Abstract
Background and objectives
Each year, Enteroviruses infect millions of people and cause different diseases. The agents are usually detected using cell culture. RD (Rhabdomyosarcoma) and L20B (L cells) are among the recommended cells by the World Health Organisation (WHO) for this purpose. Even though cell culture is the most common method used in diagnosing Enteroviruses in stool specimens, this particular method poses some problems, which include false positive or negative results, lack of a unique cell line for diagnosing all Enterovirus types in addition to being time consuming. For these reasons, an attempt was made to find better techniques of Enterovirus detection. RT-PCR (Reverse Transcriptase Polymerase Chain Reaction) is a technique used in place of the cell culture method. In this study, the cell culture method was compared with RT-PCR for detection of Enteroviruses in stool specimens.
Material and method
First, the chloroform treated stool samples were inoculated onto five cell lines, including RD, L20B, Hep-2 (Human Epidermoid carcinoma cell line), Vero (Verde Reno) and GMK (Green Monkey Kidney). The results were then compared with data from Enterovirus detection using the RT-PCR technique.
Results and conclusion
The difference between RT-PCR and cell culture results was significant. Enteroviruses were detected in 24% of specimens using RT-PCR while cell lines could isolate Enteroviruses in just 14.4% of the samples.
Keywords: Enteroviruses, RT-PCR, Cell Cultures
INTRODUCTION
Based on the latest virus classification, human Enterovirus genus is divided into five species including Poliovirus (PV-1, -2 and -3), Human Enterovirus A (HEV-A) (Coxsackievirus A2, 3, 5, 7, 8, 10, 12, 14 and 16 and Enterovirus 71), HEV-B (Coxsackievirus A9, Coxsackievirus B 1–6, Echovirus 1–7, 9, 11–21, 24–27, 29– 33 and Enterovirus 69), HEV-C (Coxsackievirus A1-3, 11, 13, 15, 17–22 and 24) and HEV-D (Enterovirus 68 and 70) which can be transferred orally and infect the intestinal tract (1–3). Infections are usually not serious, but they can sometimes pass through intestinal cells and access inner parts of the body to cause some severe illnesses such as poliomyelitis, aseptic meningitis, myocarditis, foot and mouth disease, herpangina, pleurodynia, acute hemorrhagic conjunctivitis and other diseases (4–6).
After using animal cell cultures as a perfect technique for diagnosing Enteroviruses, it has become the gold standard for detection (4, 7). Attempts have been made to improve the techniques so as to find more sensitive cell cultures for virus detection and decrease cell infection rates. Nowadays, modern methods and materials such as air laminar flow systems, nanofilters and antibiotics reduce the rate of infection in the cells used (7). Meanwhile, calf bovine serum is used as cell supporter to ensure cell growth. Furthermore, inverted microscope has facilitated observations of cells and their cytopathic effects (CPE) (7, 8).
Cell culture technique, however, has some undeniable problems, and it needs to be improved. A special cell line can not support the growth of all viruses; on the other word, each virus can grow onsome special cell lines and a combination of cell lines are needed to isolate all serotypes of a big group of viruses such as Enteroviruses. For instance, although RD (Rhabdomyosarcoma) and L20B (L cell) are used for isolating Enteroviruses from stool specimens, they can not support all the serotypes. Furthermore, using a combination of at least two cell lines for isolation of Enteroviruses makes the technique time consuming and costly. In addition, cell culture contamination is a common problem that laboratories frequently encounter (9).
PCR is considered as an efficient method for virus detection at the moment (10, 11). The high speed involved in virus detection and its improved sensitivity makes the technique a favourite method for virus detection in cell cultures, clinical specimens, biopsy and autopsy (10, 11).
This study was aimed at comparing the cell culture method and RT-PCR (Reverse Transcriptase-PCR) for detection of Enteroviruses in the stool specimens. We have tried to increase the sensitivity of virus detection by using five cell lines simultaneously: RD, L20B, Hep-2 (Human Epidermoid carcinoma cell line), Vero (Verda Reno) and GMK (Green Monkey Kidney).
MATERIALS AND METHODS
Specimens. 230 stool specimens, collected from patients with acute flaccid paralysis (AFP), were transferred to the laboratory under appropriate conditions.
Preparation of the stool specimens. Stool specimens were subjected to chloroform pre-treatment before inoculation to cell culture or genome extraction for RT-PCR. In addition to removing bacteria and fungi, chloroform pre-treatment removes potentially cytotoxic substance and dissociates virus aggregates (12).
Cell Lines. As the first step, all the cell lines used in this study (RD, L20B, Hep2, Vero and GMK) were evaluated for their sensitivity to Enteroviruses using known concentrations of vaccine Poliovirus, Echo11 and Coxsackievirus B and their sensitivity was confirmed. All the materials for cell culture, including cell culture media, fetal bovine serum and antibiotics, were prepared according to the standard procedure recommended by WHO (12). The treated samples were then inoculated onto monolayered cells prepared in cell culture tubes and were kept at 36°C (5% CO2and 80% humidity). The tubes were microscopically evaluated for 5 days to detect any evidence of cytopathic effect (CPE). To increase the sensitivity of virus isolation, blind passage was carried out on the cultures, which had remained negative, and they were checked for the next 5 days.
Serotyping by Microneutralization. In this study, microneutralization was performed based on the standard method recommended by WHO (World health organization) (12, 13).
RNA extraction and RT-PCR. RNA extraction and RT-PCR were performed as previously described (13, 14). Primers were prepared according to WHO protocol for identifying Enteroviruses (WHO, 2004) and can detect a conserved sequence in 5/ of the viral genome: EV-PCRI (5′–ACA CGG ACA CCC AAA GTA GTC GGT TCC –3′) and EV-PCR2 (5′–TCC GGC CCC TGA ATG CGG CTA ATCC-3′). The positive samples in cell cultures and neutralization test were used as positive controls.
First, a cycle was set for 20 minutes in 42°C for a reverse transcriptase reaction and 3 minutes in 95°C to deactivate this enzyme. Then, the programme contained 35 cycles (which included 45 seconds in 95°C, 45 seconds in 55°C, 45 seconds in 70°C and finally, 10 minutes in 70°C). The PCR product band (114 bp) was identified in Ethidium bromide containing agarose gel wells and by size marker with a molecular size number 8 (Roche) (12, 13).
Statistical Analysis. The data was analyzed by using SPSS, ANOVA test and Chi square tests. P-Value less than 0.05 was used to indicate statistical significance.
RESULTS
In order to determine a more effective method for diagnosing Enteroviruses from faeces, 230 stool samples (suspected to AFP) were compared by using different routine methods: cell culture and RT-PCR. Totally, 33 out of 230 specimens (14.4%) were found to be positive for Enteroviruses by cell culture (Table 1).
Table 1.
Result of cell culture for Enterovirus detection.
| Cell line Isolated virus | Hep2 | Vero | RD | L20b | GMK | Total |
|---|---|---|---|---|---|---|
| Echoviruses | 11 | 9 | 12 | 0 | 7 | 12 |
| Polioviruses | 9 | 8 | 9 | 9 | 5 | 12 |
| Coxsackieviruses | 5 | 1 | 0 | 0 | 0 | 5 |
| Unidentified Serotypes | 4 | 0 | 4 | 0 | 1 | 4 |
| Sum | 29 | 16 | 25 | 9 | 13 | 33 |
| Rate of Enterovirus Isolation | 87.9% | 48.5% | 75.8% | 27.3% | 40% | 100% |
AFP: Acute Flaccid Paralysis; RD: Rhabdomyosarcoma cell Line; L20B: L20B cell line; HEP-2: Human Epidermoid cancer cell line; vero: Verda Reno cell line; gMK: Green Monkey Kidney cell line; RT-PCR: Reverse Transcriptase Polymerase Chain Reaction; WHO: World Health Organisation.
In this study, unanimous with other studies, no cell culture has been found to be able to support growth of all Enteroviruses (15, 16). Despite being the gold standard for detecting some viruses, the cell culture technique had some limitations and it sometimes failed to detect Enteroviruses due to inhibitors which exist in the specimens, especially when the specimen is faeces (17, 18). In the case of Enteroviruses, cell cultures need at least two weeks to detect the virus in the specimen and this is a good opportunity for the virus to contaminate the surrounding environment (19, 20).
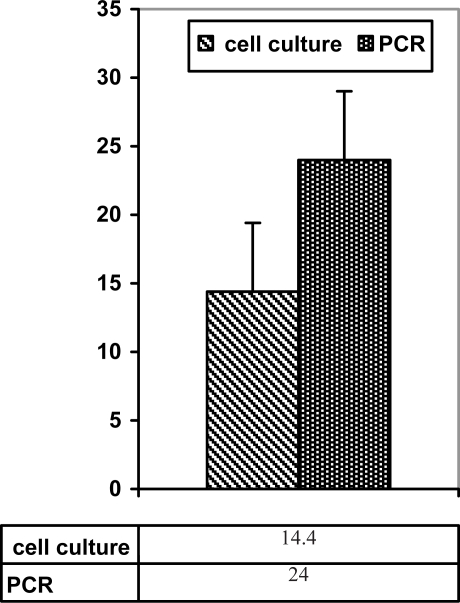
For this reason, designing a rapid and highly sensitive method for diagnosing Enteroviruses in stool specimens was crucial to help the health system in detecting the agents much faster and more accurately. After setting up and using RT-PCR for detecting Enteroviruses in the specimens, the detection rate was improved to 24% (55 out of 230 specimens) (Fig. 1). Statistical analysis showed the differences in sensitivity as meaningful.
Fig. 1.
Comparing cell culture and RT-PCR methods for isolating Enteroviruses from stool samples.
DISCUSSION
Enteroviruses are one of the most important gastric viruses that can cause some dangerous diseases, especially in children (7). Although RD and Hep-2 are efficient cell lines for detection of most Enteroviruses, they can not support growth of all Enteroviruses (7, 15, 16). Furthermore, cell maintenance and some of the materials needed for cell culture (such as serum) are quite expensive, and checking cell cultures every day makes the technique boring and time consuming (19, 20).
To detect Enteroviruses in stool specimens by molecular methods, three kinds of pre-treatment of the specimen can be used: direct RNA extraction from stool specimen and then RT-PCR (1, 11, 21, 22), extraction of RNA from the stool specimens which have been pre-treated with chloroform (1, 11, 22–24), and RT-PCR on positive cell cultures of the stool specimens (23, 25, 26).In the present study, a single-step RT-PCR method for direct detection of Enteroviruses form stool samples was used as it is more cost effective and decreases the probability of cross contamination (10).
In the cell culture step, 5 cell lines (RD, L20B, Hep-2, GMK and Vero) were used to increase the probability of Enterovirus detection because several serotypes of Enteroviruses grow only in particular cell lines. By these cell lines, 33 cases out of 230 specimens (14.4%) were positive for Enteroviruses. Among all cell lines, RD and Hep-2 were able to detect more Enteroviruses. However, RT-PCR on pre-treated stool specimens could detect 55 Enteroviruses in 230 specimens (24%); much higher than Enterovirus detection rate in cell culture.
Other studies for comparing RT-PCR and cell culture in Enterovirus detection had same outcomes (4, 27, 28). They showed that different samples (4), the chosen procedure for RT-PCR, (27) the source of samples (27) and different conditions could affect the outcomes (4, 29). Although there are some differences in the results obtained, all studies have proven that RT-PCR is more sensitive than cell culture for Enterovirus detection. The findings in this study confirm the reports by others and have shown that RT-PCR makes the researcher more confident in detecting viruses in a variety of samples.
It is important to mention that PCR is a highly sensitive method and the procedure needs to be performed in a DNA free environment (30, 31). Utilizing separated areas for PCR ionic potential of solutions and concentrations of proper primers, nucleotides and polymerases are other factors that need to be taken into consideration when molecular methods are to be used for Enterovirus detection (30, 31). Obtaining false negatives in cell culture can be due to the presence of slow growing Enteroviruses in stool specimens, lack of sensitivity of the cell line, low titre of the virus in the specimens and toxic factors (17, 18, 30–32).
ACKNOWLEDGMENTS
The project was done as a part of PhD thesis and the authors wish to thank the personnel of Iran National Polio Laboratory, Virology Division, Pathobiology Department, School of Public Health, Tehran University of Medical Sciences for their kind cooperation in this project.
CONFLICT OF INTEREST
The authors declared no conflict of interest.
REFERENCES
- 1.American Public Health Association. 19th ed. Washington, D.C.: American Public Health Association; 1995. Standard methods for the examination of water and wastewater; pp. 7–8. [Google Scholar]
- 2.Oberste MS, Maher K, Kilpatrick D R, Flemister MR, Brown BA, Pallansch MA. Typing of human Enteroviruses by partial sequencing of VP1. J Clin Microb. 1999;37:1288–1293. doi: 10.1128/jcm.37.5.1288-1293.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pöyry T, Kinnunen L, Hyypiä T, Brown B, Horsnell C, Hovi T, et al. Genetic and phylogenetic clustering of Enteroviruses. J Gen Viro. 1996;77:1699–1717. doi: 10.1099/0022-1317-77-8-1699. [DOI] [PubMed] [Google Scholar]
- 4.Terletskaia LE, Meier S, Hahn R, Leinmuller M, Schneider F, Enders M. A convenient rapid culture assay for the detection of Enteroviruses in clinical samples: comparison with conventional cell culture and RT-PCR. J Med Microb. 2008;57:1000–1006. doi: 10.1099/jmm.0.47799-0. [DOI] [PubMed] [Google Scholar]
- 5.Gajanan N, Sapkal GN, Bondre VP, Fulmali PV, Patil P, Gopalkrishna V. Enteroviruses in patients with acute encephalitis. Uttar Pradesh, India. Emerg Infect Dis. 2009;15:295–298. doi: 10.3201/eid1502.080865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fields BN, Knipe DM, Howley PM, Chanock RM, Monath TP, Melnick JL, et al. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. Enteroviruses: Polioviruses, Coxsackieviruses, Echoviruses, and newer Enteroviruses; pp. 655–712. [Google Scholar]
- 7.Leland DS, Ginocchio CC. Role of Cell Culture for Virus Detection in the Age of Technology. Clin Microb Rev. 2007;20:49–78. doi: 10.1128/CMR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dagan R, Menegus MA. A combination of four cell types for rapid detection of Enteroviruses in clinical specimens. J Med Virol. 1986;19:219–228. doi: 10.1002/jmv.1890190304. [DOI] [PubMed] [Google Scholar]
- 9.Chonmaitree T, Ford C, Sanders C, Lucia HL. Comparison of cell cultures for rapid isolation of Enteroviruses. J Clin Microb. 1988;26:2576–2580. doi: 10.1128/jcm.26.12.2576-2580.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Archimbaud C, Chambon M, Bailly JL, Petit I, Henquell C, Mirand A, et al. Impact of rapid Enterovirus molecular diagnosis on the management of infants, children, and adults with aseptic meningitis. J Med Viro. 2009;81:42–48. doi: 10.1002/jmv.21330. [DOI] [PubMed] [Google Scholar]
- 11.Hong J, Kang B, Kim A, Hwang S, Lee S, Kim J, et al. Enhanced detection of Enteroviruses in clinical samples by RT-PCR using complementary locked primer Technology. J Clin Microb. 2009;48:615–616. doi: 10.1128/JCM.01790-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.WHO. Geneva: WHO; 2004. Polio Laboratory Manual. [Google Scholar]
- 13.Shoja ZO, Tabatabai H, Sarijloo M, Shahmahmoodi S, Azad TM, Nategh R. Detection of Enteroviruses by reverse-transcriptase polymerase chain reaction in cell culture negative stool specimens of patients with acute flaccid paralysis. J Virol Methods. 2007;142(1-2):95–7. doi: 10.1016/j.jviromet.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 14.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 15.Johnston SL, Siegel CS. Presumptive identification of Enteroviruses with RD, HEp-2, and RMK cell lines. J Clin Microbiol. 1990;28:1049–1050. doi: 10.1128/jcm.28.5.1049-1050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sigel MM, Rippe DF, Beasley AR, Dorsey M. Viruses in water. Washington, D.C: American Public Health Association; 1976. Systems for detecting viruses and viral activity; pp. 139–164. [Google Scholar]
- 17.Wait D, Tai L, Sobsey MD. Methods to remove inhibitors in sewage and other fecal wastes for Enterovirus detection by the polymerase chain reaction. J Virological Methods. 1995;54:51–66. doi: 10.1016/0166-0934(95)00025-p. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt NJ, HO HH, Riggs JL, Lennette EH. Comparative sensitivity of various cell culture systems for isolation of viruses from wastewater and fecal samples. Appl Environ Microb. 1978;36:480–486. doi: 10.1128/aem.36.3.480-486.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee C, Lee S, Han E, Kim S. Use of cell culture-PCR assay based on combination of A549 and BGMK cell lines and molecular identification as a tool to monitor infectious adenoviruses and Enteroviruses in river water. Appl Environ Microbiol. 2004;70:6695–6705. doi: 10.1128/AEM.70.11.6695-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chapron CD, Ballester NA, Fontaine JH, Frades CN, Margolin AB. Detection of Astroviruses, Enteroviruses and Adenovirus types 40 and 41 in surface waters collected and evaluated by the information collection Rule and an integrated cell culture-nested PCR procedure. Appl Environ Microbiol. 2000;66:2520–2525. doi: 10.1128/aem.66.6.2520-2525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kessler HH, Santner B, Rabenau H, Berger A, Vince A, et al. Rapid diagnosis of Enterovirus infection by a new one-Step reverse transcription-PCR assay. J Clin Micro. 1997;35:976–977. doi: 10.1128/jcm.35.4.976-977.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nix WA, Oberste MS, Pallansch MA. Sensitive seminested PCR amplification of VP1 sequences for direct identification of all Enterovirus serotypes from original clinical specimens. J Clin Microb. 2006;44:2698–2704. doi: 10.1128/JCM.00542-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee C, Lee S, Han E, Kim S. Use of cell culture-PCR assay based on combination of A549 and BGMK cell lines and molecular identification as a tool to monitor infectious adenoviruses and Enteroviruses in river water. Appl Environ Microbiol. 2004;70:6695–6705. doi: 10.1128/AEM.70.11.6695-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Traore O, Arnal C, Mignotte B, Maul A, Laveran H, et al. Reverse transcriptase PCR detection of astrovirus, Hepatitis A virus and Poliovirus in experimentally contaminated mussels: comparison of several extraction and concentration method. App Environ Microb. 1998;64:3118–3122. doi: 10.1128/aem.64.8.3118-3122.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grabow WOK, Botma KL, Villiers JC, Clay CG, Erasmus B. Assessment of cell culture and polymerase chain reaction procedures for the detection of Polioviruses in wastewater. Bul WHO. 1999;12:23–24. [PMC free article] [PubMed] [Google Scholar]
- 26.Lee HK, Jeong YS. Comparison of total culturable virus assay and multiplex integrated cell culture-PCR for reliability of waterborne virus detection. App Environ Microb. 2004;70:3632–3636. doi: 10.1128/AEM.70.6.3632-3636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Straub T, Pepper IL, Gerba A. Comparison of PCR and cell culture for detection of Enteroviruses in sludgeamended field soils and determination of their transport. AppEnviron Microb. 1995;61:2066–2068. doi: 10.1128/aem.61.5.2066-2068.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khelifi H, Belghith K, Aouni M. Comparison of cell culture and RT-PCR for the detection of Enterovirus in sewage and shellfish. Pathol Biol. 2006;54:280–284. doi: 10.1016/j.patbio.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Buck GE, Wiesemann M, Stewart L. Comparison of mixed cell culture containing genetically engineered BGMK and CaCo-2 cells (Super E-Mix) with RT-PCR and conventional cell culture for the diagnosis of Enterovirus meningitis. J Clin Virol. 2002;25:13–18. doi: 10.1016/s1386-6532(02)00029-x. [DOI] [PubMed] [Google Scholar]
- 30.Kim YH, Yang I, Bae Y, Park S. Performance evaluation of thermal cyclers for PCR in a rapid cycling condition. Bio Techniques. 2008;44:495–505. doi: 10.2144/000112705. [DOI] [PubMed] [Google Scholar]
- 31.Maekawa M, Sudo K, Kanno T. Search for improved electrophoretic conditions for PCR–singlestrand conformation polymorphism analysis: is an SDS buffer condition useful. Genome Res. 1993;3:130–132. doi: 10.1101/gr.3.2.130. [DOI] [PubMed] [Google Scholar]
- 32.Beaulieux F, Berger MM, Tcheng R, Giraud P, Lina B. RNA extraction and RT-PCR procedures adapted for the detection of Enterovirus sequences from frozen and paraffin-embedded formalin-fixed spinal cord samples. J ViroMethods. 2003;107:115–120. doi: 10.1016/s0166-0934(02)00209-4. [DOI] [PubMed] [Google Scholar]