Abstract
Background
Strongyloidiasis is mostly an asymptomatic infection and diagnosis of latent infections is difficult due to limitations of current parasitological and serological methods. This study was conducted to set up a PCR-based method for molecular diagnosis of Strongyloides stercoralis infection by detection of copro-DNA in stool samples.
Methods
A total of 782 fresh stool samples were collected and examined by agar plate culture. Among those sixteen stool samples, which confirmed to be infected with S. stercoralis were examined as positive control to set up each single and nested PCR, using two primer sets designing to amplify partial ribosomal DNA of S. stercoralis genome. Since, single PCR method yielded higher efficacy in detecting positive samples, in the second step, 30 stool samples, which found negative for S. stercoralis by agar plate culture of single stool sample, were examined by single PCR. Data analysis was performed using McNemar's χ2 test, with consideration of a P-value of <0.05 as indication of significant difference.
Results
In amplification of DNA extracted from stool samples, single PCR detected S. stercoralis DNA target in all 16 positive samples, while nested PCR amplified DNA in only 75% of samples. In the second step, single PCR amplified S. stercoralis extracted DNA in 5 out of 30 samples which were negative by coproculture.
Conclusion
Single PCR method amplifying a short (100bp) target represented more efficacies for detection of S. stercoralis in faecal examination compared to agar plate culture and nested PCR, which amplified longer target.
Keywords: Strongyloides stercoralis, Diagnosis, Copro-DNA, PCR
Introduction
S trongyloidosis is an intestinal infection in humans caused by the nematode Strongyloides stercoralis, distributed in tropical and temperate areas (1, 2). In normal healthy individuals, the infection is usually asymptomatic, with low minimal and intermittent larval excretion. However, in some predisposing conditions like initiation of immunosuppressive therapy, hematologic malignancies, kidney transplant recipients and diabetics the disease may change to any forms of hyper infection or disseminated types of strongyloidosis (2–5).
Detecting latent cases of S. stercoralis decreases morbidity and mortality of the infection. The detection rate of conventional methods is low and repeated examinations of stool over a number of consecutive days is essential for diagnosis (6, 7). Several serodiagnostic tests with variable sensitivity and specificity have been studied for diagnosis of S. stercoralis (8–13). However, use of these methods has some limitations; source of antigen is necessary and if the test is positive, microscopic analysis is also necessary. These methods also do not show the level of larval excretion (5, 7).
In recent years, some PCR-based techniques have been developed and used for detection of different intestinal parasites in faecal samples (14–18). Evaluation and standardization of such techniques are necessary to overcome the limitations of the current diagnostic methods. Hence, the aim of the present study was to set up a PCR method for diagnosis of S. stercoralis infection by examination of stool samples.
Materials and Methods
Samples collection
Seven hundred and eighty two fresh stool specimens were collected from two endemic provinces of strongyloidosis in Iran, including Mazandaran Province in north and Khuzestan Province in south-west of the country, and also from patients referred to the Helminthological Laboratory of School of Public Health, Tehran University of Medical Sciences, for parasitological examinations. Sixteen culture-positive stool samples were used as positive control for setting up the PCR. Thirty samples which found negative for S. stercoralis by agar plate culture of single stool sample were randomly selected for PCR test. Furthermore, 5 stool samples, reported negative for parasites by direct smear, formalin-ether concentration technique and agar plate copro-culture on three consecutive stool samples, were used as negative controls. For molecular examinations, all stool samples were preserved in 70% ethanol at room temperature.
Coprological examination
Coprological examination for detecting S. stercoralis infected samples was conducted by copro-culture of single stool sample on agar plate medium as used by Arakaki et al. (19) and the plates were examined as explained by Kia et al. (20). A skillful parasitologist performed the morphological differentiation of the L3 larvae of S. stercoralis from other possible nematodes, especially Rhabditis spp. Filariform larvae of S. stercoralis were collected from positive agar plates by washing the surface of the agar plates with a phosphate buffer saline solution. The extracted DNA from filariform larvae was used as control DNA during molecular assays.
Extraction of genomic DNA
About 3 g of each stool sample preserved in 70% ethanol alcohol was emulsified in 4% acetic acid. The suspension passed through two layers gauze into a tube, and after adding 3ml ether, was shaken vigorously and centrifuged at 1000 rpm for 2 min. The pellet was washed twice with distilled water and then used for extraction of genomic DNA, using QIAamp® DNA stool MiniKit (QIAGEN, Hilden, Germany). In this way 1.4 ml of ASL buffer was added to the sample and put in 80°C water bath for 5 min. Later, the procedure continued according to the protocol for extraction of DNA from stool. The extracted DNA was finally eluted with 50µl AE buffer.
Single PCR
Forward (SSF: 5′ ATC GTG TCG GTG GAT CAT TC 3′) and reveres (SSR: 5′ CTA TTA GCG CCA TTT GCA TTC 3′) primer pair was designed using DNASIS software and based on alignment of rDNA sequences related to S. stercoralis, deposited In GenBank (Accession numbers: EF653266, EF653265, EF653264, EF545004) to amplify a 114bp target in rRNA gene. PCR reactions were performed using the following reaction mixture: 2X red PCR Mastermix (ROVALAB, Hauffstr, Germany), 25pmol of each primer, 1µl of template, and enough distilled water up to final volume of 25µl under following conditions: 1 cycle at 95°C for 5 min (time-delay), 30 cycle at 94°C for 30s (denaturation), 58°C for 45s (annealing) and 72°C for 45s (extension), followed by a final extension for 5 min.
The specificity of the primers was evaluated using DNA extracted from some gast-rointestinal parasites including Hymenolepis nana, Trichostrongylus colubriformis, Giardia lamblia, Entamoeba histolytica and Entamoeba coli (3 samples of each), as well as using DNA extracted from Can-dida albicans, Escherichia coli, Cytrobacter spp., and distilled water as negative controls. The in silico specificity of primers for S. stercoralis in the NCBI BLAST was 100%.
Nested PCR
PCR reactions for both rounds were performed in 25µl volumes using 2X red PCR Mastermix (Ampliqon), 25pmol of each primer and 1µl of faecal DNA sample. For the primary amplification round, primers SSF0 (Forward: 5′ ATC CTT CCA ATC GCT GTT GT 3′) and SSR0 (Reverse: 5′ TTT CGT GAT GGG CTA ATT CC 3′) (21) were used to amplify a PCR product of 750bp containing ITS-1, 5.8s and ITS-2. For each set of PCR reactions, negative controls (distilled water and DNA extracted from negative stool samples) and positive controls, were included. The cycling conditions compromised an initial denaturation step at 95°C for 7 min, 30 cycles of denaturation at 94°C for 45s, annealing at 55°C for 90s, extension at 72°C for 90s, followed by a final extension at 72°C for 5 min.
Subsequently, 1µl of 1/10 diluted of the first round amplicon was subjected to a second amplification round, using primers SSFI (Forward: 5′ GTA ACA AGG TTT TCG TAG GTG AA 3′) and SSRI (Reverse: 5′ ATT TAG TTT CTT TTC CTC CGC TT 3′). A product of 680bp was amplified under the following conditions: Initial denaturation at 94° C for 3 min, and 30 cycles of 94° C for 45s, 60° C for 45s and 72° C for 1 min, followed by a final extension for 5 min.
Electrophoresis
The products of single PCR and nested PCR were loaded on 2% and 1.5% TBE (Tris 0.09M – borate 0.09M – EDTA 0.02M) agarose gels (Bio life, Italina S.r.l, Italy), respectively. The gels contained 0.5µg/ml ethidium bromide (Roche, Germany) for staining. Electrophoresis carried out 1 hour at 80V. Ultraviolet light was used to visualize the stained DNA.
Data analysis
In order to determine whether the observed difference between the results of the tests is statistically significant McNemar's χ2 test was employed, with consideration of a P-value of <0.05 as indication of significant difference.
Results
Agar plate culture
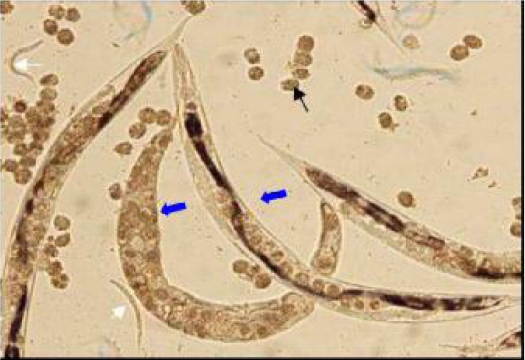
In examination of 782 stool samples by agar plate culture 16 cases were infected with S. stercoralis which all were examined as positive control to set up each single and nested PCR. During microscopic examination of the surface of agar plates of infected cases either homogonic or heterogonic life cycle of S. stercoralis (Fig.1) could be observed.
Fig. 1.
Surface of an agar plate culture showing heterogonic life cycle of Strongyloides stercoralis (black arrow: egg, white arrow: first- stage larva, center arrows: free-living female)
Single PCR
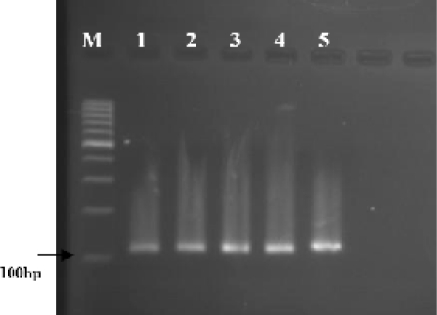
A PCR product of 114bp was amplified with designed primers using genomic DNA isolated from precipitant of S. stercoralis infected faecal samples (Fig. 2). All 16 (100%) previously confirmed S. stercoralis infected faecal samples were positive by single PCR assay. Among 30 randomly selected stool samples which found negative for S. stercoralis by agar plate culture of single stool sample, for 5 samples the PCR amplification became positive with the expected band size. No amplification was detected for any above mentioned negative control samples.
Fig. 2.
Agarose-gel electrophoresis of the single PCR products amplified by primers targeted the partial rDNA of genomic DNA extracted from precipitated stool samples infected with Strongyloides stercoralis Lanes 1-5: PCR products of 5 stool samples infected with S. stercoralis M: 100bp DNA marker ladder
Overall, in examination by single PCR, 21 out of 46 stool samples were found positive for S. stercoralis while by agar plate culture for the same stool samples the detected positive cases was lower (Table1).
Table 1.
Comparison of the results of single PCR and agar plate culture examinations for detection of Strongyloides stercoralis infection in single stool samples
| Agar plate result | Single PCR | Total | ||
|---|---|---|---|---|
| + | − | |||
| Agar plate | + | 16 | 0 | 16 |
| coproculture | − | 5 | 25 | 30 |
| Total | 21 | 25 | 46 | |
P<0.05; χ2=5
Nested PCR
Partial coding and non-coding spacer regions of rDNA were amplified using genomic DNA extracted from faecal samples. As for single PCR amplification, extracted DNA from stool precipitation was used in nested PCR assay. All the PCR amplicons produced at the second PCR round represented band of approximately 680bp. Among 16 previously confirmed S. stercoralis infected faecal samples, only 12 samples (75%) were found positive in the two-step nested PCR rounds. No amplification was detected for any negative control samples.
As the efficacy of nested PCR for the detection of S. stercoralis confirmed stool samples was lower than single PCR (χ2=4), further examinations were performed only by single PCR assay (as described above).
Statistical analysis
Statistical analysis using McNemar's χ2 test revealed that the difference between the result of single PCR and agar plate culture for the detection of S. stercoralis infected faecal samples is significant (χ2=5) and single PCR was able to detect more infected cases (Table1).
Discussion
In the majority of uncomplicated cases of strongyloidiasis, the intestinal worm load is very low and the output of larvae is minimal (22). This issue can encounter the immunocompromised infected patients to endangered conditions, leading the infection to uncontrolled disseminated forms. Therefore, development of highly sensitive diagnostic tests to detect light cases of strongyloidiasis is crucial to prevent potentially fatal infections. Among different parasitological methods, agar plate culture of stool sample has been reported as method that is more sensitive to detect S. stercoralis infections (20, 23, 24); however, this method requires multiple fresh stool sampling and experienced microscopist. Accordingly, development and evaluation of PCR based methods, as suitable alternative methods in order to increase detection rates, are needed. In several studies, PCR has been stated as highly sensitive method for detection of both protozoan (14) and helminthic (15–17) infections in faecal samples.
A multiplex real-time PCR, focusing on four target parasites including Entamoeba histolytica, Giardia lamblia, Cryptosporidium and S. stercoralis in returning travelers showed to be highly sensitive and specific technique compared with the routine approach of microscopy and antigen-based methods (18). However, PCR inhibitors are relatively common in stool samples and they can strongly influence the results of PCR assays (25). This inhibiting effect is more noticeable in samples with lower parasite DNA (15). Therefore, in the current study in order to increase the amount of faeces used for PCR assay, the stool sample was concentrated by acid-ether prior to DNA extraction. Then, nested PCR and single PCR were established for the detection of DNA of S. stercoralis in the faecal samples. Both assays were highly specific, and no amplification was detected for any negative control samples used. However, the sensitivity of single PCR was higher than nested PCR. In nested PCR only in 75% of infected samples (12 out of 16) S. stercoralis DNA were amplified; while in single PCR all positive samples were detected. The false negative results of nested PCR might be due to the size of amplified fragment which was very small in single PCR. In the study of Verweij et al. S. stercoralis real-time PCR achieved 100% specificity and high sensitivity, with a two fold increase in the detection rates compared with Baermann sedimentation method (17).
In the present study, single PCR assay not only detected all infected samples that were found positive by agar plate culture of single stool sample but also detected some infected cases that agar plate culture of single stool sample was not able to recognize them. Therefore, single PCR was more efficient in detecting S. stercoralis infected faecal samples and this difference between efficacy of these two methods was statistically significant (P<0.05). Therefore, in cases that the worm burden is too low or larvae are not alive to be detected on agar plate culture, the application of PCR will be useful.
In conclusion, the result of the current study shows that performing single PCR, complemented with concentration of larvae in stool by acid-ether technique before DNA extraction, provides highly specific and sensitive molecular method for diagnosis of S. stercoralis genome in human faeces. Further studies to find the effect of multiple stool sampling on detection rate of S. stercoralis infections by PCR based methods is recommended. In addition, the development of multiplex PCR for several target parasites can be applied for detection of infections in immunocompromised people who are at risk of disseminated strongyloidosis and other opportunistic infections.
Acknowledgments
The authors appreciate Mrs F Zahabiun and Mrs N Jalalizand from the Department of Medical Parasitology and Mycology, Tehran University of Medical Sciences for their kind assistance. This study was financially supported by the Tehran University of Medical Sciences (Project No: 88-02-27-8964). The authors declare that they have no conflict of interest.
References
- 1.Grove DI. Human strongyloidiasis. Adv Parasitol. 1996;38:251–309. doi: 10.1016/s0065-308x(08)60036-6. [DOI] [PubMed] [Google Scholar]
- 2.Viney ME, Lok JB. Strongyloides spp. WormBook. The C. elegans Research Community. 2007:1–15. doi: 10.1895/wormbook.1.141.1. doi/10.1895/1.141.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agrawal V, Agarwal T, Ghoshal UC. Intestinal strongyloidiasis: a diagnosis frequently missed in the tropics. Trans R Soc Trop Med Hyg. 2009;103:242–246. doi: 10.1016/j.trstmh.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Keiser PB, Nutman TB. Strongyloides stercoralis in the immunocompromised population. Clin Microbiol Rev. 2004;17(1):208. doi: 10.1128/CMR.17.1.208-217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marcos LA, Terashima A, DuPont HL, Gotuzzo E. Strongyloides hyperinfection syndrome: an emerging global infectious disease. Trans R Soc Trop Med Hyg. 2008;102:314–318. doi: 10.1016/j.trstmh.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 6.Concha R, Harrington Jr W, Rogers AI. Intestinal strongyloidiasis: recognition, management, and determinants of outcome. J Clin Gastroenterol. 2005;39:203. doi: 10.1097/01.mcg.0000152779.68900.33. [DOI] [PubMed] [Google Scholar]
- 7.Siddiqui AA, Berk SL. Diagnosis of Strongyloides stercoralis infection. Clin Infect Dis. 2001:1040–1047. doi: 10.1086/322707. [DOI] [PubMed] [Google Scholar]
- 8.Conway DJ, Atkins NS, Lillywhite JE, Bailey JW, Robinson RD, Lindo JF, Bundy DAP, Bianco AE. Immunodiagnosis of Strongyloides stercoralis infection: a method for increasing the specificity of the indirect ELISA. Trans R Soc Trop Med Hyg. 1993;87:173–176. doi: 10.1016/0035-9203(93)90477-8. [DOI] [PubMed] [Google Scholar]
- 9.Sato Y, Kobayashi J, Shiroma Y. Serodiagnosis of strongyloidiasis. The application and significance. Rev Inst Med Trop S Paulo. 1995;37(1):35–41. doi: 10.1590/s0036-46651995000100006. [DOI] [PubMed] [Google Scholar]
- 10.Sato Y, Toma H, Kiyuna S, Shiroma Y. Gelatin particle indirect agglutination test for mass examination for strongyloidiasis. Trans R Soc Trop Med Hyg. 1991;85(4):515–518. doi: 10.1016/0035-9203(91)90240-y. [DOI] [PubMed] [Google Scholar]
- 11.Silva LP, Barcelos ISC, Passos-Lima AB, Espindola FS, Campos DMB, Costa-Cruz JM. Western blotting using Strongyloides ratti antigen for the detection of IgG antibodies as confirmatory test in human strongyloidiasis. Mem Inst Oswaldo Cruz. 2003:687–691. doi: 10.1590/s0074-02762003000500017. [DOI] [PubMed] [Google Scholar]
- 12.Van Doorn HR, Koelewijn R, Hofwegen H, Gilis H, Wetsteyn J, Wismans PJ, Sarfati C, Vervoort T, van Gool T. Use of enzyme-linked immunosorbent assay and dipstick assay for detection of Strongyloides stercoralis infection in humans. J Clin Microbiol. 2007:438. doi: 10.1128/JCM.01735-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koosha S, Fesharaki M, Rokni MB. Comparison of enzyme-linked immunosorbent assay and indirect immunofluorescence assay in the diagnosis of human strongyloidiasis. Indian J Gastro. 2004;23:214–216. [PubMed] [Google Scholar]
- 14.Ten Hove RJ, Schuurman T, Kooistra M, Moller L, Van Lieshout L, Verweij JJ. Detection of diarrhoea-causing protozoa in general practice patients in The Netherlands by multiplex real-time PCR. Clin Microbiol Infect. 2007;13:1001–7. doi: 10.1111/j.1469-0691.2007.01788.x. [DOI] [PubMed] [Google Scholar]
- 15.Verweij JJ, Brienen EAT, Ziem J, Yelifari L, Polderman AM, van Lieshout L. Simultaneous detection and quantification of Ancylostoma duodenale, Necator americanus, and Oesophagostomum bifurcum in fecal samples using multiplex real-time PCR. Am J Trop Med Hyg. 2007;77:685–90. [PubMed] [Google Scholar]
- 16.Ten Hove RJ, Verweij JJ, Vereecken K, Polman K, Dieye L, Van Lieshout L. Multiplex real-time PCR for the detection and quantification of Schistosoma mansoni and S. haematobium infection in stool samples collected in northern Senegal. Trans R Soc Trop Med Hyg. 2008;102:179–85. doi: 10.1016/j.trstmh.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 17.Verweij JJ, Canales M, Polman K, Ziem J, Brienen EAT, Polderman AM, van Lieshout L. Molecular diagnosis of Strongyloides stercoralis in faecal samples using real-time PCR. Trans R Soc Trop Med Hyg. 2009;103(4):342–346. doi: 10.1016/j.trstmh.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Ten Hove RJ, Van Esbroeck M, Vervoort T, Van den Ende J, van Lieshout L, Verweij JJ. Molecular diagnostics of intestinal parasites in returning travellers. Eur J Clin Microbiol Infect Dis. 2009;28:1045–1053. doi: 10.1007/s10096-009-0745-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arakaki T, Iwanaga M, Kinjo F, Saito A, Asato R, Ikeshiro T. Efficacy of agar-plate culture in detection of Strongyloides stercoralis infection. J Parasitol. 1990:425–428. [PubMed] [Google Scholar]
- 20.Kia EB, Mahmoudi M, Zahabiun F, Meamar AR. An evaluation on the efficacy of agar plate culture for detection of Strongyloides stercoralis . Iranian J Parasitol. 2007;2(1):29–34. [Google Scholar]
- 21.Nilforoushan MR, Mirhendi H, Rezaie S, Rezaian M, Meamar AR, Kia EB. A DNA-Based Identification of Strongyloides stercoralis Isolates from Iran. Iranian J Publ Health. 2007;36(3):16–20. [Google Scholar]
- 22.Liu LX, Weller PF. Strongyloidiasis and other intestinal nematode infections. Infect Dis Clin North Am. 1993;7:655–82. [PubMed] [Google Scholar]
- 23.Blatt JM, Cantos GA. Evaluation of techniques for the diagnosis of Strongyloides stercoralis in human immunodeficiency virus (HIV) positive and HIV negative individuals in the city of Itajai, Brazil. Braz J Infect Dis. 2003:402–408. doi: 10.1590/s1413-86702003000600008. [DOI] [PubMed] [Google Scholar]
- 24.de Kaminsky RG. Evaluation of three methods for laboratory diagnosis of Strongyloides stercoralis infection. J Parasitol. 1993:277–280. [PubMed] [Google Scholar]
- 25.Ten Hove RJ, Schuurman T, Kooistra M, Moller L, Van Lieshout L, Verweij JJ. Detection of diarrhoea-causing protozoa in general practice patients in The Netherlands by multiplex real-time PCR. Clin Microbiol Infect. 2007;13:1001–7. doi: 10.1111/j.1469-0691.2007.01788.x. [DOI] [PubMed] [Google Scholar]