Abstract
Background and Objectives
Lipases are particularly important due to the fact that they specifically hydrolyze acyl glycerol, oils and greases, which is of great interest for different industrial applications.
Materialst and Methods
In this study, several lipase-producing bacteria were isolated from wastewater of an oil processing plant. The strain possessing the highest lipase activity was identified both biochemically and sequencing of 16S rRNA gene. Then we increase lipase activity by improving conditions of production medium. Also, lipase from this strain was preliminarily characterized for use in industrial application.
Results
The 16S rRNA sequensing revealed it as a new strain of Pseudomonas aeruginosa and the type strain was KM110. An overall 3-fold enhanced lipase production (0.76 U mL−1) was achieved after improving conditions of production medium. The olive oil and peptone was found to be the most suitable substrate for maximum enzyme production. Also the enzyme exhibited maximum lipolytic activity at 45°C where it was also stably maintained. At pH 8.0, the lipase had the highest stability but no activity. It was active over a pH range of 7.0–10.0. The lipase activity was inhibited by Zn2+ & Cu2+ (32 and 27%, respectively) at 1mM. The enzyme lost 29% of its initial activity in 1.0% SDS concentration, whereas, Triton X-100, Tween-80 & DMSO did not significantly inhibit lipase activity.
Conclusions
Based on the findings of present study, lipase of P. aeruginosa KM110 is a potential alkaline lipase and a candidate for industrial applications such as detergent, leather and fine chemical industries.
Keywords: Lipase, Pseudomonas aeruginosa, Stability
INTRODUCTION
Many attempts have been made to isolate lipase producing microorganisms since this enzyme is used in numerous biotechnological processes including food, leather, cosmetic, detergents and pharmaceutical industries and industrial wastes management (1). Lipases (triacylglycerol acylhydrolases, EC 3.1.1.3) catalyze the hydrolysis and the synthesis of esters formed from glycerol and long- chain fatty acids. Lipases occur widely in nature, but only microbial lipases are commercially significant. Microbial lipases are high in demand due to their specificity of reaction, stereo specificity and less energy consumption than conventional methods (2). Many microorganisms such as bacteria, yeast and fungi are known to secret lipases. Lipase-producing microorganisms have been found in diverse habitats such as industrial wastes, vegetable oil processing factories, dairies, soil contaminated with oil, etc (3). The oily environment (oil mill effluent) may provide a good environment for isolation of lipase producing microorganisms. Bacterial lipases are mostly extracellular and are greatly influenced by nutritional and physico- chemical factors, such as temperature, pH, nitrogen and carbon sources, inorganic salts, agitation and dissolved oxygen concentration (4). Among bacterial lipases, attention has usually been focused on particular classes of enzymes such as the lipases from the genus Pseudomonas, which are especially interesting for biotechnology. The literature reports different species of Pseudomonas from diverse environments producing lipase (5). The most important stage in a biological process is optimization to improve and increase the efficiency of the process without increasing the cost
The purpose of the present study is to isolate new bacteria from wastewater of an oil processing plant located in Tehran and increase its lipase activity by improving conditions of the production medium containing nitrogen and carbon sources. Also, Lipase from this strain was preliminarily characterized for use in industrial application.
MATERIALS AND METHODS
Screening of lipolytic bacteria. Lipase producing microbial cultures were isolated from wastewater of an oil processing plant located in Tehran, and enriched by periodic subculturing of samples in Nutrient Broth (NB) media containing 20% (v/v) and 40% (v/v) wastewater in successive. The composition of NB medium is (per liter) 5g peptone and 3g yeast extract. The pH of the medium was adjusted to 7 with 0.1M NaOH.The isolation process was performed by serial dilution of samples on tributyrin agar plates. The composition of tributyrin agar medium is (per liter) 5 g peptone, 3 g yeast extract, 10ml tributyrin and 15 g agar. Culture plates were incubated at 30°C. Colonies showing clear zones around them were picked out, purified on tributyrin agar plates and transferred to agar slants (6). Isolates having clearing zone were grown in the liquid culture and the level of lipase production was determined from the cell free culture supernatant fluid. Characterization and identification of the isolate with higher lipolytic activity was carried out both biochemically and by 16s r RNA sequencing.
Enzyme production. The composition of production medium used in this study was: (%w/v) pepton 0.2; NH4H2PO4 0.1; NaCl 0.25; MgSO4 7H2O 0.04; CaCl2.2H2O 0.04; olive oil 2.0 (v/v); pH 7.0; 1–2 drops Tween 80 as emulsifier. Overnight cultures were suspended in 5ml of sterile deionised water and used as the inoculum for pre culture to obtain an initial cell density to adjust the turbidity of 0.5 McFarland standard. Submerged microbial cultures were incubated in 500 ml Erlenmeyer flasks containing 100 ml of liquid medium on a rotary shaker (150 rpm) and incubated at 30°C. After 24 hours of incubation, the culture was centrifuged at 10,000 rpm for 20 min at 4°C and the cell free culture supernatant fluid was used as the sources of extracellular enzyme.
Assay of lipase activity. Lipase activity was deter-mined spectrophotometrically at 30°C using p-nitrophenol palmitate (pNPP) as substrate. The reaction mixture was composed of 700 µl pNPP solution and 300 µl of lipase solution. The pNPP solution was prepared by adding the solution A (0.001 g pNPP in 1ml isopropanal) into solution B (0.01 g gum arabic, 0.02 g Sodium deoxycholate, 50 µl Triton X-100 and 9 ml of 50 mM Tris-HCl buffer, pH 8) with stirring until all was dissolved. Then the absorbance measured at 410 nm for the first 2 min of reaction. One unit (1U) was defined as that amount of enzyme that liberated 1µmol of pNPP per minute (ɛ:1500l/mol cm) under the test conditions (7).
Effects of culture variable on lipase production. To investigate lipase production, olive oil was replaced by other carbon sources such as glucose and tributyrin. Each of substances (1% w/v) was used as sole carbon source. The effect of nitrogen sources on the lipase production was analyzed by supplementing production medium with different nitrogen sources (0.2% w/v) like peptone, yeast extract, ammonium dihydrogen phosphate and enzyme activity was assayed. Investigation of effect of different carbon and nitrogen source on lipase activity of Pseudomonas aeruginosa KM110 was done at pH: 7.0, 30°C and 150 rpm throughout 24 h of cultivations.
Effect of pH and temperature on lipase activity and stability. The crude enzyme used for assay was the culture broth after separation of cells and particles. The enzyme was normally stored at 4°C until used. The optimal temperature for activity was determined at different temperatures (30–70°C), at pH 8.0 for 10 min. For determination of temperature stability, the reaction mixtures containing the enzyme in 50mM Tris–HCl buffer (pH 8.0) was incubated at different temperatures (37, 45, 50, 55, 65 and 70°C) for 3 h and immediately cooled. Residual enzyme activity was measured under standard enzyme test conditions. Optimal pH was determined at 30°C in buffer solutions of pH values ranging from 5 to 11 (0.05 M citrate-phosphate pH 5-7; 0.05 M Tris–HCl pH 8-9; 0; 0.05 M Glysin – NaOH pH 11). The effect of pH on enzyme stability was analyzed by the spectrophotometric assay after pre-incubation of 300 µl of enzyme solution for 1 h at 30°C, in 700 µl of the above mentioned buffer solutions (pH 5–11). Enzymatic activity was measured according to a standard protocol with pNPP as the substrate.
Effects of different ions & detergents on lipase activity. As reported from studies on other microbial lipases, a concentration as low as 1 mM of some metal ions can affect the enzyme activity. Thus, the effect of several ions (Fe2+,,Na+, Ni+, Li+, Co2+, K+, Zn2+, Hg2+, Cu2+, Mn2+, Ca2+, Mg2+) on this P. aeruginosa lipase was determined. The enzyme solution was stored for 1 h at 30°C in the presence of 1 mM of various ions (as chloride salts) prior to the colorimetric assay for remaining lipase activity. In the case of chemical detergents, activity remaining was determined after 1 h of storage of enzyme solution at 30°C in the presence of various chemical detergents (SDS, DMSO (dimethyl sulfoxide), Tween 80 and Triton X-100) at 1% concentration. Activity was measured by the spectrophotometric assay after incubation time. Remaining enzymatic activity was determined by a standard method with pNPP. Final enzyme activity was calculated relative to control activity (a parallel enzyme reaction without additions).
Taxonomic characterization of isolated bacteria. The isolate was identified via 16S rRNA sequences. Genomic DNA of Pseudomonas aeruginosa KM110 was extracted from bacterial colonies by set buffer method. The 16S rRNA gene from the genomic DNA was amplified by PCR using the following forward and reverse primers of 16S rRNA, f (5′-AGAGTTTGATCMTGGCTCAG-3′) and r (5′- TACGGYTACCTTGTTACGAC-3′).PCRwas performed in a Thermocycler (TECHNE) using a Taq polymerase (Cinnagen, Iran). The PCR program comprised initial denaturation at 96°C for 4 min, followed by 35 cycles each of 94°C for 1 min, 61°C for 30 s, 72°C for 50 s; 72°C for 4 min; and incubation at 4°C for 10 min. PCR products were purified with DNA extraction kit (Bioneer South Korea). Both strands of the PCR product were sequenced by dideoxy chain termination method. The 16S rRNA gene sequence of the KM110 was compared with those in the NCBI/EZtaxon/ Ribosomal Database Project (RPD)/ EMBL nucleotide sequence databases by using the BLAST (blastn) program http://www.ncbi.nlm.nih.gov/BLATS/), and all of the sequences were aligned using the Clustal W program (8). A phylogenetic tree and neighbor-joining phylogeny were constructed by using the MEGA soft ware package version 4.0 (9) and bootstrapping was used to estimate the reliability of the phylogenetic reconstructions (1,000 replicates).
RESULTS AND DISCUSSION
Screening and identification of lipolytic bacteria. The almost complete 16S rRNA gene was sequenced and the (1406 bp) analysis clearly demonstrated that strain KM110 was a member of the genus Pseudomonas and exhibited maximum similarity with the 16S rRNA sequence of Pseudomonas aeruginosa LMG 1242T(Z76651) (98.94% sequence similarity). This sequence data has been submitted to the DDBJ/EMBL/GenBank databases under accession No. HQ730879.
Effects of culture variable on lipase production. The major factor for the expression of lipase activity has always been carbon, since lipases are inducible enzymes (10) and are thus generally produced in the presence of a lipid source such as oil or any other inducer, such as triacylglycerols, fatty acids, hydrolysable esters, tweens, bile salts and glycerol. However, their production is significantly influenced by other carbon sources such as sugars, polysaccharides, whey and other complex sources. Among the different carbon sources used, olive oil was found to be the most suitable source (Fig. 1). The maximum activity for olive oil was observed at 2% (v/v) (0.46U/ml). Most published experimental data have shown that lipid carbon sources (especially natural oils) stimulate lipase production (11–13). High levels of lipase production were reported from various thermophilic Bacillus sp. in the presence of olive oil as carbon source in the culture medium (14, 15). The presence of glucose in the cultivation medium depressed the production of lipase compared to olive oil. Glucose supplementation to the basal production medium inhibits lipase production, perhaps by catabolic repression. This occurred with reports for other lipase-producing organisms for which a high glucose concentration caused reduced lipase production (16–18).
Fig. 1.
Effect ofdifferent carbon sources as additives(1% w/v) to the basal medium on lipase production by Pseudomonas aeruginosa KM110 (T: 30°C, pH: 7.0, agitation rate: 150 rpm).
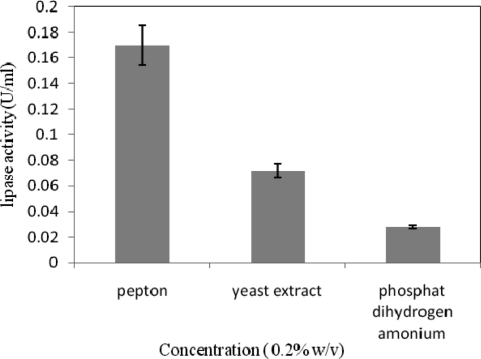
Beside carbon source, the type of nitrogen source in the medium also influenced the lipase titers in production broth. Generally, microorganisms provide high yields of lipase when organic nitrogen sources are used, such as peptone and yeast extract, which have been used for lipase production by various thermophilic Bacillus sp. and various Pseudomonads (19, 20). Among the different nitrogen sources used peptone was found to be the most suitable source (Fig. 2). In pepton (2 g/l) maximum activity was observed (0.17 U/ml).
Fig. 2.
Effect of different nitrogen sources as additives (0.2% w/v) to the basal medium on lipase production by Pseudomonas aeruginosa KM110(T: 30°C, pH: 7.0, agitation rate: 150 rpm).
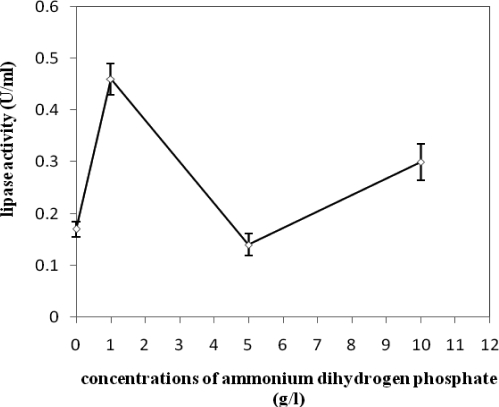
Inorganic nitrogen sources such as ammonium chloride and ammonium dihydrogen phosphate have been also reported to be effective in some microbes (21, 22). We also investigated the effect of ammonium dihydrogen phosphate as an inorganic carbon source with presence of peptone on lipase activity. Fig. 3 shows that it is effective on lipase activity so we used both peptone and ammonium dihydrogen phosphate as nitrogen sources in the medium.
Fig. 3.
Effect of different concentrations of ammonium dihydrogen phosphate (g/l) on lipase production by Pseudomonas aeruginosa KM110 (T: 30°C, pH: 7.0, agitation rate: 150 rpm).
Effect of pH on activity and stability of lipase. The effect of pH on the activity of lipase was determined in four different buffers covering the range of pH 3.0 to 12.0. The enzyme was most active at pH 6.0 & 9.0 (Fig. 4a), increasing activity from pH 7-9 can be characterized as an alkalophilic enzyme but high lipase activity at pH 6 can be made the lipase applicable at acidic pH conditions. In general bacterial lipases are stable in a wide range of pH from 4 to 11. A comprehensive review of all bacterial lipase done by Gupta, et al. (4), states that maximum activity of lipases at pH values higher than 7 has been observed in many cases. Bacterial lipases have a neutral or alkaline optimum pH. with the exeption of lipase from P. fluorescens SIK W1 that has an acidic optimum pH 4.8. Also the lipase retained over 65% of its activity at pH 8.0 (Fig. 4b). Interestingly, other Pseudomonas lipases designated as alkaline, e.g., P. fluorescens HU380 (23), P. mendocina PK-12CS (24), P. fluorescens 2D (25) and P. cepacia (26) have lower pH optima of 8.5, 8.0, 8.5 and 9.0, respectively. Lipases from P. pseudomalei 12 sm (27) and P. aeruginosa YS-7 (28) both isolated from Pseudomonas growing in different water-restricted environments, are stable within the pH ranges of 7–10.5 and 6.5–7.5, respectively. Lipase obtained in our study was stable from pH 7.0 to 10.0. However, its stability was low at acidic pH. The remarkable stability of Psudomonas aeruginosa KM110 lipase in this range has proved it to be a potential alkaline lipase.
Fig. 4.
Effects of pH on activity (a) and stability (b) of lipase.
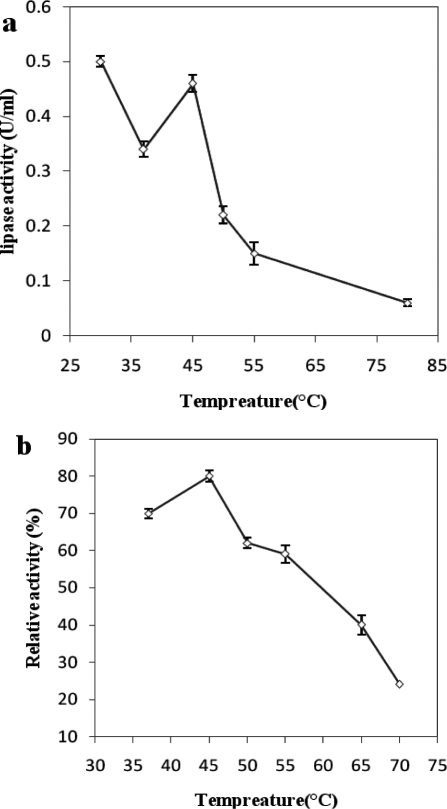
Effect of temperature on activity & thermostabi-lity of lipase. The temperature preference of this enzyme reveals higher activity values at temperature from 35 to 45°C (Fig. 5a). The lower activity at 35°C compared to 45°C, probably is due to the kinetics of enzymatic reaction as the enzyme is more conductive at 45°C. Assessment of the thermostability of lipase was performed by measuring the residual activity at various times, following incubation at different temperatures. As for the stability of the enzyme (Fig. 5b), 80% activity remained after 3 h of storage at 45°C and 70% at 37°C. At higher temperatures, the stability of the enzyme was lower; ie 40% activity remained after 3h at 65°C.The stability of the lipase decreased sharply after 1 h of incubation at high tempreatures (Fig. 5b). This indicates that Psudomonas aeruginosa KM110 lipase is a mesophilic enzyme. The optimum temperature of the lipase from P. aeruginosa EF2 was reported to be 50°C (29). The P. aeruginosa MB 5001 lipase has an optimum temperature of 55°C (30) but other Pseudomonas lipases, such as those from P. fluorescens 2D (23), P. fluorescens HU380 (23), P. fragi (31) and P. mendoncina (25) were found to be optimally active at 35–45°C. P. aeruginosa lipases seem to be more thermostable than others from this genus.
Fig. 5.
Effects of tempreature on activity (a) and stability (b) of lipase.
The effect of metal ions on activity of lipase. The effect of different metal ions on the activity of the lipase is shown in Table 1. Zn2+, Cu2+, and K+ salts decreased activity by 32, 27, and 21%, respectively, after 1 h of incubation at 30°C. Effect of the heavy metal ions Zn2+ and Cu2+ on the our lipase was similar to their effect on lipases from P.aeruginosa LP602 (17), P. fluorescens 2D (25) and HU380 (23). In contrast, activity increased around 40% in the presence of Fe2+ and 37% in the presence of Mg2+. It has been suggested that the effect of metal ions could be attributed to a change in the solubility and the behavior of the ionized fatty acids at interfaces, and from a change in the catalytic properties of the enzyme itself (32). This lipase was considered stable towards ions reported to inhibit other lipases (Fe2+ and Mg2+) (18, 32 and (33)). The metal ions Li+, Co2+, Mn2+ & Ca2+ showed a small inhibitory effect on lipase activity relative to Zn2+, Cu2+, and K+ salts and the lipase was relatively stable towards many other metal ions tested. This characteristic made the enzyme suitable for lipase reactions under various conditions without special precautions.
Table 1.
Effects of various salts on lipase activity.
| 1 mM | Remaning activity (%) |
|---|---|
| control | 100 |
| FeCl2 | 139.5 |
| MgCl2 | 136.8 |
| NaCl | 115.8 |
| HgCl | 115.8 |
| NiCl | 110.5 |
| LiCl | 92.1 |
| CoCl | 89.5 |
| MnCl2 | 86.8 |
| CaCl2 | 81.6 |
| KCl | 78.9 |
| CuCl2 | 73.7 |
| Zn(No3)2 | 68.4 |
The effects of various detergent on activity of lipase. Table 2 demonstrates the effects of various detergents on enzyme activity. P. aeruginosa KM110 lipase was sensitive to SDS than the other. In 1.0% SDS concentration, the enzymes showed 70% activity after 1 h storage. This level of stability was similar to or higher than those reported for other bacteria (32).The enzyme was relatively stable when stored with Tween-80, DMSO and Triton X-100 (i.e., the activity remaining higher than 80%). In accordance to our results, Schmidt- Dannert et al. 1994 (34) reported a total loss of lipolytic activity in the presence of Tween 20 and Tween 80, but no effect was observed when incubated with Triton X-100. Nawani et al. (1998) also found a total loss of activity in the presence of SDS but in contrast, activity was enhanced in the presence of Triton X-100, Tween 20 and Tween 80.
Table 2.
Effects of various detergents on lipase activity.
| 1% concentration | Remaining activity(%) |
|---|---|
| control | 100 |
| DMSO | 102.8 |
| Triton X-100 | 91.4 |
| Tween 80 | 82.8 |
| SDS | 71.4 |
CONCLUSION
The results obtained in this study show that olive oil and peptone were the most suitable substrate for maximum lipase production by P. aeruginosa KM110. Further studies are needed to enhance lipase production in this strain. We are currently in the process of cloning this lipase and placing it under a strong promoter to be able to determine molecular properties of the lipase as well as increasing enzyme expression and yields for future industrial applications. Also, P. aeruginosa KM110 lipase is a potential alkaline lipase. The enzyme exhibited maximum activity and stability between pH 7.0 and 10.0, but the stability was low at acidic pH. The remarkable stability of P. aeruginosa KM110 lipase in this range has proved it to be a potential alkaline lipase similar to other, and a candidate for industrial applications such as detergent, leather and fine chemical industries. Also, the lipase was relatively stable towards many metal ions and detergents tested. This characteristic made the enzyme suitable for lipase reactions under various conditions without special precautions.
ACKNOWLEDGEMENT
We thank the department of Biology of Alzahra University (Tehran, Iran) for their support of this study, which is a section of the M.S thesis of E. Mobarak-Qamsari.
REFERENCES
- 1.Sharma R, Chisti Y, Banerjee UC. Production, purification, characterization, and applications of lipases. Biotechnol Adv. 2001;19:627–662. doi: 10.1016/s0734-9750(01)00086-6. [DOI] [PubMed] [Google Scholar]
- 2.Saxena RK, Ghosh PK, Gupta R, Davinson WS, Bradoo S, Gulati R. Potential biocatalysis and future industry. Curr Sci. 1999;77:110–115. [Google Scholar]
- 3.Sztajer H, Maliszewska I, Wieczorek J. Production of exogenous lipase by bacteria, fungi and actinomycetes. Enzyme Microb Technol. 1998;10:492–7. [Google Scholar]
- 4.Gupta R, Gupta N, Rathi P. Bacterial lipases: an overview of production, purification and biotechnological properties. Appl Microbiol Biotechnol. 2004;64:763–781. doi: 10.1007/s00253-004-1568-8. [DOI] [PubMed] [Google Scholar]
- 5.Arpigny JL, Jaeger KE. Bacterial lipolytic enzymes: classification and properties. Biochem J. 1999;343:177–13. [PMC free article] [PubMed] [Google Scholar]
- 6.Ertugrul S, Donmez G, Takac S. Isolation of lipase producing Bacillus sp. From olive mill waste water and improving its enzyme activity. J Hazard Mater. 2007;149:720–724. doi: 10.1016/j.jhazmat.2007.04.034. [DOI] [PubMed] [Google Scholar]
- 7.Karadzic I, Masui A, Lidija Izrael Zivkovic LI, Fujiwara N. Purification and characterization of an alkaline lipase from Pseudomonas aeruginosa isolated from putrid mineral cutting oil as component of metal working fluid. J Biosci Bioeng. 2006;102:82–89. doi: 10.1263/jbb.102.82. [DOI] [PubMed] [Google Scholar]
- 8.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 9.Thompson JD, Higgins DG, Gibson TJ, Clustal W. improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lotti M, Monticelli S, Montesinos JL, Brocca S, Valero F, Lafuente J. Physiological control on the expression and secretion of Candida rugosa lipase. Chem Phys Lipids. 1998;93:143–148. doi: 10.1016/s0009-3084(98)00038-3. [DOI] [PubMed] [Google Scholar]
- 11.Abdel-Fattah YR. Optimization of thermostable lipase production from a thermophilic Geobacillus sp. using Box-Behnken experimental design. Biotechnol Lett. 2002;24:1217–1222. [Google Scholar]
- 12.Kaushik R, Saran S, Isar J, Saxena RK. Statistical optimization of medium components and growth conditions by response surface methodology to enhance lipase production by Aspergillus carneus . J Mol Catal B-Enzym. 2006;40:121–126. [Google Scholar]
- 13.He YQ, Tan TW. Use of response surface methodology to optimize culture medium for production of lipase with Candida sp. J Mol Catal . B-Enzym. 2006;43:99–125. [Google Scholar]
- 14.Eltaweel MA, Rahman RNZRA, Salleh AB, Basri M. An organic solvent-stable lipase from Bacillus sp. strain 42. Ann Microbiol. 2005;55:187–192. [Google Scholar]
- 15.Lee DW, Koh YS, Kim KJ, Kim BC, Choi HJ, Kim DS, et al. Isolation and characterization of a thermophilic lipase from Bacillus thermoleovorans ID-1. FEMS Microbiol Lett. 1999;179:393–400. doi: 10.1111/j.1574-6968.1999.tb08754.x. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Ishii T, Shimura S, Kirimura S, Usami S. Lipase production of Tricosporon fermentans WUC12 a newly isolated yeast. J Ferment Bioeng. 1992;73:412–414. [Google Scholar]
- 17.Dharmsthiti S, Kuhasuntisuk B. Lipase from Pseudomonas aeruginosa LP602: Biochemical Properties Application for Wastewater Treatment. J Ind Microbiol Biotechnol. 1998;21:75–80. [Google Scholar]
- 18.Lotrakul P, Dharmsthiti S. Lipase production by Aeromonas sobria LP004 in a medium containing whey and soybean meal. World J Microbiol Biotechnol. 1997;13:163–166. [Google Scholar]
- 19.Sharma R, Soni SK, Vohra RM, Jolly RS, Gupta LK, Gupta JK. Production of extracellular alkaline lipase from a Bacillus sp. RSJ1 and its application in ester hydrolysis. Ind J Microbiol. 2002;42:49–54. [Google Scholar]
- 20.Sugihara A, Tani T, Tominaga Y. Purification and characterization of a novel thermostable lipase from Bacillus sp . J Biochem. 1991;109:211–215. [PubMed] [Google Scholar]
- 21.Dong H, Gao S, Han SP, Cao SG. Purification and characterization of a Pseudomonas sp. lipase and its properties in non-aqueous media. Biotechnol App Biochem. 1999;30:251–256. [PubMed] [Google Scholar]
- 22.Rathi P, Saxena RK, Gupta RA. Novel alkaline lipase from Bulkholderia cepacia for detergent formulation. Process Biochem. 2001;37:187–192. [Google Scholar]
- 23.Kojima Y, Shimizu S. Purification and characterization of the lipase from Pseudomonas flurescens HU380. J Biosci Bioeng. 2003;96:219–226. [PubMed] [Google Scholar]
- 24.Jinwal U, Roy U, Chowdhury A, Bhaduri A, Roy PK. Purification and characterization of an alkaline lipase from a newly isolated Pseudomonas mendoncina PK-12Cs and chemoselective hydolysis of fatty acid ester. Bioorgan Med Chem. 2003;11:1041–1046. doi: 10.1016/s0968-0896(02)00516-3. [DOI] [PubMed] [Google Scholar]
- 25.Makhzoum A, Owusu-Apenten RK, Knapp JS. Purification and properties of lipase from Pseudomonas fluorescens strain 2D. Int Diary J. 1996;6:459–472. [Google Scholar]
- 26.Svendsen A, Borch K, Barfoed M, Nielsen T, Gormsen E, Patkar S. Biochemical properties of cloned lipases from Pseudomonas family. Biochim Biophys Acta. 1995;1259:9–17. doi: 10.1016/0005-2760(95)00117-u. [DOI] [PubMed] [Google Scholar]
- 27.Kanwar L, Goswami P. Isolation of Pseudomonas lipase produced in pure hydrocarbon substrate and its application in the synthesis of isoamyl acetate using membrane immobilized lipase. Enzyme Microb Technol. 2002;31:727–735. [Google Scholar]
- 28.Shabtai Y, Daya-Mishne N. Production, purification and properties of lipase from a bacetrium (Ps. aeruginosa YS-7) capable of growing in water-restricted environments. Appl Environ Microbiol. 1992;58:174–180. doi: 10.1128/aem.58.1.174-180.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilbert EJ, Cornish A, Jones C. Purification and proper-ties of extracellular lipase from Pseudomonas aeruginosa EF2. J Gen Microbiol. 1991b;137:2223–2229. doi: 10.1099/00221287-137-9-2223. [DOI] [PubMed] [Google Scholar]
- 30.Chartrain M, Katz L, Marcin C, Thien M, Smith S, Fisher E, et al. Purification and characterization of a novel bioconverting lipase from Pseudomonas aeruginosa MB 5001. Enzyme Microb Technol. 1993;15:575–580. [Google Scholar]
- 31.-Mencher JR, Alford JR. Purification and characterization of the lipase from Pseudomonas fragi . J Gen Microbiol. 1967;48:317–328. doi: 10.1099/00221287-48-3-317. [DOI] [PubMed] [Google Scholar]
- 32.Matsumae H, Shibatani T. Purification and characterization of lipase from Serratia marcescens Sr41 8000 responsible for asymmetric hydrolysis of 3-phenylglycidic esters. J Ferment Bioeng. 1994;77:152–158. [Google Scholar]
- 33.Lelie D. Screening, purification and characteriza-tion of the thermoalkalophilic lipase produced by Bacillus thermoleovorans CCR11. Enzyme Microb Tech. 2005;37:648–654. [Google Scholar]
- 34.Schmidt-Dannert C, Sztajer H, Stocklein W, Menge U, Schmid RD. Screening, purification and properties of a thermophilic lipase from Bacillus thermocatenulatus . Biochim Biophys Acta. 1994;1214:43–53. doi: 10.1016/0005-2760(94)90008-6. [DOI] [PubMed] [Google Scholar]