Abstract
Background
Coccidiosis is an intestinal disease of chickens caused by various species of protozoan parasites within the genus Eimeria. Diagnosis and genetic characterization of different species of Eimeria are central to the prevention, surveillance, and control of coccidiosis. The aim of this study was to detect different chicken Eimeria species from several areas in Khuzestan, southwest Iran.
Methods
From February to September 2008, PCR assay as well as parasitological examinations was applied for the identification of field isolates of Eimeria parasites around Ahvaz, center of Khuzestan, southwest Iran. Data were analyzed by the Kappa statistic test.
Results
Eimeria maxima, E. necatrix, E. tenella, E. acervulina and E. mitis were detected in this study. The prevalence of Eimeria spp. was 31.5% (126 of 400) and E. tenella was the most prevalent species in Khuzestan. Based on the Kappa statistical test, a good correlation between the results of PCR and traditional biometrical methods was only observed for E. maxima.
Conclusion
The present study is the first on the prevalence of Eimeria species in Khuzestan, based on the molecular findings. We believe that traditional methods are not sufficiently reliable for specific diagnosis of Eimeria species in chickens and PCR based amplification of DNA sequence of parasite, could resolve this problem.
Keywords: Eimeria, Poultry coccidiosis, PCR, ITS1, Microscopic examinations, Iran
Introduction
Protozoan parasites of the genus Eimeria (Coccidia: Eimeriidae) are highly successful organisms which inhabit and multiplies in the intestinal tract. These parasites cause chickens coccidiosis, an enteric disease of major economic importance worldwide (1). Economic importance of the disease is due to production losses and high morbidity resulting from an acute, bloody enteritis and mortality rates (2). However, intestinal lesions of the infection vary, depending on the species of coccidian. About 1800 Eimeria spp. affect the intestinal mucosa of different mammals and birds, but seven species of Eimeria including E. tenella, E. necatrix, E. acervulina, E. maxima, E. brunetti, E. mitis, and E. praecox are the causative agents of coccidiosis in chickens (3).
Diagnosis of coccidiosis is based on clinical features and gut pathology of host, parasite characteristics such as morphology at different stages of parasitism, and the pre-patent period (4, 5). Analysis of these characteristics is labor intensive for diagnosis and does not provide accurate data for identification of the Eimeria species (6).
Identification and genetic characterization of different species of Eimeria genus are central to prevention, surveillance, and control of coccidiosis. This is particularly important with regard to the appearance of a widespread anticoccidial resistance of Eimeria species and the problems associated with drug residues.
Due to difficulties in the morphologic identification of some of chicken Eimeria spp., diagnostic laboratories are increasingly utilizing DNA-based technologies for the specific identification of the parasite (7).
So far, there is limited knowledge on the epidemiology of Eimeria infections under different rearing conditions in Iran. In The present study, together with morphometric diagnosis, PCR assay, based on the amplification of internal transcribed spacer 1 (ITS1) regions of ribosomal DNA (8) was used for identification of chicken Eimeria species in Khuzestan Province, southwest Iran.
Materials and Methods
Sample collection preparation
From February to September 2008, 400 samples of fresh fecal droppings were collected by cluster sampling from 40 broiler chickens flocks without previous exposure to anticoccidial vaccines. The sampled flocks were located in different areas within a radius of 20 to 100 km from the center of Khuzestan Province. Samples of each flock were collected from different locations in the poultry house. One hundred samples, containing a high number of Eimeria and representing all areas of the province were selected and preserved in 2.5% potassium dichromate at 28°C.
After sporulation of oocysts, potassium dichromate was removed from the master stock by repeated centrifugation and resuspension in water. Oocysts present in feces were then purified by a saturated saline solution and washed by centrifugation at 1000 x g for 5 min with distilled water. The sediment containing oocysts was resuspended in phosphate buffered saline (PBS) pH 7.4 with 0.1% Tween 80 and homogenized by vortexing and subsequent centrifugation at 800 x g for 5 min. Thereafter, a 6% solution of sodium hypochlorite was added, 20 minutes incubation was performed at 4°C and the oocysts were finally washed 4 times, with distilled water (6).
Parsitologic examination
A modified saturated salt flotation technique was used to isolate oocysts for length measurements using a calibrated ocular micrometer at 400x magnification (4). Fifty random oocysts from each sample were examined by measuring their length and width with light microscopy, armed with calibrate ocular lens as well as determination of the oocysts shape and index (Length/Width). For more accurate diagnosis, sporocysts’ diameters were also determined (9).
PCR protocol
Sporozoites were released from oocysts and sporocysts wall by sonication and DNA was extracted from sporozoites by Stool Mini Kit extraction (Qiagen) according to manufacturer's recommendation. Single PCR assays targeting ITS-1 regions of ribosomal DNA were performed for each of the seven chicken Eimeria species. Forward and reverse species-specific primer sequences used in this study have been reported previously by Haug et al. 2007 (10) (Table 1). Amplification of the ITS-1 sequences of genomic rDNA was carried out in 20 µl reaction volumes containing 2 µl of DNA template, 8 pmol of reverse and forward primers, 3.0 mM MgCl2, 2.0 µl 10 X PCR Buffer, 200 µM of each dNTP and 0.4 U Taq DNA Polymerase. Thermal program of PCR was as follows: denaturation step at 95°C for 5 min, 35 cycles of denaturation at 95°C for 30 s, annealing at 58°C or 65°C for 30 s and extension at 72°C for 1 min. A final prolonged extension step at 72°C for 3 min completed the PCR process. A commercial vaccine (Paracox® 8, UK) which included all the eight pathogenic Eimeria species of chicken was used for positive controls and fecal samples without any oocyst examined by floatation method used as negative control in PCR. To verify the results, 10 µl of each PCR product was electrophoresed in a 1.5% agarose gel, stained with ethidium bromide, and visualized on a UV transilluminator. The PCR products were identified by size using a 100 base pair ladder.
Table 1.
Sequences of the PCR primer pairs used (10)
| Eimeria species | Amplicon size (bp) | Primer sequence 5′ to 3′ |
|---|---|---|
| E. maxima | 205 | 5′-GTGGGACTGTGGTGATGGGG-3′ 5′-ACCAGCATGCGCTCACAACCC-3'′ |
| E. acevulina | 145 | 5′-GGGCTTGGATGATGTTTGCTG-3′ 5′-GCAATGATGCTTGCACAGTCAGG-3′ |
| E. brunetti | 183 | 5′-CTGGGGCTGCAGCGACAGGG-3′ 5′-ATCGATGGCCCCATCCCGCAT-3′ |
| E.mitis | 330 | 5′-GTTTATTTCCTGTCGTCGTCTCGC-3′ 5′-GTATGCAAGAGAGAATCGGGATTCC-3′ |
| E. praecox | 215 | 5′-CATCGGAATGGCTTTTTGAAAGCG-3′ 5′-GCATGCGCTAACAACTCCCCTT-3′ |
| E. tenella | 278 | 5′-AATTTAGTCCATCGCAACCCTTG-3′ 5′-CGAGCGCTCTGCATACGACA-3′ |
| E. necatrix | 160 | 5′-AGTATGGGCGTGAGCATGGAG-3′ 5′-GATCAGTCTCATCATAATTCTCGCG-3′ |
Statistical analysis
The Kappa statistic test (κ) was used to test the level of agreement between the PCR and the parsitologic examination for the detection of Eimeria species. Kappa and its 95% Confidence Interval (CI), was used further to measure the degree of agreement between the two procedures after taking into account the probability of agreement by chance alone. Strength of agreement based on κ was judged according to the following guidelines: <0.2=slight; 0.2–0.4=fair; 0.4– 0.6=moderate; 0.6–0.8=good; >0.8=very good (11).
Results
Out of 400 specimens collected, 126 (31.5%) fecal droppings were positive for oocysts of Eimeria spp. in parasitological examination (E. tenella=31%, E. maxima=24.6%, E. acervulina=23%, E. mitis=12.7% and 8.73%).
The data showed that E. tenella, E. necatrix, E. acervulina, E. mitis and E. maxima exist in poultry farms of Khuzestan by both molecular examination, using species-specific PCR (Fig. 1 and 2) and parasitological methods. E. tenella was the most prevalent species and E. brunette and E. praecox were not detected at all. PCR analyses of 100 collected samples showed that all the identified species were present in different areas, around the center of Khuzestan Province.
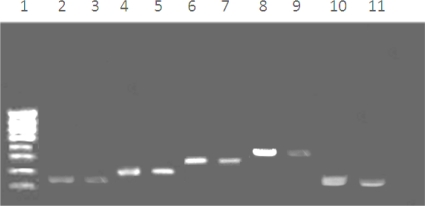
Fig. 1.
Agarose gel electrophoresis of Eimeria species-specific PCR products. Polymerase chain reaction based on amplification of the species-specific ITS-1 sequences of the genomic rDNA Positive reactions: E. acervulina (145bp) (2-Control, 3-Sample), E. maxima (205 bp) (4-Control, 5-Sample), E. tenella (278 bp) (6-Control, 7-Sample), E. mitis(330bp)(8-Control, 9-Sample) and E. necatrix(160bp)(10-Control, 11-Sample). 1=100 bp ladder
Statistical analyses using Kappa test revealed a poor agreement between PCR and the traditional biometrical identification for diagnosis of detected Eimeria species, except for E. maxima and to some extent for E. acervulina (κ=0.35, 030, 0.56, 0.22 and 0.84 for E. tenella, E. necatrix, E. acervulina, E.mitis and E. maxima respectively).
Discussion
The specific diagnosis of Eimeria infections in chickens is clearly central to a better understanding of epidemiology and dynamics of the disease in intensive and extensive chicken establishments. This is particularly important for planning an effective prevention and control program of coccidiosis. Traditionally, diagnosis has been achieved by detecting Eimeria oocysts excreted in the feces of chickens by measuring oocyst and sporocyst dimensions or assessing the site and extent of the pathological lesions in the intestine of chickens. Although the microscopic examinations can absolutely show the negative fecal samples, such traditional methods have generally had major limitations in the specific diagnosis of coccidiosis and identification of Eimeria species. These approaches are unreliable, particularly when multiple species of Eimeria simultaneously infect a single host and there is overlap in the size and shape of oocysts and the sites of infection in the intestines (5).
During recent years, there have been significant advances in the development of molecular-diagnostic tools. Several PCR based assays targeting different regions of the Eimeria genome have been described, such as the 5S rRNA (, the small subunit rRNA (12, 13), the sporozoite antigen gene EASZ240/160 (14) and ITS-1 (8, 15–17) and ITS-2 (18–20) genomic regions. Since the ITS regions are less conserved than the rRNA genes, detecting variations in this region of DNA sequence, makes the design of primers straightforward and reduces the risk of cross reactions among different species (21). Apart from an accurate identification of Eimeria species, molecular methods can also be helpful in epidemiological study of the parasite, an aspect that has been less investigated to date.
At yet, there has not been any documentary report related to the occurrence and epidemiological pattern of the pathogenic Eimeria species of domestic chickens, in Khuzestan. Therefore, the results of the present study are the first on the prevalence of Eimeria species in the region, based on the molecular findings.
Out of 400 specimens collected, 126 (31.5%) samples were positive for Eimeria spp. Nowzari et al. in a large study including 5 provinces of Iran showed that E. maxima, E. mitis, E. brunetti, E. tenella and E. acervulina were distributed all over Iran. They identified that E. mitis and E. brunetti for the first time by PCR (22). E. brunetti has been found uncommon in broiler flocks (15). This is in accordance with our finding in Khuzestan by PCR. E. necatrix has also been considered as uncommon in broiler flocks, but we identified this species in 11 samples of seven farms. In our study, E. tenella was the dominant species. This finding suggests that in poor management conditions, poultry houses may encounter acute coccidiosis in Khuzestan due to highly pathogenic species, E. tenella.
Razmi et al, reported that prevalence of subclinical coccidiosis was 38% in Mashad, northeast of Iran and E. acervulina was the most prevalent species in broiler chicken farms (23). In north-west of Iran, Tabriz, five Eimeria spp., E. acervulina, E. tenella, E. necatrix, E. maxima and E. mitis, were identified by morphometric study and E. acervulina was the most prevalent species (23.58%) (24).
Poor evaluation of parsitologic method to identify the Eimeria species has been reported by Anita Haug et al., previously. They reported that PCR and morphometric identification were in complete agreement in only 49% of the cases (25). The present study also showed that there was poor agreement between PCR and traditional identification for diagnosis of Eimeria species. However, there was a good agreement between PCR and morphometric diagnosis only for E. maxima. This finding can be due to the large size of the organism. Hence, as showed by low agreement with Kappa test, traditional methods are not sufficiently reliable for specific diagnosis of Eimeria species in chickens. Moreover, occurrence of multiple infections in a single bird and the fact that, Eimeria species with low oocysts frequency in the mixture maybe missed, indicates that PCR based amplification of DNA sequence of parasite, could resolve this problem and overcame the limitation in analysis of small amounts of oocysts in mixed infections. On the other hand, this protocol can even identify strains of Eimeria species, characterized by different drug-resistance phenotypes (15, 26).
In conclusion, we believe that traditional methods are not sufficiently reliable for specific diagnosis of Eimeria species in chickens and PCR based amplification of DNA sequence of parasite, could resolve this problem.
Acknowledgments
This study was supported by Shahid Chamran University for project number 666, and the authors wish to thank Vice-Chancellor for Research of the Shahid Chamran University for the research. The authors declare that they have no conflicts of interest.
References
- 1.McDougald LR, Reid WM. Coccidiosis. In: Calnek BW, Barnes HJ, Beard CW, McDougald LR, Saif MY, editors. Diseases of poultry. Ames, IA: Iowa State University Press; 1997. pp. 865–883. [Google Scholar]
- 2.Shirley MW, Smith AL, Tomley FM. The biology of avian Eimeria with an emphasis on their control by vaccination. Adv Parasitol. 2005;60:285–330. doi: 10.1016/S0065-308X(05)60005-X. [DOI] [PubMed] [Google Scholar]
- 3.McDougald LR. Protozoal Infections. In: Saif YM, Barnes HJ, Glisson JR, Fadly AM, McDougald LR, Swayne DE, editors. Diseases of poultry. Iowa State University Press; 2003. pp. 973–1023. [Google Scholar]
- 4.Long PL, Reid WM. A guide for the diagnosis of coccidiosis in chickens. 1982;404:1–17. The University of Georgia, College of Agriculture Experiment Stations, Research report. [Google Scholar]
- 5.Long PL, Joyner LP. Problems in the identification of species of Eimeria . J Protozool. 1984;31(4):535–41. doi: 10.1111/j.1550-7408.1984.tb05498.x. [DOI] [PubMed] [Google Scholar]
- 6.Meireles MV, Roberto LO, Riera RF. Identification of Eimeria mitis and Eimeria praecox in Broiler Feces Using Polymerase Chain Reaction. Brazilian J Poult Sci. 2004;6(4):249–252. [Google Scholar]
- 7.Morgan JA, Morris GM, Wlodek BM, Byrnes R, Jenner M, Constantinoiu CC, Anderson GR, Lew-Tabor AE, Molloy JB, Gasser RB, Jorgensen WK. Real-time polymerase chain reaction (PCR) assays for the specific detection and quantification of seven Eimeria species that cause coccidiosis in chickens. Mol Cell Probes. 2009;23(2):83–89. doi: 10.1016/j.mcp.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Schnitzler BE, Thebo P, Mattson JG, Tomley F, Shirley MW. Development of a diagnostic PCR assay for the detection and discrimination of four pathogenic Eimeria species of the chicken. Avian Pathol. 1998;27(5):490–497. doi: 10.1080/03079459808419373. [DOI] [PubMed] [Google Scholar]
- 9.Conwa DP, McKenzie ME. Poultry coccidiosis, Diagnostic and testing procedures. 3th. Blackwell publishing; 2007. pp. 7–11. [Google Scholar]
- 10.Haug A, Thebo P, Mattsson JG. A simplified protocol for molecular identification of Eimeria species in field samples. Vet Parasitol. 2007;146(1–2):35–45. doi: 10.1016/j.vetpar.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 11.Dohoo I, Martin W, Stryhn H. 1st. Charlottetown, Canada: AVC Inc; 2003. Veterinary Epidemiologic Research; pp. 85–120. [Google Scholar]
- 12.Stucki U, Braun R, Roditi I. Eimeria tenella: characterization of a 5S ribosomal RNA repeat unit and its use as a species specific probe. Exp Parasitol. 1993;76(1):68–75. doi: 10.1006/expr.1993.1008. [DOI] [PubMed] [Google Scholar]
- 13.Tsuji N, Kawazu S, Ohta M, Kamio T, Isobe T, Shimura K, Fujisaki K. Discrimination of eight chicken Eimeria species using the two-step polymerase chain reaction. J Parasitol. 1997;83(5):966–970. [PubMed] [Google Scholar]
- 14.Molloy JB, Eaves FW, Jeston PJ, Minchin CM, Stewart NP, Lew AE, Jorgensen WK. Detection of Eimeria acervulina using the polymerase chain reaction. Avian Dis. 1998;42(1):119–123. [PubMed] [Google Scholar]
- 15.Lew AE, Anderson GR, Minchin CM, Jeston PJ, Jorgensen WK. Inter and intra-strain variation and PCR detection of the internal transcribed spacer 1 (ITS-1) sequences of Australian isolates of Eimeria species from chickens. Vet Parasitol. 2003;112(1–2):33–50. doi: 10.1016/s0304-4017(02)00393-x. [DOI] [PubMed] [Google Scholar]
- 16.Schnitzler BE, Thebo P, Tomley F, Uggla A, Shirley MW. PCR identification of chicken Eimeria: a simplified readout. Avian Pathol. 1999;28(1):89–93. doi: 10.1080/03079459995091. [DOI] [PubMed] [Google Scholar]
- 17.Su YC, Fei AC, Tsai FM. Differential diagnosis of five avian Eimeria species by polymerase chain reaction using primers derived from the internal transcribed spacer 1 (ITS-1) sequence. Vet Parasitol. 2003;117(3):221–227. doi: 10.1016/j.vetpar.2003.07.028. [DOI] [PubMed] [Google Scholar]
- 18.Gasser RB, Woods WG, Wood JM, Ashdown L, Richards G, Whithear KG. Automated, fluorescence-based approach for the specific diagnosis of chicken coccidiosis. Electrophoresis. 2001;22(16):3546–3550. doi: 10.1002/1522-2683(200109)22:16<3546::AID-ELPS3546>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 19.Lien YY, Sheu SC, Liu HJ, Chen SC, Tsai MY, Luo SC, Wu KC, Liu SS, Su HY. Cloning and nucleotide sequencing of the second internal transcribed spacer of ribosomal DNA for three species of Eimeria from chickens in Taiwan. Vet J. 2007;173(1):186–191. doi: 10.1016/j.tvjl.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 20.Woods WG, Whithear KG, Richards DG, Anderson GR, Jorgensen WK, Gasser RB. Single-strand restriction fragment length polymorphism analysis of the second internal transcribed spacer (ribosomal DNA) for six species of Eimeria from chickens in Australia. Int J Parasitol. 2000;30(9):1019–1023. doi: 10.1016/s0020-7519(00)00084-9. [DOI] [PubMed] [Google Scholar]
- 21.Holmdahl OJM, Mattsson JG. Rapid and sensitive identification of Neospora caninum by in vitro amplification of the internal transcribed spacer 1. Parasitology. 1996;112(2):177–182. doi: 10.1017/s0031182000084742. [DOI] [PubMed] [Google Scholar]
- 22.Nowzari N, Yakhchali B, Rahbari S, Mouazeni Jula GH. Identification of Eimeria spp. isolated from poultry breeder farms in Iran by PCR. J fac Vet Med. Univ Tehran. 2004;59(2):125–130. [Google Scholar]
- 23.Razmi GR, Kalideri A. Prevalence of subclinical coccidiosis in broiler-chicken farms in the municipality of Mashhad, Khorasan, Iran. Prevent Vet Med. 2000;44(3–4):247–253. doi: 10.1016/s0167-5877(00)00105-7. [DOI] [PubMed] [Google Scholar]
- 24.Nematollahi A, Moghaddam GH, Farshbaf Pourabad R. Prevalence of Eimeria species among broiler chicks in Tabriz (Northwest of Iran) Mun Ent Zool. 2009;4(1):53–58. [Google Scholar]
- 25.Haug A, Gjevre AG, Thebo P, Mattsson JG, Kaldhusdal M. Coccidial infections in commercial broilers: epidemiological aspects and comparison of Eimeria species identification by morphometric and polymerase chain reaction techniques. Avian Pathol. 2008;37(2):161–70. doi: 10.1080/03079450801915130. [DOI] [PubMed] [Google Scholar]
- 26.Nowzari N, Dinparast Djadid N, Rahbari S, Yakchali B, Kazemi B, Moazeni Jula G. Inter and intra-specific genetic variation of avian Eimeria isolated from Iran by random amplified polymorphic DNA- PCR. Vet Parasitol. 2005;128(1–2):59–64. doi: 10.1016/j.vetpar.2004.11.015. [DOI] [PubMed] [Google Scholar]