Abstract
Background and Objectives
Bartonella species are being recognized as increasingly important bacterial pathogens in veterinary and human medicine. These organisms can be transmitted by an arthropod vector or alternatively by animal scratches or bites. The objectives of this study were to identify contamination of cat's saliva and nail with B. henselae as a causative role to infect human in a sample of the cat population in Iran.
Materials and Methods
Blood, saliva and nail samples were collected from 140 domestic cats (stray and pet) from Tehran and Shahrekord and analyzed for the presence of B. henselae with cultural and polymerase chain reaction (PCR) methods and DNA sequencing.
Results
In this study B. henselae was detected in 10.9% of saliva samples (12/110) from pet cats. B. henselae was not detected in nail samples of pet cats (n=110), and in any feral cats’ saliva and nail samples (n=30).
Conclusion
Our data suggest that pet cats are more likely than stray cats to infect human with B. henselae after a bite and also stray cats can play a role as a reservoir for this bacteria. This is the first report that investigates the presence of B. henselae in cats oral cavity in Iran.
Keywords: Bartonella henselae, cat, cat scratch disease, Iran
INTRODUCTION
Bartonella species are being recognized as increasingly important bacterial pathogens in veterinary and human medicine. These organisms can be transmitted by an arthropod vector or alternatively by animal scratches or bites (1). Among the 11 species or subspecies known or suspected to be pathogenic in humans, 8 have been detected in or isolated from pet dogs or cats, thereby highlighting the zoonotic potential of these bacteria (2). Although cat scratch disease (CSD) was recognized in 1930 and first reported by Debrè et al., the etiological agent was identified only in 1992 when Bartonella henselae was definitely associated with CSD in humans (3–5). Bartonella henselae, the etiologic agent of CSD, has been identified as a cause of bacillary angiomatosis in immunocompromised persons (6, 7). Pet ownership is an extremely common phenomenon worldwide, with a tradition that dates back 15,000 years or more. Pet- associated zoonoses can include skin and soft tissue syndromes secondary to bites, scratches, and other direct contact, septicemia from contamination of intravascular and other indwelling medical hardware, parasitic syndromes, gastroenteritis, viral pathogens, and other zoonoses. An emerging pathogen among dogs and cats is Bartonella henselae. CSD is quite common and affected approximately 25000 people annually in the United States (8). The objectives of this study were to identify contamination of cats’ saliva and nail with B. henselae as a causative role to infect human in a sample of the cat population of Tehran and Shahrekord, Iran.
MATERIALS AND METHODS
Blood, saliva and nail samples were collected from healthy pet cats (n=110) and stray cats (n=30) at the Veterinary Medical Teaching Clinic of Tehran and Shahrekord University, Iran. All samples were collected from June 2005 to November 2007. Stray cats were caught by automatic box trap. All cats were examined physically, and healthy cats without any infectious diseases or antibiotic therapies during one month before of the study, were selected. None of these cats were infested with fleas at the time of sampling.
Isolation of B. henselae from cat's blood (culture method). EDTA tubes were filled with 2 ml of blood from the external jugular vein of each cat under aseptic condition and they were stored at −70°C for 3 to 4 weeks. After this time, samples were defrosted, tubes were centrifuged at 3000×g for 30 min at room temperature and the pellets were inoculated onto a 5% fresh sheep blood agar plates. The Brucella growth supplement (Mast, Merseyside, UK) containing polymixin B, bacitracin, natamycin, nalidixic acid, nystatin, and vancomycin was added to the plates to inhibit the growth of other micro organisms. The plates were then incubated at 35°C under conditions of 5–7% CO2 and humidity of>40% for 4 to 8 weeks (9, 10).
Isolation of B. henselae from cat's mouth and paw (culture method). An oral swab was collected using a sterile cotton applicator. The swab was placed against the inside surface of the cat's cheek. Saliva was collected by rolling the swab against the cheek and to satisfy sampling from the paw; swab was tainted with nutrient broth. Specimen from the paw was cultured directly on chocolate agar plate. Subsequently, the oral swabs were suspended in 1ml of nutrient broth, and the broth was diluted by a factor up to 10−9. From each dilution, 0.1 ml was inoculated onto chocolate agar plate and the plates were incubated as described above.
After gram staining and microscopic examination of the colonies, differential characteristics of commonly pathogenic Bartonella spp. were investigated using standard methods. Finally, the strains were subjected to PCR to identify the species of the isolated organisms (9).
DNA extraction for PCR amplification. The strains were grown on chocolate or blood agar plates. Total genomic DNA was extracted from samples using the commercial kit (Qiagen, Hilden, Germany, genomic DNA purification kit). The procedures provided by the manufacturer were followed. The extracted DNA was used as a template in the PCR assay. Purified DNA from B. henselae ATCC 49793 provided by Giladi (11) was used as a positive control in PCR experiments.
PCR Assay and sequencing. Primers CAT1 (5′- GATTCAATTGGTTTGAAGGAGGCT-3′) and CAT2 (5′- TCACATCACCAGGACGTATTC-3′) were used to amplify a 414-bp fragment of htrA gene as described by Anderson et al. and Sander et al. (12, 13). The htrA DNA gene amplification was carried out in 25 µl reaction volumes containing; 5 µl of the extracted DNA sample, 2.5 µl AMS buffer 5x, 50 mM MgCl2, 100µM dNTP, 10 to 20 pmol of each primer (CAT1 and CAT2), 12.5 µl distilled water, and 0.5 U of Taq DNA polymerase (Fermentas, Vilnius, Lithuania).
The DNA amplification was performed using the following cycling conditions: initial denaturation at 95°C (5 min), followed by 30 cycles of 94°C (1min), 57°C (1min), and 72 °C (1.5 min), with a single final extension step at 72°C (7min). Standard procedures were taken to prevent contamination of the sample DNA (14).
PCR products were separated on a 1.5% agarose gel and visualized after staining with ethidium bromide. All PCR products were analyzed by sequencing with an automated sequencer ABI 3730XL Genetic Analyzer (Macrogen, Seoul, Korea).
RESULTS
In this study B. henselae was detected in 10.9% of saliva samples (12/110) from pet cats. B. henselae was not detected in nail samples of pet cats (n=110), and in any stray cats’ saliva and nail samples (Table 1). B. henselae was not detected in blood samples of pet cats; in contrast it was detected in 16.6% of blood sample (5/30) from stray cats. All of the 10.9% of positive saliva samples were culture positive for Bartonella spp. according to morphological and biochemical characteristics.
Table 1.
Characteristics of pet cats with B. henselae salivary infection.
| Cat number | Age (month) | Gendera | Outdoor | indoor | |
|---|---|---|---|---|---|
| 1 | 51 | 8 | F | outdoor | |
| 2 | 53 | 12 | F | outdoor | |
| 3 | 57 | 2 | M | outdoor | |
| 4 | 63 | 4 | M | outdoor | |
| 5 | 64 | 4 | M | indoor | |
| 6 | 65 | 4 | F | indoor | |
| 7 | 73 | 36 | F | outdoor | |
| 8 | 75 | 15 | M | outdoor | |
| 9 | 76 | 36 | M | outdoor | |
| 10 | 84 | 3 | M | indoor | |
| 11 | 93 | 18 | M | outdoor | |
| 12 | 94 | 24 | M | outdoor | |
M, male; F, female
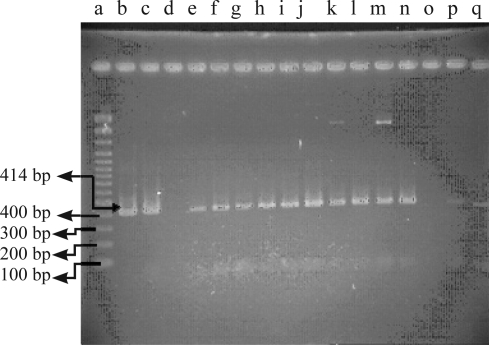
PCR amplification of the extracted DNA from 12 culture positive specimens for htrA gene produced an amplicons with 414-bp in size (Fig. 1). The htrA sequences obtained for B. henselae were submitted to the GenBank under DQ874333 and DQ874334 accession numbers.
Fig. 1.
Agarose gel electrophoresis of the PCR products. a. Marker (Fermentas); b. Positive control; c. Isolate from cat 51; d. Negative control; e-n. Isolates from cats 53, 57,63,64,65, 73, 75, 76, 84, 93, 94; o,p. Negative controls; q. Blank
DISCUSSION
This is the first study that documents isolation of B. henselae from cats’ saliva in Iran. Up to 10.9% (12 of 110) of healthy pet cats in our study showed the presence of B. henselae DNA in oral swabs obtained from saliva, however B.henselae was not detected from the saliva of stray cats. Furthermore, our results indicated that stray cats had 16.6% (5 of 30) bacteremia with B. henselae. All 12 pet cats in this study lacked bacteriologic evidence of B. henselae infection. These data suggest that pet cats are more likely than stray cats to infect human with B. henselae after a bite and also stray cats can play a role as a reservoir for this bacteria.
The risk of human Bartonella infection from stray cats can be direct and indirect. Stray cats do not allow themselves to be stroked, and hence; their contacts with human (scratching and biting) are relatively limited and the risk of direct infection should be low. The risk of indirect infection is greater, because pet cats, which are occasionally outside, can be infected by Bartonella spp. either if they are scratched or bitten by stray cats (15).
Several investigators have reported a high seroprevalence and asymptomatic B. henselae bacteremia among naturally affected cat population (14, 16, 17). In other studies a low prevalence of bacteremia has been reported in pet cats from France (8.1%) and Japan (9.1%), whereas prevalences higher than 60% have been reported in the United States, Europe and Southeast Asia (3, 18–21). Chomel et al. reported the percentage of bacteremic cats harbouring B. henselae as 89% (17 of the 19 culture positive cats) in the Philippines with the 68% (73 of 107) of them showed 1:64≤ titers of antibodies to B. henselae (18). The percentage of B. henselae seroprevalence have been reported 23% (23 of 100) in domestic cats from Tehran, Iran (22). This study also confirmed that indoor pet cats are less frequently seropositive than outdoor pet cats or stray cats.
Koehler et al. found in 41% of their cats a B. henselae bacteremia, many of these animals had close contact to each other, and this could be a reason for the high prevalence of B. henselae in that study (14). However, low detection of B. henselae bacteremia in our study appears to be due to the fact that few of these animals had close contact to each other and no ectoparasite infestation especially flea infestation was observed.
In general, cats are implicated in the transmission of B. henselae, typically resulting in cat-scratch disease; however, there have also been sporadic reports of Bartonella transmission by dogs (17, 18, 23). B. henselae DNA in dogs and cats saliva was detected from the USA and Korea; however Sander et al. could not demonstrate Bartonella species from gingival swab (10, 19) (20). Recently, Bartonella DNA has been amplified from peripheral lymph nodes of healthy dogs (21). B. henselae was also amplified from salivary gland tissues from a dog with sialadenitis (22). There are several plausible routes by which a Bartonella spp. could gain entry to the oral cavity. Future studies should determine if the tonsillar lymphoid tissues, salivary glands, or periodontal, gingival, or other oral tissues can serve as sources of Bartonella spp. contamination of canine saliva. As there is no information about canine B. henselae infection in Iran and Bartonella infection may represent an occupational risk for veterinary professionals and others with extensive animal contact (23), further investigation is recommended to elucidate the role of this organism as threats to humans and animals health in Iran.
ACKNOWLEDGMENTS
We acknowledge Dr. B. Ziaei, Mr. Ghafari and Mr. Ashrafi from the Faculty of Veterinary Medicine, University of Shahrekord and University of Tehran for their technical assistance during sampling collection, bacteriologic study and PCR assay with this work.
REFERENCES
- 1.Boulouis HJ, Chang CC, Henn JB, Kasten RW, Chomel BB. Factors associated with the rapid emergence of zoonotic Bartonella infections. Vet. Res. 2005;36:383–410. doi: 10.1051/vetres:2005009. [DOI] [PubMed] [Google Scholar]
- 2.Chomel BB, Boulouis HJ, Maruyama S, Breitschwerdt EB. Bartonella spp. in pets and effect on human health. Emerg Infect Dis. 2006;12:389–394. doi: 10.3201/eid1203.050931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Debrè R, Lamy M, Jammet M, Costil LL, Mozziconacci P. La maladie des griffes de chat. Bulletins et Mémoires de la Société Médicale des Hôpitaux de Paris. 1950;66:76–79. [PubMed] [Google Scholar]
- 4.Regnery RL, Anderson BE, Clarridge JED, Rodriguez MC, Jons DC, Carr JH. Characterization of a novel Rochalimaea species, R. henselae sp. Nov., Isolation from blood of a febrile human immunodeficiency virus- positive patient. J Cli Microbiol. 1992;30:265–274. doi: 10.1128/jcm.30.2.265-274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Regnery RL, Martin M, Olson JG. Naturally occurring Rochalimaea henselae infection in domestic cats. Lancet. 1992;340:557–558. doi: 10.1016/0140-6736(92)91760-6. [DOI] [PubMed] [Google Scholar]
- 6.Marstone EL, Finkel B, Regnery RL, Winoto IL, Ross GR, Wignal S. Prevalence of Bartonella henselae and Bartonella Clarridgeiae in an urban Indonesian cat population. Clin Diag Lab Immun. 1999;6:41–44. doi: 10.1128/cdli.6.1.41-44.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LeBolt PE. Bacillary angiomatosis. Mod Path. 1995;8:218–222. [PubMed] [Google Scholar]
- 8.Jackson LA, Perkins BA, Wenger JD. Cat scratch disease in the United States. Am J Public Health. 1993;83:1707–1717. doi: 10.2105/ajph.83.12.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mandell GL, Bennett JE, Dolin R. 6th ed. Churchill: Livingstone; 2005. Principles and practice of infectious diseases. [Google Scholar]
- 10.Sander A, Buhler C, Pelz K, Cramm EV, Bredt W. Detection and identification of two Bartonella henselae variants in domestic cats in Germany. J Cli Microbiol. 1997:584–587. doi: 10.1128/jcm.35.3.584-587.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giladi M, Kletter Y, Avidor B, Metzokor-Cotter E, Varon M, Golan Y, et al. Enzyme immunoassay for the diagnosis of cat scratch disease defined by PCR. Clin Infect Dis. 2001;33:1852–1858. doi: 10.1086/324162. [DOI] [PubMed] [Google Scholar]
- 12.Anderson B, Sims K, Regnery RL, Robinson I, Schmidt MJ, Goral S, et al. Detection of Rochalimaea henselae DNA in specimens from cat scratch disease patients by PCR. J Cli Microbiol. 1994;32:942–948. doi: 10.1128/jcm.32.4.942-948.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sander A, Posselt M, Bohm N, Ruess M, Altwegg M. Detection of Bartonella henselae DNA by two different PCR assay and determination of the genotypes of strains involved in histological defined cat scratch disease. J Cli Microbiol. 1999:993–997. doi: 10.1128/jcm.37.4.993-997.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koehler JE, Glaser CA, Tappero JW. Rochalimae henselae infection: a.new zoonosis with the domestic cat as a reservoir. JAVMA. 1994;271:531–535. doi: 10.1001/jama.271.7.531. [DOI] [PubMed] [Google Scholar]
- 15.Heller R, Artois M, Xemar V, Briel DD, Gehin H, Jaulhac B, et al. Prevalence of Bartonella henselae and Bartonella clarridgeiae in Stray Cats. J Cli Microbiol. 1997;35:1327–1331. doi: 10.1128/jcm.35.6.1327-1331.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breitschwerdt EB, Kordick DL. Bartonellosis. J Am Vet Med Asso. 1995;06:1928–1931. [PubMed] [Google Scholar]
- 17.Childs JE, Olson JG, Wolf A, Cohen N, Fakile Y, Rooney JA, et al. Prevalence of antibodies to Rochalimaea species in cat. Vet Rec. 1995;136:519–520. doi: 10.1136/vr.136.20.519. [DOI] [PubMed] [Google Scholar]
- 18.Chomel BB, Carlos ET, Kasten RW, Yamamoto K, Chang C, Carlos RS, et al. Bartonella henselae and bartonella clarridgeiae infection in domestic cats from the Philippines. Am J Trop Med Hyg. 1999;60:593–597. doi: 10.4269/ajtmh.1999.60.593. [DOI] [PubMed] [Google Scholar]
- 19.Chomel BB, Boulouis HJ, Peterson H, Kasten RW, Yamamoto K, Change CC, et al. Prevalence of Bartonella infection in domestic cats in Denmark. Vet Res. 2002;33:205–221. doi: 10.1051/vetres:2002008. [DOI] [PubMed] [Google Scholar]
- 20.Maruyama S, Nogami S, Inoue I, Namba S, Asanome K, Katsube Y. Isolation of Bartonella henselae from domestic cats in Japan. J Vet Med Sci. 1996;58:81–83. doi: 10.1292/jvms.58.81. [DOI] [PubMed] [Google Scholar]
- 21.Rolain JM, Locatelli C, Chabanne L, Davoust B, Raoult D. Prevalence of Bartonella clariddgeiae and Bartonella henselae in domestic cats from France and detection of the organism in erythrocytes by immunofluorescence. Clin Diag Lab Immun. 2004:423–425. doi: 10.1128/CDLI.11.2.423-425.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oskouizadeh K, Zahraei Salehi T, Aldavood SJ, Majlesi B, Ghaffari H, Ashrafi Tamami I, et al. Study in prevalence of Bartonella henselae infection in domestic cats from Tehran. JVR. 2008;63:183–189. [Google Scholar]
- 23.Kerkhoff FT, Ossewaarde JM, Loos WS, Rothova A. Presumed ocular Bartonellosis. Br J Ophthalmol. 1999;83:270–572. doi: 10.1136/bjo.83.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keret D, Giladi M, Kletter Y, Wientroub S. Cat-scratch disease osteomyelitis from a dog scratch. J Bone Joint Surg Br. 1998;80:766–767. [PubMed] [Google Scholar]
- 25.Tsukahara M, Tsuneoka H, Iino H, Ohno K, Murano I. Bartonella henselae infection from a dog. Lancet. 1998;352:1682. doi: 10.1016/s0140-6736(05)61455-9. [DOI] [PubMed] [Google Scholar]
- 26.Duncan AW, Maggi RG, Breitschwerdt EB. Bartonella DNA in Dog Saliva. Emerg Infect Dis. 2007;13:12. doi: 10.3201/eid1312.070653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim Y, Seo K, Lee J, Choi E, Lee H, Hwang CY, et al. Prevalence of Bartonella henselae and Bartonella clarridgeiae in cats and dogs in Korea. J Vet Sci. 2009;10:85–87. doi: 10.4142/jvs.2009.10.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duncan AW, Birkenheuer AJ, Maggi RG, Breitschwerdt EB. Bartonella DNA detected in the blood and lymph nodes of healthy dogs; 2006. Sep 27, Abstract 110. [DOI] [PubMed] [Google Scholar]
- 29.Saunders GK, Monroe WE. Systemic granulomatous disease and sialometaplasia in a dog with Bartonella infection. Vet Path. 2006;43:391–392. doi: 10.1354/vp.43-3-391. [DOI] [PubMed] [Google Scholar]
- 30.Breitschwerdt EB, Maggi RG, Duncan AW, Nicholson WL, Hegarty BC, Woods CW. Bartonella species in blood of immunocompetent persons with animal and arthropod contact. Emerg Infect Dis. 2007;13:938–941. doi: 10.3201/eid1306.061337. [DOI] [PMC free article] [PubMed] [Google Scholar]