Abstract
Background
Leishmaniasis is a protozoan disease cause by Leishmania genus. Anthroponotic and zoonotic cutaneous leishmaniasis are endemic in Iran. The aim of this study was to identify the causative agent of cutaneous leishmaniasis by mini-exon gene in five regions of Khuzestan Province, southwest of Iran.
Methods
From 2007 to 2008 in this cross-sectional study, cutaneous samples were collected from patients referred to Health Centers and Hospitals of the Khuzestan Province for cutaneous leishmaniasis diagnosis and cultured in Novy-MacNeal-Nicolle (NNN) and RPMI 1640. The propagated promastigotes were harvested and Leishmania species of cutaneous leishmaniasis were identified by RFLP and DNA sequencing of the PCR generated fragments.
Results
L. major and L. tropica were the causative agents of cutaneous leishmaniasis by predominantly of L. major species. The alignment of the mini-exon sequencing isolates with reported sequencing of L. major and L. tropica revealed 92%-99% identity.
Conclusion
Our study showed that mini-exon PCR-RFLP was useful method to identify the causative species of cutaneous leishmaniasis.
Keywords: Cutaneous leishmaniasis, Mmini-exon, RFLP, Sequencing, Iran
Introduction
Leishmaniasis has a variety of clinical manifestations ranging from self-curing cutaneous lesions to fatal visceral form that is caused by parasitic protozoan Leishmania genus. Leishmaniasis is endemic in many parts of the world and remains a serious public health problem. Two species of Leishmania are involved in cutaneous leishmaniasis (CL) in Iran. L. tropica and L. major are the causative agents of anthroponotic cutaneous leishmaniasis (ACL) and zoonotic cutaneous leishmaniasis (ZCL), respectively (1). Based on conducted studies, Khuzestan Province is one of the important endemic areas of CL in Iran (2–5). In Iran–Iraq war lasting from September 1980 to August 1988, thousands of ZCL cases appeared among soldiers and paramilitary men who were sent to the war front in the south-west in the first two years (6). Correct identification of the Leishmania species is important for appropriate species-specific therapeutic as well as epidemiologic and genetic studies (7).
In the past decade, DNA-based methods have been used for diagnosis and identification of Leishmania species (8–11). Detection of the Leishmania parasites by PCR methods is highly sensitive and specific (12). Identifying Leishmania isolates using kinetoplast sequences as a target revealed extensive intraspecific polymorphism among strains of one species hence this method is less reliable (13). Among targets applied in DNA-based methods, mini-exon is highly specific and sensitive because the gene is present in all Kinetoplastida, whereas absent from the vertebrate host or invertebrate vector. Moreover, different Leishmania species have distinct length of the non-transcribed intergenic spacer region (14, 15).
Prior to this work, the majority of DNA-based molecular studies in Iran used internal transcribed spacer (ITS) in the ribosomal operons and kinetoplast DNA, such as minicircle sequences (2, 3) (16–22). To our knowledge, there have been no studies based on mini-exon gene in Iran. Consequently, we applied mini-exon PCR-RFLP and sequencing tools for identification of Leishmania species, in Khuzestan Province.
Materials and Methods
Samples collection
From August 2007 to May 2008 in a cross-sectional study, 128 cutaneous samples were collected from patients referred to Health Centers and Hospitals of the Khuzestan Province for cutaneous leishmaniasis diagnosis. The patients had given their informed consent for including their samples in our study. Geographically the Khuzestan Province is in the southwest of Iran, bordering Iraq's Basra Province, and the Persian Gulf. Its capital is Ahvaz and covers an area of 63,238 km2. The province has an estimated population of 4,277,998 inhabitants.
Five regions of the province were considered for sampling; North (Shush), South (Hendijan), West (Dashteazadegan: Susangerd, Hovizeh, Bostan), East (Ramhormoz) and Center (Ahvaz).
For each patient, a questionnaire including gender, age, patient location, and lesion type information was filled and then three samples were taken by scraping the internal border of skin lesions with a surgical blade.
Specimens were used for microscopical examination, culture in RPMI 1640 medium supplemented with 5% fetal bovine serum (FBS) and 200 IU/ml penicillin-streptomycin and culture in NNN (Novy-MacNeal-Nicolle) medium. Primary Leishmania isolates were subcultured in RPMI 1640 media with 10% FBS. Culture tubes were incubated at 25°C. The cultures were checked for promastigotes every 3 days for 4 weeks.
DNA extraction
The mass cultured promastigotes were harvested by centrifugation (4000 rpm at 4°C for 15min) and washed three times in cold sterile PBS (pH 7.2). DNA was extracted by QIA DNeasy blood and tissue kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. Purified DNA was eluted in 100 µl of elution buffer and stored in −20°C until use. PCR control DNA preparations were also extracted from L. major MHOM/IR/75/ER, L. tropica MHOM/IR/02/Mash 10 and L. infantum MCAN/IR/97/LON 49 (Iranian standard strains).
PCR amplification
Amplification of the mini-exon gene was performed as a single PCR with forward (5'-TATTGGTATGCGAAACTTCCG-3') and reverse (5'-ACAGAAACTGATACTTATATAGCG-3') primers as described before (23). Two-seven µl of template DNA (75-100 ng/reaction) were amplified in 20 µl of modified Taq DNA polymerase Master Mix RED reaction (Bioneer Korea) containing 75 mM Tris-HCl (PH 8.5), 20 mM (NH 4)2SO4, 1.5 mM MgCl2, 0.1% Tween20, 0.2 mM dNTPs, 0.25 unit Amplicon Taq DNA polymerase, 12% DMSO (78.13 g/mol Cinnagen Iran), 10 pM of each primer, inert red dye and a stabilizer.
DNA was amplified using thermal cycler (Eppendorf AG 22331, Hamburg, Germany) under the following conditions: 5 min at 94°C followed by 35 cycles of 30 sec at 94°C, 30 sec at 51.5°C, 45 sec at 72°C and a final elongation at 72°C for 10 min. For each sample, one positive control and one negative control were included.
The PCR products were separated on a 1.5% (W/V) agarose gel and visualized by staining with ethidium bromide.
RFLP and sequencing
Fifteen µl of the PCR products were digested with 1.5 µl of Eae I (Fermentase life science, Germany) or 1.5 µl of Hae III (Takara Bio Inc, Japan) at 37°C for 2h (23). Digestion products were separated by gel electrophoresis in 2.5% agarose and visualized with ethidium bromide staining.Three Iranian reference strains and randomly 14 of 60 samples from five different parts of the province were selected based on the RFLP results. The PCR products of 17 samples were purified using an Accuprep Gel Purification kit (Bioneer, Deajeon, Korea) then sequenced (MWG-Biotech, Ebersberg, Germany) by the primers employed in the PCR.
Sequence alignments were constructed using the program CLUSTAL W version 1.83 (http://www.ddbj.nig.ac.jp/search/clustalw-e.html).
Results
One hundred twenty eight patients with wet and dry skin lesions were enrolled in the study; 58 were male and 70 female. The median age was 11.25 years (range, 3 months to 75 years).
Only 60 (47%) culture tubes grew promastigotes after incubation at 25°C.
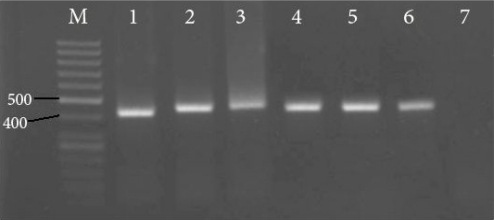
The PCR products of L. tropica, L. major and L. infantum reference strains and samples were around 410–440 bp (Fig. 1).
Fig. 1.
PCR products of Iranian reference strains and samples on 1.5% agarose gel. M: 50 bp molecular weight marker; lane 1: Leishmania tropica (MHOM/IR/02/Mash10); lane 2: L. major (MHOM/IR/75/ER); lane 3: L. infantum (MCAN/IR/97/LON 49); lanes 4-6: samples; lane 7: negative control (Distilled water instead of DNA template).
The PCR products of 60 isolates and 3 Iranian reference strains were purified and digested by Eae I restriction enzyme.
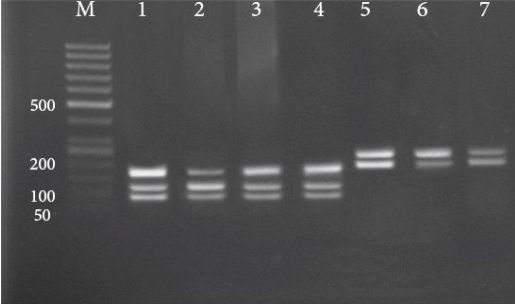
RFLP analysis of most L. major samples and of L. major reference strain showed DNA fragments of 175, 120 and 94 bp (Fig. 2). The purified PCR products of both L. tropica samples and of the L. tropica reference strain however were failed to digest by Eae I restriction enzyme, even after overnight incubation at 37°C. Subsequently, these samples were digested using Hae III restriction enzyme and produced two fragments around 230 and 180 bp after incubation for 2h at 37°C (Fig. 2). Totally, based on RFLP profiles two different Leishmania species, L. major (96.6%) and L. tropica (3.4%), were identified from 60 samples. The two L. tropica species were belonged to Ahvaz and Dashteazadegan cities. To confirm the RFLP results, the amplified mini-exon gene of 14 isolates and 3 Iranian reference strains were sequenced. The results were submitted to DDBJ/Genbank at accession nos. AB470338, AB470339 and AB465575 for Iranian reference strains of L. tropica, L. infantum and L. major, respectively and AB494686-AB494697 (L. major) and AB494698-AB494699 (L. tropica) for the samples.
Fig. 2.
RFLP patterns of Leishmania species after digestion with Eae I (for L. major) and Hae III (for L. tropica) on 2.5% agarose gel. M: 50 bp molecular weight marker; lane 1: L. major (reference HOM/IR/75/ER); lane 2: L. major (center region); lane 3: L. major (south region); lane 4: L. major (west region); lane 5: L. tropica (reference MHOM /IR/02/Mash10); lane 6: L. tropica (center region); lane 7: L. tropica (west region)
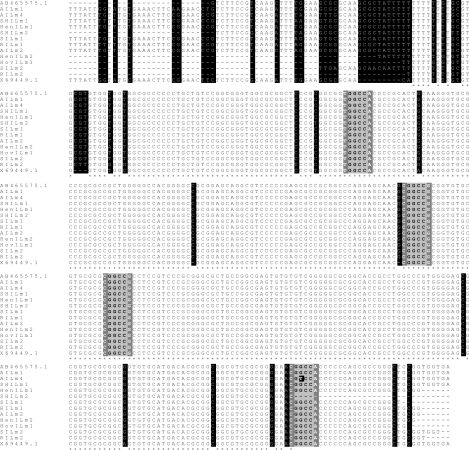
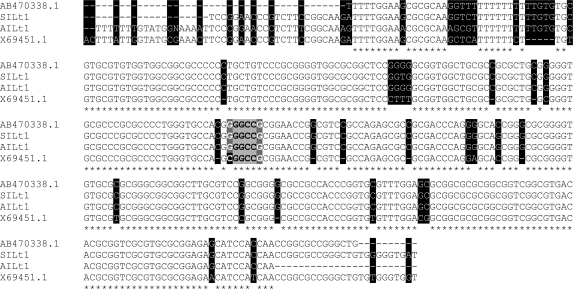
The nucleotide sequences of mini-exon gene from isolates were aligned with nucleotide sequences of L. major (GenBank accession nos. X69449.1 and AB465575) (Fig. 3) and L. tropica (GenBank accession Nos. X69451.1 and AB470338) (Fig. 4). The Fig. 3 showed that L. major, which isolated from samples showed 2 to 8 (97%-99% homology) and 13 to 35 (92%-97% homology) nucleotide substitutions as compared to Iranian reference strain and Israeli isolate (X69449.1), respectively (Fig. 3). Furthermore, the Fig. 4 showed comparison substituted nucleotides of two L. tropica isolated with Iranian reference strain (4 and 6) by 97% and 98% identity and Sudanese isolate (AB470338)(34 and 46) by 88% and 91% homology.
Fig. 3.
Multiple alignments of nucleotide sequences of mini-exon gene from 12 isolates, Iranian L. major reference strain (AB465575.1) and Israeli isolate (X69449.1). L. major isolates sequences are AILm1: AB494695; AILm4: AB494690; SHILm1: AB494693; HenILm1: AB494688; SHILm2: AB494696; SILm1: AB494686; RILm1: AB494687; AILm2: AB494689; HenILm2: AB494692; HovILm1: AB494694; SILm2: AB494697; RILm2: AB494691. Dashes indicate computer-generated gaps. Asterisks indicate identical nucleotides. Substitutions are shaded in black and the Eae I restriction sites are highlighted by gray boxes.
Fig. 4.
Multiple alignments of nucleotide sequences of mini-exon gene from 2 isolates, Iranian L. tropica reference strain (AB470338.1) and Sudanese isolate (X69451.1). L. tropica isolates sequences are SILt1: AB494699; AILt1: AB494698. Dashes indicate computer-generated gaps. Asterisks indicate identical nucleotides. Substitutions are shaded in black and the Eae I recognition site (5'-Py▼GGCCPu-3') are highlighted by gray boxes, dark gray boxes show Pyrimidine (C, T and U) and Purine (A and G) nucleotides where Guanine substitution instead of Cytosine in Iranian L. tropica are tinted with black.
Discussion
Morphological discrimination of Leishmania species is not usually possible. The isoenzyme electrophoresis method requires cultured promastigotes but culture is time consuming, labor intensive and its sensitivity is suboptimal, as shown in our own study where only 47% of strains cultured grew promastigotes (24, 25). Over the last few years, several DNA-based molecular assays have been developed for species identification (26–28).
In the present study, we report the first application of mini-exon PCR and sequencing in order to characterize the Leishmania species causing cutaneous leishmaniasis in Khuzestan Province of Iran. The results showed that both L. major and L. tropica were occurring in this endemic area, L. major being by far the most prevalent. These findings are in agreement with previous studies. Maraghi et al. showed that 90% of cutaneous leishmaniasis isolates were L. major and the remaining were L. tropica (2), in Tashkori et al. (3) and Moatazadian et al. (4) studies, all cutaneous leishmaniasis isolates from Khuzestan Province were identified as L. major.
The mini-exon amplicon fragments yielded by both L. tropica and L. major were of 410-440 pb. Our findings are similar to those reported by Fernandes et al. on old world dermotropic and viscerotropic Leishmania species (14). Hence, mini-exon PCR is not discriminatory enough and for an accurate characterization, additional methods are needed. Our results suggest that mini-exon PCR-RFLP and sequencing to be very adequate for this purpose.
According to Manfurt et al., it is expected that all Leishmania species of the subgenus Leishmania can be distinguished by mini-exon PCR-RFLP, with Eae I being the most informative restriction enzyme (29). However, in both L. tropica Iranian samples and the reference L. tropica strain used in our study, Eae I failed to cut the mini-exon PCR products. The sequencing analyses revealed that our observation was due to one nucleotide substitution exactly in the Eae I recognition site. The nucleotide sequences alignment of Iranian L. tropica reference and isolates with Sudanica L. tropica isolate demonstrated Guanine (G) substitution instead of Cytosine (C) nucleotide in Iranian L. tropica causing this ineffectiveness of Eae I (Fig. 4).
Multiple alignments of mini-exon gene nucleotide sequences from isolates, Iranian reference strain and other sequences reported in GenBank revealed some nucleotides substitutions (Fig. 3 and 4). Despite these variations in mini-exon gene, Eae I restriction enzyme RFLP scheme in Iranian L. major reference and isolates were identical to those reported by Manfurt et al. (29) except in one isolate (AB494690) that substitution in restriction site altered the number of the fragments generated.
Therefore, in order to obtain unequivocal RFLP results, digestion of PCR products with Hae III along with Eae I is recommended.
In conclusion, our finding confirms that L. major is the predominant causative agent of cutaneous leishmaniasis in Khuzestan Province and mini-exon PCR-RFLP is a very practical method to identify the causative species of cutaneous leishmaniasis; and would therefore be very useful for identification of Leishmania in both vectors and reservoir hosts for further epidemiological studies in this endemic region.
Acknowledgements
The authors would like to thank Dr. H. Hajjaran and Dr. R. Hadighi to kindly providing us Leishmania reference strains, Dr. Shoshizadeh, Dr. Obaidavi, Mr. Rashti, Mr. Meripur, Mr. Rodbari, Mr. Sadri, Mrs. Moradi, Mrs. Amiri and all managers and staff of the district Health Centers and Hospitals in Khuzestan Province for their precious collaboration. This work was supported by the grants of Iran University of Medical Sciences, with code No P-446. The authors declare that they have no conflicts of interest.
References
- 1.Nadim A, Seyedi-Rashti MA. A brief review of the epidemiology of various types of leishmaniasis in Iran. Acta Med Iran. 1971;8:99–106. [Google Scholar]
- 2.Maraghi S, Samarbaf Zadeh A, Sarlak AA, Ghasemian M, Vazirianzadeh B. Identification of cutaneous leishmaniasis agents by nested polymerase chain reaction (Nested-PCR) in Shush city, Khuzestan Province, Iran. Iranian J Parasitol. 2007;2(3):13–5. [Google Scholar]
- 3.Tashakori M, Kuhls K, Al-Jawabreh A, Mauricio IL, Schönian G, Farajnia S, et al. Leishmania major: genetic heterogeneity of Iranian isolates by single-strand conformation polymorphism and sequence analysis of ribosomal DNA internal transcribed spacer. Acta Trop. 2006;98(1):52–8. doi: 10.1016/j.actatropica.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 4.Motazedian MH, Nomanpoor B, Ardehali S. Characterization of Leishmania parasites isolated from provinces of the Islamic Republic of Iran. East Mediterr Health J. 2002;8:2–3. [PubMed] [Google Scholar]
- 5.Javadian E, Mesghali A. Studies on cutaneous leishmaniasis in Khuzestan, Iran. Part I. The leptomonad infection of sandflies. Bull Soc Pathol Exot Filiales. 1974;67(5):513–6. [PubMed] [Google Scholar]
- 6.Nadim A, Javadian E, Mohebali M. The experience of leishmanization in the Islamic Republic of Iran. East Mediterr Health J. 1997;3(2):284–9. [Google Scholar]
- 7.NOA ET. Molecular approaches to direct diagnosis and characterization of Leishmania donovani in clinical samples. M Sc Biologie. 2004:31–70. [Google Scholar]
- 8.Oliveira JG, Novais FO, de Oliveira CI, da Cruz AC, Junior, Campos LF, da Rocha AV, et al. Polymerase chain reaction (PCR) is highly sensitive for diagnosis of mucosal leishmaniasis. Acta trop. 2005;94(1):55–9. doi: 10.1016/j.actatropica.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Degrave W, Fernandes O, Campbell D, Bozza M, Lopes U. Use of molecular probes and PCR for detection and typing of Leishmania-a mini-review. Mem Inst Oswaldo Cruz. 1994;89(3):463–9. doi: 10.1590/s0074-02761994000300032. [DOI] [PubMed] [Google Scholar]
- 10.Chargui N, Bastien P, Kallel K, Haouas N, Akrout FM, Masmoudi A, et al. Usefulness of PCR in the diagnosis of cutaneous leishmaniasis in Tunisia. Trans R Soc Trop Med Hyg. 2005;99(10):762–8. doi: 10.1016/j.trstmh.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Wilson SM. DNA-based methods in the detection of Leishmania parasites: field applications and practicalities. Ann Trop Med Parasitol. 1995;89(Suppl 1):95–100. [PubMed] [Google Scholar]
- 12.Garcia AL, Kindt A, Quispe-Tintaya KW, Bermudez H, Llanos A, Arevalo J, et al. American tegumentary leishmaniasis: antigen-gene polymorphism, taxonomy and clinical pleomorphism. Infect Genet Evol. 2005;5(2):109–16. doi: 10.1016/j.meegid.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Rogers WO, Wirth DF. Kinetoplast DNA minicircles: regions of extensive sequence divergence. Proc Natl Acad Sci U S A. 1987;84(2):565–9. doi: 10.1073/pnas.84.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandes O, Murthy VK, Kurath U, Degrave WM, Campbell DA. Mini-exon gene variation in human pathogenic Leishmania species. Mol Biochem Parasitol. 1994;66(2):261–71. doi: 10.1016/0166-6851(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 15.Hassan MQ, Ghosh A, Ghosh SS, Gupta M, Basu D, Mallik KK, et al. Enzymatic amplification of mini-exon-derived RNA gene spacers of Leishmania donovani: primers and probes for DNA diagnosis. Parasitology. 1993;107(Pt 5):509–17. doi: 10.1017/s0031182000068086. [DOI] [PubMed] [Google Scholar]
- 16.Baghaei M, Mehdizadeh M, Afarid M, Attarzadeh A, Farzin D, Hosseini SH, et al. Intraspecific variation in Leishmania major isolated from different forms of zoonotic cutaneous Leishmaniasis. Iran J Med Sci. 2005;30(2):51–4. [Google Scholar]
- 17.Fazaeli A, Fouladi B, Sharifi I. Emergence of cutaneous leishmaniasis in a border area at south-east of Iran: an epidemiological survey. J Vector Borne Dis. 2009;46(1):36–42. [PubMed] [Google Scholar]
- 18.Karamian M, Motazedian MH, Fakhar M, Pakshir K, Jowkar F, Rezanezhad H. Atypical presentation of Old-World cutaneous leishmaniasis, diagnosis and species identification by PCR. J Eur Acad Dermatol Venereol. 2008;22(8):958–62. doi: 10.1111/j.1468-3083.2008.02674.x. [DOI] [PubMed] [Google Scholar]
- 19.Mahboudi F, Abolhassani M, Tehrani SR, Azimi M, Asmar M. Differentiation of old and new world Leishmania species at complex and species levels by PCR. Scand J Infect Dis. 2002;34(10):756–8. doi: 10.1080/0036554021000026930. [DOI] [PubMed] [Google Scholar]
- 20.Safaei A, Motazedian MH, Vasei M. Polymerase chain reaction for diagnosis of cutaneous leishmaniasis in histologically positive, suspicious and negative skin biopsies. Dermatology. 2002;205(1):18–24. doi: 10.1159/000063150. [DOI] [PubMed] [Google Scholar]
- 21.Parvizi P, Ready PD. Nested PCRs and sequencing of nuclear ITS-rDNA fragments detect three Leishmania species of gerbils in sandflies from Iranian foci of zoonotic cutaneous leishmaniasis. Trop Med Int Health. 2008;13(9):1159–71. doi: 10.1111/j.1365-3156.2008.02121.x. [DOI] [PubMed] [Google Scholar]
- 22.Alborzi A, Pourabbas B, Shahian F, Mardaneh J, Pouladfar GR, Ziyaeyan M. Detection of Leishmania infantum kinetoplast DNA in the whole blood of asymptomatic individuals by PCR-ELISA and comparison with other infection markers in endemic areas, southern Iran. Am J Trop Med Hyg. 2008;79(6):839–42. [PubMed] [Google Scholar]
- 23.Marfurt J, Nasereddin A, Niederwieser I, Jaffe CL, Beck HP, Felger I. Identification and differentiation of Leishmania species in clinical samples by PCR amplification of the miniexon sequence and subsequent restriction fragment length polymorphism analysis. J Clin Microbiol. 2003;41(7):3147–53. doi: 10.1128/JCM.41.7.3147-3153.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boggild AK, Miranda-Verastegui C, Espinosa D, Arevalo J, Adaui V, Tulliano G, et al. Evaluation of a microculture method for isolation of Leishmania parasites from cutaneous lesions of patients in Peru. J Clin Microbiol. 2007;45(11):3680–4. doi: 10.1128/JCM.01286-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravel S, Cuny G, Reynes J, Veas F. A highly sensitive and rapid procedure for direct PCR detection of Leishmania infantum within human peripheral blood mononuclear cells. Acta Trop. 1995;59(3):187–96. doi: 10.1016/0001-706x(95)00079-t. [DOI] [PubMed] [Google Scholar]
- 26.Berzunza-Cruz M, Cabrera N, Crippa-Rossi M, Sosa Cabrera T, Perez-Montfort R, Becker I. Polymorphism analysis of the internal transcribed spacer and small subunit of ribosomal RNA genes of Leishmania mexicana . Parasitol Res. 2002;88(10):918–25. doi: 10.1007/s00436-002-0672-x. [DOI] [PubMed] [Google Scholar]
- 27.Cupolillo E, Brahim LR, Toaldo CB, de Oliveira-Neto MP, de Brito ME, Falqueto A, et al. Genetic polymorphism and molecular epidemiology of Leishmania (Viannia) braziliensis from different hosts and geographic areas in Brazil. J Clin Microbiol. 2003;41(7):3126–32. doi: 10.1128/JCM.41.7.3126-3132.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smyth AJ, Ghosh A, Hassan MQ, Basu D, De Bruijn MH, Adhya S, et al. Rapid and sensitive detection of Leishmania kinetoplast DNA from spleen and blood samples of kala-azar patients. Parasitology. 1992;105(Pt 2):183–92. doi: 10.1017/s0031182000074096. [DOI] [PubMed] [Google Scholar]
- 29.Marfurt J, Niederwieser I, Makia ND, Beck HP, Felger I. Diagnostic genotyping of Old and New World Leishmania species by PCR-RFLP. Diagn Microbiol Infect Dis. 2003;46(2):115–24. doi: 10.1016/s0732-8893(03)00040-3. [DOI] [PubMed] [Google Scholar]