Abstract
Background and Objectives
Streptococcosis/lactococcosis is a hyperacute systemic disease that can occur in marine and fresh waters of many species of fish. The aim of this work was to study the disease outbreak in the major rainbow trout (Oncorhynchus mykiss) production of Iran.
Materials and Methods
108 Gram positive cocci isolates were obtained from diseased trout in seven provinces with major trout production during 2008 till 2009. These bacterial isolates were characterized using phenotypic and molecular studies. The isolates were also analysed phylogeneticaly and compared with the available data.
Results
49 samples (45.37%) were identified as Streptococcus iniae, 37 samples (35.2%) matched with Lactococcus garvieae; and 22 samples (19.43%) were identified as members of Streptooccus genus by culture-based and biochemical tests of API 50 CH, API 20 STREP and rapid 32 STREP systems. Using universal primers for differentiation of Streptococcus sp. and Enterococcus sp, all 108 samples were identified as Streptococcus sp. with a target region of 500 bp. Single specific PCR resulted in identification of 64 (59.2%) isolates as S. iniae and 44 (40.8%) isolates as L. garvieae. The phylogenetic analysis of the S. iniae isolates resulted in maximal similarity to some strains reported from Taiwan and to all Brazilian strains. Also, one strain showed less sequence similarity values with other tested strains although this strain has high similarity with ATCC 29178 strain, all reported Chinese, and some Taiwanian strains. Also, analysis of S. iniae LctO gene sequence showed that this isolate clustered within the S. iniae group. The sequence analysis of L. garvieae strains also showed that they have maximum similarity to all Japanese and Chinese strains, but one strain has lower sequence similarity values with all other recorded strains.
Conculsion
The results of this study clearly show that trout farming in Iran is severely affected by both species of S. Iniae and L. garvieae and requires serious preventive criteria.
Keywords: Streptococcosis, lactococcosis, rainbow trout, Iran
INTRODUCTION
Streptococcosis/lactococcosis was described as a hyperacute systemic disease that can occur in marine and fresh waters of many species of fish including rainbow, tilapia, sea bass, eel and yellow tail (1–6). The disease also known as pop-eye disease, is now one of the most important bacterial diseases in farmed rainbow trout in almost all countries having trout aquaculture activity (1, 2, 7). Several species of Streptococcus and Lactococcus bacteria including S. iniae, S. agalactiae, S. dysagalactiae, S. parauberis, S. feacalis, L. garvieae and L. lactis have been so far discriminated as the cause of streptoococcosis/ lactococcosis (2, 4, 8–10).
Iran is now one of the leading countries in trout production in freshwater with a total production of about 60000 tons in 2008 (Iranian Fisheries Organization, 2008). Since the first reports of a presumptive streptococcosis (11), S. iniae and L. garvieae were identified as causative agents of the disease during 2005–2008 (12, 13). Despite significant losses due to this zoonotic bacterial disease in trout aquaculture in Iran, little information is available particularly on the epizootiology and the causative agents involved. In the present study, the disease epidemiology has been assessed in seven major trout- producing provinces. Conventional bacteriology and polymerase chain reaction (PCR) were used to compare the accuracy of disease detection. Also, isolated bacteria were phylogenetically characterized and compared with available data.
MATERIALS AND METHODS
Source of bacterial isolates. Total of 108 isolates of Gram positive cocci bacteria were used. These bacterial isolates were obtained from farmed rainbow trout in seven provinces of Iran including Mazandran (36 isolates), Tehran (17 isolates.), Gilan (13 isolates), Kermanshah (2 isolates), Lorstan (14 isolates), Fars (18 isolates) and Charmahal- va-Bakhteyari (8 isolates). The bacterial isolates were recovered from kidney or spleen tissues of diseased fish on blood agar medium incubated at 25–30°C for 72 h. During the sampling time,clinical observations and water quality parameters were also recorded.
Phenotypic characterization. The pure colonies of fresh cultures were subjected to morphological and biochemical tests for phenotypic characterization (3, 14–16). Biochemical tests including acidification of carbohydrates and enzymatic tests were performed with API 50 CH, API 20 STREP and Rapid 32 STREP (Biomerieux, France). Manufacturers? instructions were followed except for the incubation temperature for API 50 CH and API 20 STREP, which was maintained at 24±1°C instead of the recommended 36 ± 1°C. Final results were read at 4 h for API Rapid 32 STREP and at 72 h for API 50 CH and API 20 STREP after incubation.
Extraction of bacterial DNA. DNA was extracted from pure colonies using the rapid genomic DNA isolation kit (MBST Company, Iran) based on extraction by proteinase K according to the manufacturer's instructions. Extracted DNA was dissolved in 100 µl of distilled water and stored at −20°C until used.
PCR amplification of the Streptococcus sp. and Enterococcus sp. 16S rRNA gene. Initially two pairs of universal primers of Streptococcus sp and Entrococcus sp were used to amplify the 16S rRNA gene for diagnosis of Streptococcus and Entrococcus genera (8, 17, 18) (Table 1). The PCR amplification were performed in 25 µl reaction mixture containing 1.5 µl of template DNA, 100 pmol concentration each primer (all primers were synthesized by DNA technology A/S, Arhus, Denmark), 2.5 mM MgCl, 10 mM concentration of each dNTP and 2U of Taq DNA polymerase (Promega, USA) in 5X reaction flexi buffer. After a denaturation step at 94°C for 5 min, 30 serial cycles consisting of a denaturation step at 92°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 90 s were run followed with a final extension step at 72°C for 5 min. A negative control (no template DNA) and positive control consisting of S. iniae (ATCC29178) L. garvieae (TKS KG+), S. parauberis (NCDO2020), and E. faecalis strains (CCUG19916) were included in each run.
Table 1.
Oligonucleotid primers used for single PCR assays.
| Primer pairs | Sequence (5′-3′) | Target gene | PCR Amplicon (bp) | Pathogen | Reference |
|---|---|---|---|---|---|
| Strep.sp. | AGAGTTTGATCCTGGCTCAG | 16S rRNA | 500 | .Streptococcus sp | Conrads et al., 1997 |
| FW/BW | GTACCGTCACAGTATGAACTTTCC | ||||
| Entero.sp | TAC TGA CAA ACC ATT CAT GAT G | 16S rRNA | 112 | .Enterococcus sp | Ke et al., 1999 |
| FW/BW | AAC TTC GTC ACC AAC GCG AAC | ||||
| Spa2152 | TTTCGTCTGAGGCAATGTTG | 23S rRNA | 718 | S. parauberis | Riffon et al., 2001 |
| Spa2870 | GCTTCATATATCGCTATACT | ||||
| LOX-1 | AAGGGGAAATCGCAAGTGCC | IctO | 870 | S. iniae | Mata et al., 2004 |
| LOX-2 | ATATCTGATTGGGCCGTCTAA | ||||
| PlG-1 | CATAACAATGAGAATCGC | 16S rRNA | 1100 | L. garvieae | Mata et al., 2004 |
| PlG-2 | GCACCCTCGCGGGTTG | ||||
| FW/BW (V1/V2) | V1:5′-TTTGGTGTTTACACTAGACTG-3′ | 16SrRNA | 120 | S.agalactiae | Meiri-Bendek et al., 2002 |
| V2: 5′-TGTGTTAATTACTCTTATGCG-3′ | |||||
| FW/BW (Strd-dyl/Dys-16s-23s-2) | 5′-TGGAACACGTTAGGGTCG-3′ | 16S-23S rDNA | 300 | S.dysgalactiae | Forsman et al.,1997; Hassan et al., 2003 |
| 5′CTTAACTAGAAAAACTCTTGATTATTC-3′ |
PCR amplification of S. iniae lactate oxidase (lctO), S. parauberis 23 rRNA, S. dysagalctiae 16S-23S rDNA and S. agalactiae 16SrRNA and L. garvieae 16S rRNA, genes. The oligonucleotid primers used for PCR amplification of S. iniae, L.garvieae, S. parauberis, S. agalactiae and S. dysagalactiae genes are given in Table 1. At the first step all bacterial strains were subjected to PCR for identification of S. iniae. At the second step, those bacterial strains that were negative for S. iniae were subjected to PCR for identification of L. garvieae. Based on the results of phenotypic features, 22 bacterial strains showed variable reactions particularly to tested sugars. Therefore, these bacterial strains were also subjected to PCRs for S. agalactiae and S. dysgalactiae.
The PCR amplification for S. iniae, L. garvieae and S. parauberis were performed in 25 µl reaction mixture containing 1.5 µl of template DNA, 100 pmol concentration of each primer (all primers were synthesized by ISOGEN Bioscience BV, polymerase buffer (1.5 mM MgCl); 1.0 µl of each forward and reverse primers (10 µM each); 0.2 µl of dNTP (25 mM), 0.1 µl of Taq polymerase (0.25 u); 5 µl of DNA (50 to 100 ng/µl); add ddH2O (sterile) to total volume 25 µl. The reaction was carried out in a PCR thermocycler as follows: 94°C for 4 min; five cycles of 94°C, Tm°C and 72°C for 45 s each step; 20 cycles of 94°C, 72°C for 45 s each step; and a step of 72°C for 5 min, at the end of the reaction. Also, to amplify part of the 16S–23S rDN intergenic spacer region that is specific to S. dysgalactiae, the oligonucleotide forward and reverse primers (dys-16S-23S-2) recommended by Forsman et al. (18) and Hassan et al. (19) were used (Table 1). The PCR assay was performed according to Hassan et al. (2003) using a thermal cycler (Biorad). PCR products were run on agarose gel (1.8 to 2.0%) and visualized by Etidium Bromide 0.005%. A negative control (no template DNA) and positive controls of these bacterial strains were also included in each PCR run.
16S rRNA and lctO genes sequence analysis. The PCR products of 16S rRNA of L. garvieae and lctO of S. iniae were sequenced. Sequencing of each PCR product was undertaken using DNA technology A/S analyzer. The forward and reverse nuclide acid sequence data were used to construct a continuous sequence of inserted DNA. Further comparison of the continuous sequences was then made with previously available sequences in the NCBI data base using BLAST (Basic Local Alignment Search Tool). Multiple sequence alignment analysis and construction of a phylogenetic tree were performed using MEGA 4 software via FASTA algorithms. The phylogenic trees were then constructed on the basis of the UPGMA method and the evolutionary distances were estimated using MEGA 4 (16). Maarssen, The Netherlands), 2.5 mM MgCl2,10 mM concentration of each dNTP and 5U of Taq DNA polymerase (Cinagene, Iran) in 10X reaction buffer. After a denaturation step at 94°C for 5 min,30 serial cycles consisting of a denaturation step at 92°C for 1 min, annealing at 58.6°C for S. iniae;52.7°C for L. garvieae and 52.5°C for S. parauberis for 1 min, and extension at 72°C for 90 s were used. The final extension step was performed at 72°C for 5 min. For S. agalactiae we used the PCR procedure recommended by Meiri-Bendek et al.(15). Brifely, the PCR reaction mixture contained 2.5 µl of 10 ×Taq
RESULTS
Clinical observations. Sluggish movement, darkening of body, bilateral exophthalmia sometimes together with cataract and hemorrhage, abdominal distention and prolaps of anal area with hyperemia/ hemorrhage were observable in most affected fish. Also, accumulation of bloody fluids in abdominal cavity, hemorrhage in intestinal lumen, pale liver and precarditis (in brood fish) were seen in dissection examination. In most cases, the affected fish farms were using rivers as the main source of their water with water temperature in the range of 14-20°C, dissolved oxygen of 6-8 mg/l, carbon dioxide of 4–15 mg/l, nitrite of 0.0–0.1 mg/l, and unionized ammonia of 0.06-0.1 mg/l.
Biochemical features. Biochemical analyses showed that 45.37% of bacterial isolates matched with S. iniae isolates i.e. positive reactions on the Esculin (ESC), pyrrolidonyl arylamidase (PYRA), β glucuronidase (ß GUR), L-leucine arylamidase (LAP), trehalose (TRE), starch (STA), sucrose (SUC), maltose (MAL), galactose (GAL), D-glucose (GLU), D-fructose (FRU), D-manose (MNE), arbutin (ARB), salicin (SAL), cellobiose (CEL) and N-acetyl glucosamine (NAG) tests and negative reactions on the Voges-Proskauer (VP), hippurate hydrolysis (HIP), α-galactosidase (α GAL), β-galactosidase (ß-GAL), N-acetyl-β-glucosaminidase (ß-NAG), glycyl- tryptophan-arylamidase (GTA), Lactose (LAC), L-arabinose (LARA), sorbitol (SOR), inulin (INU), cyclodextrin (CDEX), melibiose (MEL), tagatose (TAG), erythritol (ERY), D-arabinose (DARA), D-xylose (DXYL), L-xylose ( LXYL), adonitol (ADO), L-sorbose (SBE), rhamnose (RHA), dulcitol (DUL), inositol (INO), xylitol (XLT), D-turanose (TUR), D-lyxose (LYX), D-tagatose (D-TAG), D-fucose (D-FUC), L-fucose (L-FUC), L-arabitol (L-ARL), gluconate (GNT), 2-keto-gluconate (2KG), 5-keto- gluconate (5KG), α-methyl-D-glucoside (MDG) and glycerol (GLY) tests. In addition, variable reactions were observed on the arginin dihydrolase (ADH), β glucosidase (ß-GLU), alkaline phosphatase (PAL), β-manosidase (ß-MAN), ribose (RIB), mannitol (MAN), raffinose (RAF), glycogen (GLYG), D-arabitol (DARL), melezitose (MLZ), puliulane (PUL), amygdalin (AMY) and ß-gentiobiose (GEN) tests.
Also 35.2% of bacterial isolates matched with L. garvieae i.e. positive reactions to VP, ESC, ADH, ß-GLU, ß-NAG, RIB, MAN, TRE, MAL, methyl-Bd glucopyranoside acidification (MßDG), TAG, GAL, GLU, FRU, MNE, AMY, ARB, SAL,CEL, GEN, GNT and NAG and negative reactions to HIP, PYRA, ß-GLU, α-GAL, PAL, ß-GAL, GTA, ß-MAN, LAP, RAF, LARA, SOR, GLYG, INU, DARL, CDEX, MLZ, PUL, MEL, ERY, DARA, DXYL, LXYL, ADO, SBE, RHA, DUL, INO, XLT, TUR, LYX, DFUC, LFUC, LARL, 2KG, 5KG, α-methyl-D- mannoside (MDM), MDG and GLY. However, some L. garvieae isolates displayed variable reactions to LAC, STA and SUC.
The remaining 22 isolates were phenotypically identified as the members of Streptococcus genus having variable reactions for some tested sugars (data not shown).
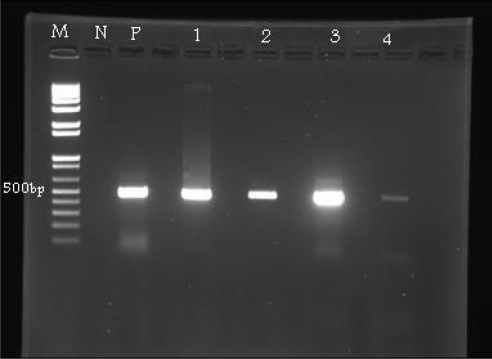
PCR amplification of the Streptococcus sp. and Enterococcus sp. 16S Rrna. A 500bp band was detected in all 108 bacterial isolates that confirms Streptococcus sp., while none of these samples revealed a 112bp band that is matched with Enterococcus sp (Fig. 1). Regional distribution of these isolates of Streptococcus sp. are given in Table 2. The highest and lowest infected trout farms were Mazandaran (33.3%) and Kermanshah (1.9%) regions, respectively.
Fig. 1.
Representative amplification of PCR products using universal primers of Streptococcus sp. Lane M=Marker 100bp; Lane N=Negative control (Entrococcus faecalis CCUG19916); Lane P=positive control (S. iniae); Lanes 1, 2 and 3=test samples.
Table 2.
Regional distribution (%) of L. garvieae, S. iniae and Streptococcus sp based on traditional and molecular works in seven states with major trout production in Iran. Numbers in parentheses indicating the numbers of bacterial strains. LG=L. garvieae, SI=S. iniae, SP=S. parauberis, SD=S. dysagalactiae, SA=S. agalactiae.
| State | Traditional bacteriology | PCR with universal primers | PCRwith specific primers | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LG | SI | Strep. sp | Strept. sp | Ent. sp | LG | SI | SP | SA | SD | |
| Gilan | 0 (0) | 8.2 (4) | 40.9 (9) | 12 (13) | 0 | 0 (0) | 20.3 (13) | 0 | 0 | 0 |
| Mazandran | 29.7 (11) | 38.8 (19) | 27.3 (6) | 33.3 (36) | 0 | 39 (17) | 29.7 (19) | 0 | 0 | 0 |
| Tehran | 18.9 (7) | 14.3 (7) | 13.6 (3) | 15.7 (17) | 0 | 18.2 (8) | (9) 14 | 0 | 0 | 0 |
| Kermansha | 5.4 (2) | 0 (0) | 0 (0) | 1.9 (2) | 0 | 4.4 (2) | 0 (0) | 0 | 0 | 0 |
| Charmahal va Bakhteyari | 10.8 (4) | 4.1 (2) | 9.1 (2) | 7.4 (8) | 0 | 9 (4) | 6.3 (4) | 0 | 0 | 0 |
| Lorestan | 29.7 (11) | 6.1 (3) | 0 (0) | 13 (14) | 0 | 25 (11) | 4.7 (3) | 0 | 0 | 0 |
| Fars | 5.4 (2) | 28.6 (14) | 9.1 (2) | 16.7 (18) | 0 | 4.4 (2) | 25 (16) | 0 | 0 | 0 |
| Total | 100 (37) | 100 (49) | 100 (22) | 100 (108) | 0 | 100 (44) | 100 (64) | 0 | 0 | 0 |
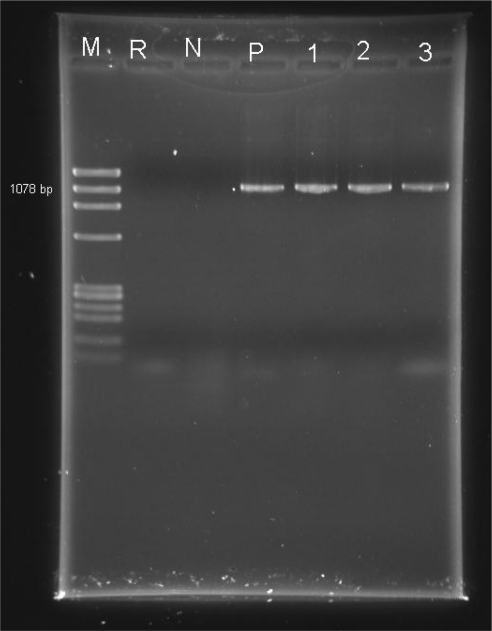
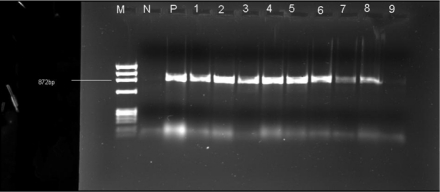
Specific single pCRs amplification. Each of the three pairs of primers exclusively amplified the targeted gene of the specific bacteria. From 108 bacterial isolates, 37 isolates showed 1100bp that is identical to L. garviea (Fig. 2) and 49 isolates revealed 870bp which is identical to S. iniae (Fig. 3). None of the samples produced a band of 718 bp which is identical to S. parauberis. The regional distribution of infection by S. iniae shows that trout farming in Mazandaran (29.7%) and Fars (25%) states were more affected than other examined states. Also, no infection by S. iniae was detected in Kermanshah region (Table 2). Furthermore, infection by L. garvieae was higher in Mazandran (39%) and Lorestan (25%) regions than other studied states, while no infection by L. garvieae was detected in Gilan region (Table 2).
Fig. 2.
Amplification of the PCR products for detection of L. garviae (1100bp). Lane M=Marker; Lane B=distilled water; Lane N=Negative control (S. iniae); Lane P=Positive control (L. garvieae); Lanes 1, 2 and 3=test samples.
Fig. 3.
Amplification of the PCR products for detection of S. iniae (870bp). Lane M=Marker; Lane N=Negative control (L. garvieae); Lane P=Positive control (S. iniae); Lanes 1-9=test samples.
Sequence analysis. The results of sequencing of the representative bacterial strains of S. iniae and L.garvieae showed 746bp and 856- 852bp, respectively. These sequences were recorded in Gene bank under accession number ATCC numbers GQ850377, GQ850376, GQ850375, FJ870987 and HM055571-4 (Table 2).
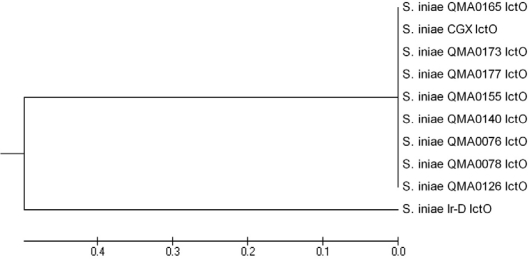
Phylogenetic analysis and genetic distance of S. iniae16s rRNA and lctO genes. Partial 16S rRNA gene fragment of S. iniae strains LG3, LHK2, 0141 and SF2 were sequenced (Accession numbers HM055572, HM055573, HM055574 and FJ870987). Data for the phylogenetic analysis were obtained from sequences contained in the Gen Bank nucleotide sequences database. Strains LG3, LHK2 and 0141 from Iran have maximum similarity to strains SCC104, SCC106, SCC103 and SCC107 reported from Taiwan and all Brazilian strains. Lower sequence similarity values were found between strain SF2 and all other three Iranian strains. Strain SF2 has also high similarity with the ATCC 29178, all reported Chinese strains and some Taiwanian strains (Fig. 4). Also, LctO gene of the Ir-D strain was examined. Phylogenetic analyses inferred from LctO gene sequence comparisons using the neighbor-joining showed that Ir-D strain clustered within the S. iniae group (Fig. 5).
Fig. 4.
Phylogenetic tree based on 16S rRNA gene sequences constructed according to UPGMA method, showing the posi- tion of Iranian strains of S. iniae and the.isolates from other regions. (140×50 mm (400×400 DP).
Fig. 5.
Phylogenetic tree based on lctO gene sequences, constructed according to UPGMA method, showing the position of Iranian strains of S. iniae and other isolates of S. iniae. 140×50 mm (400×400 DP).
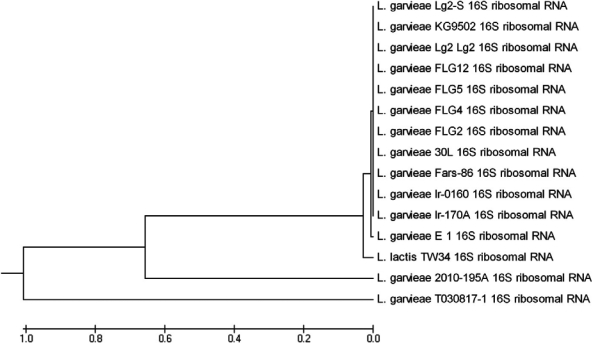
Phylogenetic analysis and genetic distance of L. garvieae. A 856 bp 16S rRNA gene fragment of L. garvieae strains 195A, Ir-0160 and Ir-170A were sequenced (Accession numbers HM055571, GQ850376 and GQ850375). Phylogenetic tree for these strains along with the other reported sequences contained in the Gen Bank nucleotide sequences database is shown in Fig. 6. The Ir-0160 and Ir-170A strains have maximum similarity to all Japanese and Chinese strains, but strain 195A has lower sequence similarity values with all other recorded strains.
Fig. 6.
Phylogenetic tree based on 16S rRNA gene sequences constructed according to UPGMA method, showing the position of Iranian strains of L. garvieae and the isolates other regions. (140×50mm (400×400 DP).
DISCUSSION
Streptococcosis/lactococcosis have become one of the most serious bacterial pathogens causing significant losses in many farmed marine and freshwater fish species of both cold and warm water environments. Data obtained on the clinical observations as well as traditional and molecular bacteriology provides adequate information on the epizootiology of streptococcosis and lactococcosis outbreaks in trout aquaculture in Iran. Clinically, in most cases the affected farms showed a chronic to subacute disease and the most diseased fish showed bilateral exophthalmia together with cataract and in some cases complete loss of the eyes. Sluggish movement, darkening of body, mild abdominal distention, prolaps of anal area, hemorrhage in the intestine and accumulation of bloody fluid in the abdominal cavity were also clinically observable signs. Total mortality was varied from 5 to 50% during a period of approximately 3 month of fish farming. In most cases the affected fish were above 100 g. The water temperature of all affected farms were in the range of 13-19°C. The river water was the main source of water for those fish farms with more severe disease outbreaks.
Conventional bacteriology resulted in the isolation and characterization of 108 Gram positive cocci bacterial strains identical to S. iniae (45.35%) and L. garvieae (35.18%). The remaining isolates (19.43%) were identified as members of Streptococcus genus based on the phenotypic and PCR analysis using universal primers. When these bacterial isolates were subjected to the specific PCR analysis, about 60% of them were characterized as S. iniae and about 40% as L. garvieae . Therefore, these data show that S. inae and L. garvieae are the main causative agents of streptococcosis/lactocococcis outbreaks in the major trout production states of Iran.
For the phylogenic analysis and investigation of relationship between Iranian isolates and the other isolates in the world, we initially searched in NCBI and found several isolates of S. iniae in different regions such as Australia (8 strains), Brazil (7 strains), China (18 strains), Taiwan (10 strains) Singapore (1 strain), Thailand (6 strains), USA (1 strain) and Middle East (4 strains) (Table 3). Also, 10 isolates of L. garvieae were found in different countries including Japan (4 strains), China (5 strains) and Iran (1 strain). Result of sequencing of the representative strains of S. Iniae 16S rRNA gene shows that Iranian strains are closer to Taiwan and Brazilian strains than other reported strains (Fig. 3). Also, phylogenetic analysis of S. iniae LctO gene shows that the Iranian strains are clustered within S. iniae group having more genetic distance with other recorded strains reported from other regions (Fig. 4). In addition, the sequencing of the representative strains of L. garvieae shows that Iranian strains are closer to isolates reported from both China and Japan than other strains (Fig. 5). However, strain 195A showed lower similarity values to other reported strains.
Table 3.
Data on S. iniae and L. garvieae strains analyzed in phylogenic analysis.
| Country | Bacterial species | Accession number | Source (year) | Target gene |
|---|---|---|---|---|
| Argentina | L. garvieae TW34 (1398bp) | GQ845022 | Odontesthes platensis (2010) | 16S rRNA |
| Australia | S. iniae QMA0078 (1180 bp) | EU086698 | Lates calcarifer (2007) | LctO |
| S. iniae QMA0076 (1228bp) | EU086697 | L. calcarifer (2007) | LctO | |
| S. iniae QMA00126 (1279bp) | EU086699 | L. calcarifer (2007 | LctO | |
| S. iniae QMA00140 (1228bp) | EUO86700 | L. calcarifer (2007) | LctO | |
| S. iniae A00155 (1228bp) | EUO86701 | L. calcarifer (2007) | LctO | |
| S. iniae QMA00177 (1248bp) | EU086704 | L. calcarifer (2007) | LctO | |
| S. iniae QMA00173 (1185bp) | EU086703 | L. calcarifer (2007) | LctO | |
| S. iniae QMA00165 (1176bp) | EU086702 | L. calcarifer (2007) | LctO | |
| Brazil | S. iniae S122-06 (398bp) | FJ803994 | Oreochromis niloticus (2010) | 16S rRNA |
| S. iniae S123-06 (522bp) | FJ803995 | O. niloticus (2010) | 16S rRNA | |
| S. iniae S124-06 (522bp) | FJ803996 | O. niloticus (2010) | 16S rRNA | |
| S. iniae S125-06 (702bp) | FJ803997 | O. niloticus (2010) | 16S rRNA | |
| S. iniae S127-06(696bp) | FJ803998 | O. niloticus (2010) | 16S rRNA | |
| S. iniae S128-06 (500bp) | FJ803999 | O. niloticus (2010) | 16S rRNA | |
| S. iniae S129-06 (688bp) | FJ804000 | O. niloticus (2010) | 16S rRNA | |
| Chile | L. garvieae 30L (817bp) | FJ151399 | Aplodactylus punctatus (2008) | 16S rRNA |
| China | S. iniae CGX (870bp) | EF126045 | Tilapia (2006) | |
| S. iniae CGX (1447bp) | DQ985468 | Tilapia (2006) | LctO | |
| S. iniae SF1 (501bp) | GQ891547 | Japanese flunder (2010) | 16S rRNA | |
| S. iniae DGX01 (1500bp) | HM053435 | Channel cat fish (2010) | 16S rRNA | |
| S. iniae YG1 (1168bp) | GQ169798 | Selenotoca multifasciata (2009) | 16S rRNA | |
| S. iniae DGX070902 (1497bp) | FJ951434 | Channel cat fish (2009) | 16S rRNA | |
| S. iniae CMS004 (1465bp) | EU620577 | Unknown fish (2008) | 16S rRNA | |
| S. iniae CMS005 (1463bp) | EU620578 | Unknown fish (2008) | 16S rRNA | |
| S. iniae CMS006 (1464bp) | EU620579 | Unknown fish (2008) | 16S rRNA | |
| S. iniae CMS003 (1464bp) | EU620580 | Unknown fish (2008) | 16S rRNA | |
| S. iniae CMS007 (1464bp) | EU622508 | Unknown fish (2008) | 16S rRNA | |
| S. iniae CMS008 (1464bp) | EU622509 | Unknown fish (2008) | 16S rRNA | |
| S. iniae CMS009 (1463bp) | EU622510 | Unknown fish (2008) | 16S rRNA | |
| S. iniae CMS0010 (1463bp) | EU622511 | Unknown fish (2008) | 16S rRNA | |
| S. iniae CMS0011 (1463bp) | EU622512 | Unknown fish (2008) | 16S rRNA | |
| S. iniae CMS0012 (1463bp) | EU622513 | Unknown fish (2008) | 16S rRNA | |
| S. iniae CMS0013 (1463bp) | EU622514 | Unknown fish (2008) | 16S rRNA | |
| S. iniae CMS0014 (1463bp) | EU622515 | Unknown fish (2008) | 16S rRNA | |
| L. garvieae T030817-1 (1427bp) | DQ010113 | Paralichthys olivaceus (2005) | 16S rRNA | |
| L. garvieae FLG2 (1544bp) | AF352163 | Mugil cephalus (2002) | 16SrRNA | |
| L. garvieae FLG4 (1544bp) | AF352164 | M. cephalus (2002) | 16SrRNA | |
| L. garvieae FLG5 (1544bp) | AF352165 | M. cephalus (2002) | 16SrRNA | |
| L. garvieae FLG12 (1544bp) | AF352166 | M. cephalus (2002) | 16SrRNA | |
| Iran | S. iniae Ir-D (746bp) | GQ850377 | Oncorhynchus mykiss (2009) | LctO |
| S. iniae SF2 (1384bp) | FJ870987 | O. mykiss (2009) | 16SrRNA | |
| S. iniae 0141-4 (344bp) | HM055574 | O. mykiss (2010) | 16SrRNA | |
| S. iniae LHK2 (369bp) | HM055573 | O. mykiss (2010) | 16SrRNA | |
| S. iniae LG3 (407bp) | HM055572 | O. mykiss (2010) | 16SrRNA | |
| L. garvieae 195A (409bp) | HM055571 | O. mykiss (2010) | 16SrRNA | |
| L. garvieae Ir-170A (856bp) | GQ850376 | O. mykiss (2009) | 16SrRNA | |
| L. garvieae Ir-0160 (852bp) | GQ850375 | O. mykiss (2010) | 16SrRNA | |
| L. garvieae Fars (1007bp) | EU727199 | O. mykiss (2008) | 16SrRNA | |
| Israel | S. iniae Dan1 (1490bp) | AF335573 | O. mykiss (2001) | 16SrRNA |
| S. iniae S41 (654bp) | AY260834 | O. mykiss (2003) | 16S rRNA | |
| S. iniae ATCC29178 (1536bp) | AF335572 | O. mykiss (2001) | 16S rRNA | |
| S. iniae ATCC29178 (1536bp) | NR-025148 | O. mykiss (2009) | 16S rRNA | |
| Japan | L. garvieae E1 (1506bp) | AB018211 | Cyprinus carpio (2000) | 16S rRNA |
| L. garvieae Lg2 (1471bp) | AB267897 | Seriola quinqueradiata (2006) | 16S rRNA | |
| L. garvieae Lg2-S(1471bp) | AB267898 | S. quinqueradiata (2006) | 16S rRNA | |
| L. garvieae KG9502 (1471bp) | AB267899 | S. quinqueradiata (2006) | 16S rRNA | |
| Singapore | S. iniae DB39299/02 (533bp) | DQ193527 | Red tilapia (2005) | 16S rRNA |
| Taiwan | S. iniae SCCS101 (510bp) | AY465111 | Rachycentron canadum (2004) | 16S rRNA |
| S. iniae SCCS102 (517bp) | AY480053 | R. canadum (2004) | 16S rRNA | |
| S. iniae SCCS103 (513bp) | AY480054 | R. canadum (2004) | 16S rRNA | |
| S. iniae SCCS104 (506bp) | AY737430 | R. canadum (2005) | 16S rRNA | |
| S. iniae SCCS106 (497bp) | AY737432 | R. canadum (2005) | 16S rRNA | |
| S. iniae SCCS107 (497bp) | AY737433 | R. canadum (2005) | 16S rRNA | |
| S. iniae SCCS108 (497bp) | AY737434 | R. canadum (2005) | 16S rRNA | |
| S. iniae SCCS109 (497bp) | AY737435 | R. canadum (2005) | 16S rRNA | |
| S. iniae SCCS110 (522bp) | AY489403 | R. canadum (2004) | 16S rRNA | |
| S. iniae SCCS111 (520bp) | AY489404 | R. canadum (2004) | 16S rRNA | |
| Thailand | S. iniae JW1 (1120bp) | GQ169769 | Oreochromis niloticus (2009) | 16S rRNA |
| S. iniae JW3 (1130bp) | GQ338313 | O. niloticus (2009) | 16S rRNA | |
| S. iniae JW4 (1118bp) | GQ169770 | O. niloticus (2009) | 16S rRNA | |
| S. iniae JW6 (1120bp) | GQ338314 | O. niloticus (2009) | 16S rRNA | |
| S. iniae JW7 (1114bp) | GQ169771 | O. niloticus (2009) | 16S rRNA | |
| S. iniae JW9 (1141bp) | GQ338315 | O. niloticus (2009) | 16S rRNA | |
| USA | S. iniae ATCC29178 (534bp) | AY577823 | Unknown fish (2004) | 16S rRNA |
According to the geographical distribution of the identified bacterial strains shown in Table 2, it is clear that the trout farming in the states of Mazandran, Tehran, Charmahal-va-Bakhteyri, Lorstan and Fars are affected with both species of S. iniae and L. garvieae, while fish farms of Gilan and Kermanshah regions are infected with either S. iniae or L. garvieae. Also, it seems that infection by S. iniae is more dominant in Fars region than other investigated areas, while the outbreaks by L. garvieae was more in Lorestan state. In previous studies by Soltani et al. (13, 19), infections by either S. inaie or L. garvieae was detected as the cause of the disease outbreak in some trout farming in Iran. However, it is possible that infection by other members of Streptococcus genus may be involved in some farmed trout located in other regions of the country and therefore, warranted further investigations.
Most of disease outbreaks were detected during the warm seasons, late spring till mid autumn, and the time that water temperature of trout farming increases up to 20C particularly in those fish farms that use rivers as the source of water. Increase in water temperature together with impact of polluted water sources will cause a significant decline in water quality parameters resulting in outbreaks by infectious diseases including streptococcosis/lactococcosis.(1, 2, 10).
It is notable that the owners of most affected fish farms have no adequate training related to health management criteria. Such training is nowadays; very important particularly in the case of streptococcosis/ lactococcosis that is a human and terrestrial animal zoonotic disease (1, 2, 10, 20), providing it easy transportation to the fish farms through sewage of terrestrial animals. In Iran, transportation of eyed- eggs, larvae and broodstock between the fish farms is currently undertaken by many trout hatcheries providing an easy way for disease spreading inside the country.
Annual losses by streptococcosis/lactococcosis has been estimated at 100 million USD. In fact, the economic impact due to this bacterial disease is quite higher than 100 million USD. For instance, Iran is one of the leading countries in production of trout in freshwater having above 60000 ton per year and our annual estimated losses due to this zoonotic disease is about 15 million USD (26). This is a reason why the Iran veterinary organization has recently established the national committee of trout streptococcosis to reduce losses due to this highly devastating zoonotic contagious disease.
In conclusion, clinical observations plus molecular studies show that both S. iniae and L. garvieae are the causative agents involved in disease outbreaks in major trout production regions of Iran. Also, some other members of Streptococcus sp may be involved in disease production in Iranian trout aquaculture. Therefore, further investigations are warranted. Also, poor water quality, high water temperature and poor health management criteria, e.g. quarantine and other protective measures such as lack of vaccination, are the main reasons for disease spread inside the country.
ACKNOWLEDGEMENT
This work was financially supported by a grant from research council of University of Tehran. Authors are grateful to the assistance made by the staff of Department of Aquatic Animal Health, Faculty of Veterinary Medicine, University of Tehran and the trout farmers in the investigated regions.
REFERENCES
- 1.Agnew W, Barnes AC. Streptococcus iniae: an aquatic pathogen of global veterinary significance and a challenging candidate for reliable vaccination. J Vet Microbiol. 2007;122:1–15. doi: 10.1016/j.vetmic.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Austin B, Austin DA. Chichester: Springer Praxis Publication; 2007. Bacterial Fish Pathogens: Disease of Farmed and Wild Fish; pp. 58–63.pp. 156pp. 238–239. [Google Scholar]
- 3.Colorni A, Diamant A, Eldar A, Kvitt H, Zlotkin A. Streptococcus iniae infections in Red Sea cage-cultured and wild fishes. Dis Aquat Org. 2002;49:165–170. doi: 10.3354/dao049165. [DOI] [PubMed] [Google Scholar]
- 4.Eldar A, Horovitcz A, Bercovier H. Development and efficacy of a vaccine against Streptococcus iniae infection in farmed rainbow trout. Vet Immunol Immunopathol. 1997;56:175–183. doi: 10.1016/s0165-2427(96)05738-8. [DOI] [PubMed] [Google Scholar]
- 5.Plumb JA, Schachte JH, Gaines JL, Pltier W, Carrol B. Streptococcus sp. from marine fishes along the Alabama and northwest Florida coast of the Gulf of Mexico. Tran Am Fish Soc. 1975;103:358–361. [Google Scholar]
- 6.Toranzo AE, Devesa S, Heinen P, Riaza A, Nunez S, Barja JI. Sterprococcosis in cultured turbout caused by an Enterococcus-like bacterium. Bull Eur Ass Fish Pathol. 1994;14:19–23. [Google Scholar]
- 7.Carson J, Munday B. Streptococcosis: An emerging disease in aquaculture. Aust Aquaculture. 1990;5:32–33. [Google Scholar]
- 8.Conrads G, Gharbia SE, Gulabivala K, Lampert F, Shah HN. The use of a 16s rDNA directed PCR for the detection of endopathogenic bacteria. J Endod. 1997;23:433438. doi: 10.1016/S0099-2399(97)80297-X. [DOI] [PubMed] [Google Scholar]
- 9.Muzquiz JI, Roya FM, Orgega C, Deblas I, Ruiz I, Alonso JL. Pathogenicity of streptococcosis in rainbow trout (Onchorhynchus mykiss): dependence on age of diseased fish. Bull Eur Ass. Fish Pathol. 1999;19:114–119. [Google Scholar]
- 10.Vendrell D, Balcázar JL, Ruiz-Zarzuela I, Blas I, Olivia-Gironés O, Múzquiz JL. Lactococcus garvieae in fish: A review. Com Immunol Microbiol Inf Dis. 2006;29:177–198. doi: 10.1016/j.cimid.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Riffon R, Sayasith K, Khalil H, Dubreuil P, Drolet M, Lagace J. Development of a rapid and sensitive test for identification of major pathogens in bovine mastitis by PCR. J Clin Microbiol. 2001;39:2584–2589. doi: 10.1128/JCM.39.7.2584-2589.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shoemaker CA, Klesius PH, Evans JJ. Prevalence of Streptococcus iniae in tilapia, hybrid striped bass, and channel catfish on commercial fish farms in the United States. Am J Vet Res. 2001;62:174–177. doi: 10.2460/ajvr.2001.62.174. [DOI] [PubMed] [Google Scholar]
- 13.Soltani M, Jamshidi S, Sharifpour I. Streptococcosis caused by Streptococcus iniae in farmed rainbow trout (Onchorhynchus mykiss) in Iran: Biophysical characteristics and pathogenesis. Bull Eur Ass Fish Pathol. 2005;25:95–106. [Google Scholar]
- 14.Baeck GW, Kim JH, Gomez DK, Park SC. Isolation and characterization of Streptococcus sp. from diseased flounder (Paralichthys olivaceus) in Jeju Island. J Vet Sci. 2006;7:53–58. doi: 10.4142/jvs.2006.7.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meiri-Bendek I, Lipkin E, Friedmann A, Leitner G, Saran A, Friedman S, et al. A PCR-Based method for the detection of Streptococcus agalactiae in milk. J Dairy Sci. 2002;85:1717–1723. doi: 10.3168/jds.S0022-0302(02)74245-8. [DOI] [PubMed] [Google Scholar]
- 16.Tamura K, Dudley J, Nei M, Kumar S. 2007. Molecular evolutionary genetics analysis(MEGA) p. 229. Center of evolutionary functional genomics biodesign institute, Arizona state University. [DOI] [PubMed] [Google Scholar]
- 17.Fouad AF, Barry J, Caimano M, Clawson M, Zhu Q, Carver R, et al. PCR-Based identification of bacteria associated with Endod infections. J Clin Microbiol. 2002;40:3223–3231. doi: 10.1128/JCM.40.9.3223-3231.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ke D, Picard FJ, Martineau F, Menard C, Roy PH, Ouellette M, et al. Development of a PCR assay for rapid detection of enterococci. J Clin Microbiol. 1999;37:3497–3503. doi: 10.1128/jcm.37.11.3497-3503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forsman P, Tilsala-Timisjarvi A, Alatossava T. Identification of staphylococcal and streptococcal causes of bovine mastitis using 16S-23S rRNA spacer regions. Microbiol. 1997;143:3491–3500. doi: 10.1099/00221287-143-11-3491. [DOI] [PubMed] [Google Scholar]
- 20.Hassan AA, Khan IU, Lammler C. Identification of Streptococcus dysgalactiae strains of Lancefield's group C, G and L by polymerase chain reaction. J Vet Med. 2003;B50:161–165. doi: 10.1046/j.1439-0450.2003.00650.x. [DOI] [PubMed] [Google Scholar]
- 21.Soltani M, Nikbakht-Borojeni GH, Ebrahimzadeh- Moussavi HA, Ahmadzadeh N. Epizootic outbreaks of Lactoccoccosis caused by Lactococcus garvieae in farmed rainbow trout (Onchorhynchus mykiss) in Iran. Bull Eur Ass Fish Patholo. 2008;5:209–214. [Google Scholar]
- 22.Goh SH, Driedger D, Gillett S, Low DE, Hemmingsen SM, Amos M, et al. Streptococcus iniae, a human and animal pathogen: specific identification by the chaperonin 60 gene identification method. J Clin Microbiol. 1998;36:2164–2166. doi: 10.1128/jcm.36.7.2164-2166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colorni A, Ravelo C, Romalde JL, Toranzo AE, Diamant A. Lactococcus garvieae in wild Red Sea wrasse Coris aygula (Labridae) Dis Aqu Org. 2003;56:275–278. doi: 10.3354/dao056275. [DOI] [PubMed] [Google Scholar]
- 24.Eldar A, Ghittino C. Lactococcus garvieae and Streptococcus iniae infection in rainbow trout Oncorhynchus mykiss: similar, but different disease. Dis Aqu Org. 1999;36:227–231. doi: 10.3354/dao036227. [DOI] [PubMed] [Google Scholar]
- 25.Mata AI, Gibello A, Casamayor A, Blanco MM, Dominguez L, Fernandez-Garayzabal JF. Multiplex PCR assay for detection of bacterial pathogens associated with warm-water streptococcosis in fish. App Env Microbiol. 2004;70:3183–3187. doi: 10.1128/AEM.70.5.3183-3187.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nomoto R, Munasinghe LI, Jin DH, Shimahara Y, Yasuda H, Nakamura A, et al. Lancefield group C Streptococcus dysgalactiae infection responsible for fish mortalities in Japan. J Fish Dis. 2004;27:679–686. doi: 10.1111/j.1365-2761.2004.00591.x. [DOI] [PubMed] [Google Scholar]
- 27.Payghan R, Esmaeilli F, Moazedi J. Study of skin lesions of Epinephelus bleekeri cultured in cage. Iranian J Fish. 1996;2:37–42. [Google Scholar]
- 28.Plumb JA. Iowa State University Press; 1999. Health Manitenance and Principle microbial disease of cultured fishes; pp. 297–301. [Google Scholar]
- 29.Soltani M, Mousavi HA, Mirzargar S. Status of aquaculture health management in the Islamic Republic of Iran; Islamic Republic of Iran, Tehran: 1th international congress on aquatic animal; 2009. pp. 27–28. [Google Scholar]
- 30.Yung W, Li A. Isolation and characterization of Streptococcus dysagalactiae from diseased Acipenser schrenckii. Aquaculture. 2009;294:14–17. [Google Scholar]
- 31.Zlotkin A, Eldar A, Ghittino C, Bercovier H. Identification of Lactococcus garvieae by PCR. J Clin Microbiol. 1998;36:983–985. doi: 10.1128/jcm.36.4.983-985.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]