.1016/j.vetmic.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Gale KR, Dimmock CM, Gartside M, Leatch G. Anaplasma marginale: detection of carrier cattle by PCR. Int J Parasitol. 1996;26:1103–1109. [PubMed] [Google Scholar]
- 13.Carelli G, Decaro N, Lorusso A, Elia G, Lorusso E, Mari V, et al. Detection and quantification of Anaplasma marginale DNA in blood samples of cattle by real-time PCR. Vet Microbiol. 2007;124:107–114. doi: 10.1016/j.vetmic.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 14.Ge NL, Kocan KM, Murphy GL, Blouin EF. Detection of Anaplasma marginale DNA in bovine erythrocytes by slot-blot and in situ hybridization with a PCR-mediated digoxigenin-labeled DNA probe. J Vet Diagn Invest. 1995;7:465–472. doi: 10.1177/104063879500700407. [DOI] [PubMed] [Google Scholar]
- 15.Bekker CP, de Vos A, Taoufik A, Sparagano OA, Jongejan F. Simultaneous detection of Anaplasma and Ehrlichia species in ruminants and detection of Ehrlichia ruminantium in Amblyomma variegatum ticks by reverse line blot hybridisation. Vet Microbiol. 2002;89:223–238. doi: 10.1016/s0378-1135(02)00179-7. [DOI] [PubMed] [Google Scholar]
- 16.Molad T, Mazuz ML, Fleiderovitz L, Fish L, Savitsky I, Krigel Y, Leibovitz B, et al. Molecular and serological detection of A. centrale- and A. marginale-infected cattle grazing within an endemic area. Vet Microbiol. 2006;113:55–62. doi: 10.1016/j.vetmic.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 17.Torioni de Eschaide S, Bono MF, Lugaresi C, Aguirre N, Mangold A, Moretta R, et al. Detection of antibodies against Anaplasma marginale in milk using a recombinant MSP5 indirect ELISA. Vet Microbiol. 2005;106:287–292. doi: 10.1016/j.vetmic.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 18.Coetzee JF, Apleya MD, Kocan KM, Rurangirwac FR, Donkersgoed JV. Comparison of three oxytetracycline regimens for the treatment of persistent Anaplasma marginale infections in beef cattle. Vet Parasitol. 2005;127:61–73. doi: 10.1016/j.vetpar.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 19.Bradway DS, Torioni de Echaide S, Knowles DP, Hennager SG, McElwain TF. Sensitivity and specificity of the complement fixation test for detection of cattle persistently infected with Anaplasma marginale . J Vet Diagn Invest. 2001;13(1):79–81. doi: 10.1177/104063870101300117. [DOI] [PubMed] [Google Scholar]
- 20.Dreher UM, Hofmann-Lehmann R, Meli ML, Regula G, Cagienard AY, Stark KDC, et al. Seroprevalence of anaplasmosis among cattle in Switzerland in 1998 and 2003: No evidence of an emerging disease. Vet Microbiol. 2005;107:71–79. doi: 10.1016/j.vetmic.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 21.de la Fuente J, Lew A, Lutz H, Meli M, HofmannLehmann R, Shkap V, et al. Genetic diversity of Anaplasma species major surface proteins and implications for anaplasmosis serodiagnosis and vaccine development. Anim Health Res Rev. 2005;6:75–89. doi: 10.1079/ahr2005104. [DOI] [PubMed] [Google Scholar]
- 22.Torina A, Caracappa S. Anaplasmosis in cattle in Italy. Vet Res Commun. 2007;31(Suppl 1):78–73. doi: 10.1007/s11259-007-0072-x. [DOI] [PubMed] [Google Scholar]
- 23.Scoles GA, Goff WL, Lysy TJ, Lewis GS, Knowles DP. Validation of an Anaplasma marginale cELISA for use in the diagnosis of A. ovis infections in domestic sheep and Anaplasma spp. in wild ungulates. Vet Microbiol. 2008;130:184–190. doi: 10.1016/j.vetmic.2007.12.020. [DOI] [PubMed] [Google Scholar]