Abstract
Leishmania is a protozoan parasite belonging to the family Trypanosomatidae, which is found among 88 different countries. The parasite lives as an amastigote in vertebrate macrophages and as a promastigote in the digestive tract of sand fly. It can be cultured in the laboratory using appropriate culture media. Although the sexual cycle of Leishmania has not been observed during the promastigote and amastigote stages, it has been reported by some researchers. Leishmania has eukaryotic cell organization. Cell culture is convenient and cost effective, and because posttranslational modifications are common processes in the cultured cells, the cells are used as hosts for preparing eukaryotic recombinant proteins for research. Several transcripts of rDNA in the Leishmania genome are suitable regions for conducting gene transfer. Old World Leishmania spp. has 36 chromosomes, while New World Leishmania spp. has 34 or 35 chromosomes. The genomic organization and parasitic characteristics have been investigated. Leishmania spp. has a unique genomic organization among eukaryotes; the genes do not have introns, and the chromosomes are smaller with larger numbers of genes confined to a smaller space within the nucleus. Leishmania spp. genes are organized on one or both DNA strands and are transcribed as polycistronic (prokaryotic-like) transcripts from undefined promoters. Regulation of gene expression in the members of Trypanosomatidae differs from that in other eukaryotes. The trans-splicing phenomenon is a necessary step for mRNA processing in lower eukaryotes and is observed in Leishmania spp. Another particular feature of RNA editing in Leishmania spp. is that mitochondrial genes encoding respiratory enzymes are edited and transcribed. This review will discuss the chromosomal and mitochondrial (kinetoplast) genomes of Leishmania spp. as well as the phenomenon of RNA editing in the kinetoplast genome.
Keywords: Leishmania, Kinetoplast, Genome, RNA editing, Trans-splicing
Introduction
Leishmania spp. lives in the gastrointestinal tract of the sand fly vector, and can be cultured using appropriate laboratory culture media as promastigotes. They can also exist in the vertebrate host macrophages in the amastigote form (1–3). While asexual reproduction is known to occur in this species (4), their sexual forms have not yet been discovered (5). Clonal reproduction is believed to occur among the protozoan parasites of the family Trypanosomatidae (6), considering that nuclear fusion occurs in some forms of this parasite that may give rise to sexual reproduction (7, 8). Researchers have been unable to confirm sexual reproduction and identify sexual gametes of these microorganisms by using classical methods (9). It should be noted that the exchange of genetic material in Trypanosomatidae has been proven (10–12).
Leishmania is used as an intracellular molecular model for research in microbiology, immunology, and biochemistry (1, 2, 13–20). This article will discuss the genomic organization of this parasite.
Genomic organization of Leishmania
The haploid genome of Leishmania spp. has 32,816,678 bp organized into 36 chromosomes (21), with a total of 911 RNA genes and 39 pseudo-genes (21, 22). A total of 8272 genes are known to encode proteins. Producer protein genes are encoded as long polycistronic genes lacking transcription factors in L. major, Trypanosoma brucei, and T. cruzi (Tritryp) (Fig. 1). The Old World Leishmania spp. has 36 chromosomes, while the New World Leishmania spp. has 34 or 35 chromosomes. L. mexicana has linkage groups of chromosomes 8 and 29 as well as of chromosomes 30 and 36, and L. braziliensis has a linkage group of chromosomes 20 and 34 (23). The general pattern of nucleotide sequences of genes in 30 Leishmania spp. is conserved (24–26).
Fig. 1.
Organization of chromosomes of Leishmania genes: clusters of genes on chromosomes 1, 2, 3, 4, and 35 are shown as thick lines. The direction of mRNA transcription is indicated. Vertical lines indicate the right side of the chromosome 1 repeated sub-telomeric sequence. The arrows indicate chromosome 2 splice leader categories. The arrows between the individual genes in chromosome 3 genes indicate tRNA. The space on chromosome 35 indicates an area of undetermined sequence (Source Ref. 30).
Chromosome 1 of L. major is the smallest Leishmania spp. chromosome, and contains 79 protein-encoding genes. Its genes have been organized into 2 converted polycistronic clusters, and mRNA transcription is directed to the telomeres (27–30).
Leishmania spp. chromosome 3 has about 79 genes, and is organized as 2 convergent polycistronic transcripts. These transcripts encode 2 protein clusters and tRNA genes are located between them. They remain at the end of a gene that is transcribed in contrast to the previous clusters (28, 29, 31).
Leishmania spp. proteins are expressed during translation or after completion of replication (32). The mechanism involves regulation of transcription of the eukaryotic RNA polymerase II. This mechanism differs from the other mechanisms, although they have a chromatin remodeling process (21).
In contrast to other members of Trypanosomatidae, the Leishmania genome does not have a sub-telomeric region (species-specific genes) and a transposable element. There have been no reports of RNAi in this organism (22).
The Leishmania spp. genome is organized in the nucleus, which contains chromosomal and episomal DNA, and in the kinetoplasts, which comprise independently replicating DNA molecules. Furthermore, virus-like particles are contained in the cytoplasm. The kinetoplasts have been separated and studied by ultracentrifugation, whereas the chromosomes have been studied by pulsed-field gel electrophoresis (PFGE). There are questions regarding the changes occurring in karyotype species, sexual reproduction in Leishmania spp., and the number of copies of each gene in each chromosome (33).
The electrophoretic patterns of Leishmania spp. chromosomes investigated by PFGE indicate haploid, diploid, and polyploid arrangements. Isolation by hybridization that parts of chromosomes can be common, but the genes Hsp70, Hsp80, adenylate cyclase, glyceraldehyde phosphate dehydrogenase, beta tubulin, phosphofructokinase, pho-sphoenolpyruvate carboxymethyl pyruvate kinase, pyruvate kinase, and ubiquitin are conserved. The chromosomes range from 400 to 900 kbp in size and contain mini-exons (5′-spliced leader genes). Chromosomal changes that occurred during the evolution of Leishmania spp. have been confirmed, and the molecular karyotypes in the promastigote and amastigote forms have been found to be identical. Three molecular karyotypes have been identified in Leishmania spp.: (1) The L. major karyotype is completely conserved, even in different geographical regions, (2) The members of the L. braziliensis panamensis group have more than one karyotype, and (3) L. mexicana amazonensis has highly diverse molecular karyotypes, even among those isolated from the same clinical samples. The mechanism of chromosomal polymorphism in Leishmania spp. does not include removal or translocation. Among the genes amplified in Leishmania spp. are genes that confer resistance against sodium arsenate and methotrexate drugs. This phenomenon of increasing the number of copies of genes involved in metabolic phenomena and environmental response appears to be important. Regions of genes that are involved in drug resistance are increased by 2–20 folds in copy number. Two genomic regions, namely, H-DNA and R-DNA (encoding dihydrofolate reductase and thymidylate synthase), are chromosomal derivatives, which are surrounded by inverted repeats. The inverted repeats are involved in the supercoiling of amplified gene products (33).
Methotrexate and arsenate drugs induce gene amplification in Leishmania spp. Methotrexate induces amplification of the R-DNA and H-DNA genomic regions. In a methotrexate-resistant L. tarentolae mutant, the H region is amplified as linear or circular DNA. The dihydrofolate reductase, thymidylate synthase, and ltdh genes in the H region are resistant to drugs (34, 35). Gene amplification in the amphotericin B-resistant L. tarentolae occurs in the circular form in different chromosomes (36). Drug resistance to sodium stibogluconate (pentostam) in L. tarentolae is due to the amplification of a gene described by Haimeur and Ouellett, which encodes a 770-amino acid-long protein (37).
Gene expression control among the members of the parasitic Trypanosomatidae family involves unusual antigenic shifts, involving DNA rearrangements, generation of polycistronic transcripts from multi-copy genes, and post-transcriptional modification by trans-splicing and RNA editing (38).
Gene transcription in Leishmania spp.
The genetic information of most organisms has been discovered in cDNA sequences known as expressed sequence tags (EST) (39). It should be noted that most Leishmania genes have no introns (40), and that chromosomal DNA is used as the template for cloning by PCR (41–45).
Gene transcription to produce proteins in eukaryotes involves RNA polymerase II and transcription by RNA polymerase I to produce ribosomal RNA. In kinetoplastids, gene transcription involves RNA polymerase I and a trans-splicing mechanism (46).
Discontinuous mRNA synthesis is a process occurring in the kinetoplastids. In this process, a 35-nucleotide sequence is placed at the 5′-end of all mRNAs. This sequence is encoded by a gene duplication cluster, 1.35 kb in length, which is known as a mini-exon or a trans-splice. Mini-exon mRNA was first identified as being related to the trypanosome variable surface glycoprotein (47).
Martınez-Calvillo et al. analyzed the sequence of Leishmania chromosome 1, which is the smallest of all the chromosomes. A total of 39 genes were transcribed from a strand of DNA and 50 other genes in a polycistronic transcript (48). Martınez-Calvillo et al. also analyzed Leishmania chromosome 27 and indicated that the organization of transcription of Leishmania genes is a complex process. It was determined that chromosome 27 of Leishmania spp. is transcribed by RNA polymerase II (49). This is contrary to the findings of Ploeg and Lee (46).
Non-coding RNAs, about 300–600 nucleotides long, are known to be expressed only in the amastigotes; these RNAs are transcribed by RNA polymerase II. Both sense and antisense transcripts are processed by trans-splicing and polyadenylation, but the antisense transcripts are transcribed 10 folds lesser than the sense transcripts. It is possible that these antisense transcripts play a role in RNA stability. It should be noted that these molecules are not transcribed in promastigotes, and that RNA stability in promastigotes is less than in amastigotes (50).
Trans-splicing of the Leishmania mRNA transcript
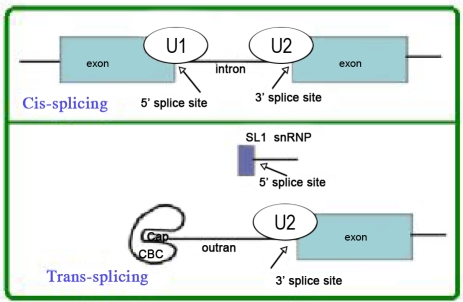
There is a 35-nucleotide-long sequence known as a spliced leader (SL) or 5′-mini exon at the 5′ end of Leishmania mRNA transcripts. The SL sequence is at the 5′-end of a preliminary transcript about 85 nucleotides in length that contains a 5′-exon-intron connection adjacent to the 3′-spliced leader (Fig. 2 and 3).
Fig. 2.
Comparison of cis- and trans-splicing: In cis-splicing, pair bases U1 small nuclear ribonucleoprotein (snRNP) are in the 5′ SL [?] and U2 snRNPs are in the break point, while intron breaks two exons are connected. In trans-splicing, a 5′-splice site on the mRNA for binding to U1 snRNP is absent. Instead, a 5′-splice site produced by the donor SL snRNP interacts with U2 in the 3′-splice site. The splice leader connects to the next exon. (http://www.wormbook.org/chapters/www_transsplicingoperons/transsplicingoperons.pdf)
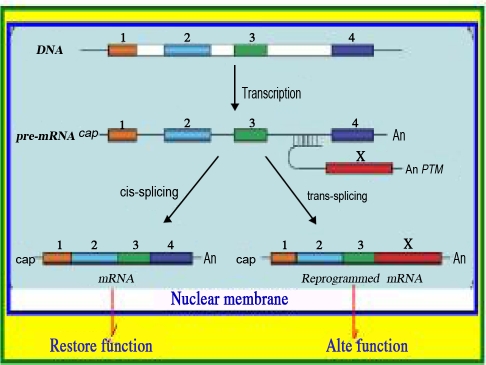
Fig. 3.
Cis-splicing and trans-splicing: There are 4 exons in the initial transcript, which contains both exons and introns. In the cis-splicing phenomenon, the mRNA contains 4 exons and 3 introns. The 3 introns are removed, and the exons are connected. In trans-trans-splicing, a pre-trans-splicing molecule attaches exon X to intron 3. The 5'-splice donor is attached to the 3'-splice acceptor (Source Ref. 52).
The sequence of the SL connects the 3′-end of the genes encoding proteins. Previous reports have indicated the possibility of such intermediaries in mRNA processing. There is a 50-nucleotide-long interval at the 3′-end of SL, which is known as the SL intron sequence (SLIS). The SLIS and SL are connected to the 5′-end of RNA. Density centrifugation analyses have shown that SL mRNA is in the 60S rRNA, but SLIS is in the 40S rRNA. It is likely that the observed nucleoprotein particles are the same spliceosomes that can be observed in other microorganisms (51, 52).
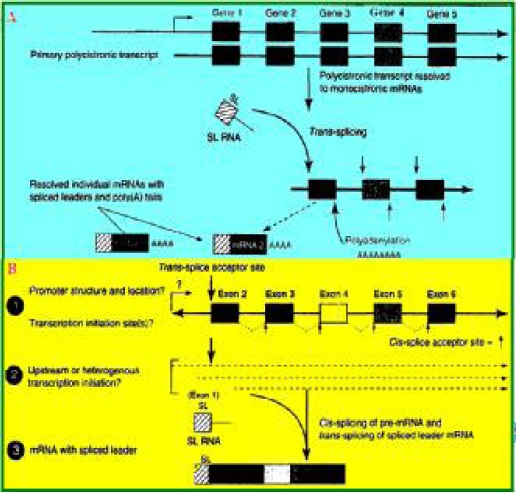
Since the discovery of trans-splicing in Leishmania spp., it has also been observed in other microorganisms (53). Trans-splicing is an essential stage of eukaryotic precursor mRNA and is not observed in mammals, insects, yeast, and plants (54). This phenomenon is observed in rotifera (55), dinoflagellates (56), nematodes, and protozoan parasites (57–60) as shown in Fig. 4.
Fig. 4.
Trans-splicing in metazoan parasites: A) Transcription occurs via a polycistronic transcript and trans-splicing. The initial transcript contains mRNAs with 5′-trans-splicing and polyadenylation. Each box represents 1 gene with an exon and an intron. The bent arrows indicate the promoter and the transcription start site. B) The phenomenon of transcription and trans-splicing in metazoan genes (worms). The solid squares indicate genes with an intron between them. 1) Promoter and possible transcription start site. 2) The position of transcription initiation. 3) mRNA molecules with SL (Source Ref. 60).
Table 1.
Comparison of the characteristics of the genomes of 3 species of Leishmania (22)
| Leishmania spp. Characteristics | L. major | L. infantum | L. braziliensis |
|---|---|---|---|
| Chromosome | 36 | 34 | 35 |
| Contigs | 36 | 562 | 1041 |
| G+C percent | 89.7 | 59.3 | 57.76 |
| Size ( No nucleotide; bp) | 32,816,678 | 32,134,935 | 32,005,207 |
| Coding genes | 8298 | 8154 | 8153 |
| Pseudo genes | 97 | 41 | 161 |
| G+C content (%) in coding region | 52.5 | 52.45 | 60.38 |
The organization and regulation of gene expression in trypanosomatid parasites differs from that of other cells. Collected information has led to advances in effective disease control (61).
The genes in the parasites of family Trypanosomatidae are organized as long polycistronic transcripts (more than 100–300 kb) on the same DNA strand. The genes encoding proteins are transcribed from unknown promoters, and precursor polycistronic RNA is produced. Monocistronic mRNA is produced by trans-splicing and polyadenylation of RNA. The trans-splicing mechanism includes a mini-exon containing 39 nucleotides, which is not translated. This mini-exon is connected to the 5′-end of the mRNA molecule. There are some similarities between cis-splicing and trans-splicing mechanisms. The AG is at the 3′ of the splice acceptor site downstream of a polypyrimidine tract. Polyadenylation in Leishmania spp. requires trans-splicing and differs from that of other eukaryotes. There are no introduced polyadenylation signals in kinetoplastidae undefined and instead choose to place poly A site depends on positions upstream acceptor site (61.) Gopta et al. analyzed chromosomes 1 and 3 of L. major and predicted the positions of trans-splicing with 92% accuracy. Computer analyses were performed to identify elements involved in trans-splicing. The following components are present: (1) nucleotide A, (2) a polypyrimidine rich stretch of T and C, varying in size from 5 to 100 nucleotides with purine bases occasionally located between the T and C, (3) a variable spacer, and (4) a 3′-acceptor site consisting of AG (62).
Synthesis of nucleic acids in Leishmania spp.
Leishmania spp. generates pyrimidine nucleic acids via de novo biosynthesis, but obtains purine nucleic acids via a salvage process (63–66).
Cunningham and Beverley have studied the amastigote stage of pathogenic species of Leishmania. This study indicated that salvage activities in the amastigote stage would limit the effectiveness of chemotherapy in patients infected with Leishmania spp. Salvage activities do not involve RNA transcripts and likely occur via posttranscriptional modifications (67).
Structure of the kinetoplast
The kinetoplast or mitochondrion is the energy-producing organelle of Leishmania spp. (68, 69). If DNA replication is inhibited in the kinetoplast by ethidium bromide, energy production will reduced in the parasite (70).
The extra-chromosomal DNA is located in the kinetoplast organelle in an arrangement similar to that in the mitochondria of the eukaryotes. The kinetoplast has a particular DNA topology, which is not found in other eukaryotic cells (Fig. 5), and is composed of large circular molecules up to about 50,000 nucleotides that are known as maxicircles. They are not present in large numbers. These circular molecules carry the genes encoding the enzymes and coenzymes involved in the Krebs cycle (71–74).
Fig. 5.
Structure of the kinetoplast disk and the proteins involved in its replication SSE1, Structure -specific endonuclease 1; UMSBP, Universal minicircle sequence-binding protein (http://www.pnas.org/content/101/13/4333/F2.expansion.html)
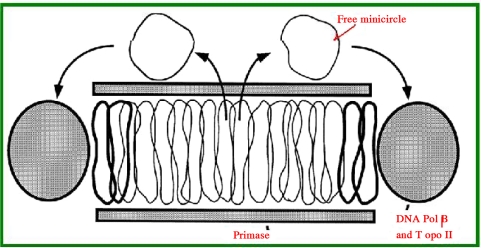
Other DNA molecules that are present in larger numbers but have fewer nucleotides (600–2,500 bp) are known as minicircles. Chritidia fasciculate has 25 maxicircles (each containing 37,000 nucleotides) and 5,000 minicircles (each containing 2,500 nucleotides) (72–74), some of which have been identified as free-form molecules (73). A minicircle DNA sequence “GGGGTTGGTGTAA” is conserved among all members of the family Kinetoplastidae (75), and some believe that this sequence is the origin of replication of the minicircle (76). Other parts of the minicircle sequence vary among the minicircles. One region known as the variable region is used for parasite genotyping. Large and small circles can exist inside each other so that each loop intercalates with 2 other minicircle loops and eventually “maxicircles and minicircles” become intertwined with each other (interlocked or catenated) and a heavy molecule (about 400S) is formed during the extraction of parasite DNA that is distinct from the chromosomal DNA sediment. When minicircles replicate, some are released (Fig. 6) as opened loops. When replication is completed, a replicated minicircle will become connected to the kinetoplast (75, 77, 78). The blank section of the kinetoplast is restored by DNA topoisomerase II (79).
Fig. 6.
In vivo replication of a kinetoplast shown as a disk section with catenated minicircles surrounded by DNA polymerase beta and DNA topoisomerase II. Primase is located at the top and bottom. During replication, the minicircles are released and connected to the network after replication is complete. Two newly synthesized minicircles are shown in bold (http://www.jbc.org/content/272/33/20787.full.pdf+html)
The origin of replication of the kinetoplast is recognized by a protein known as UMSBP (universal minicircle sequence-binding protein). This protein is responsible for initiating replication (80). This reaction is regulated in vivo by an oxidation-reduction reaction (80, 81). The inhibition of UMSBP halts the growth of the parasite (82). A zinc ion (Zn) is involved in this process, and is essential for connecting UMSBP to DNA (83). It should be noted that replication of the members of Kinetoplastidae occurs via different mechanisms (84).
The functions of minicircles were not clarified until recently, and the genes of some of the enzymes involved in Krebs cycle were not observed. The discovery of the RNA editing phenomenon was an exciting new finding (85–87). It was found that the parasites have copies of RNA molecules, which are altered because of posttranscriptional modification. This is accompanied by deletion or insertion of a number of nucleotide residues (mostly uracil). RNA editing emits signals by guide RNA-derived transcripts of minicircles (88, 89) or maxicircles (90) (Fig. 7). A gRNA-binding complex is involved in the processing of a gRNA, which includes polyadenylation and stabilization of the edited mRNA transcript (89). The kinetoplast of L. tarentolae has a 9S rRNA (91–94) and a 12S rRNA (92–95), but does not have supercoiled circles (96).
Fig. 7.
RNA editing of cytochrome oxidase B of Leishmania tarantula (http://dna.kdna.ucla.edu/trypanosome/index.html)
The RNA editing phenomenon produces deletions, replacements, and insertions in mRNA transcripts (86, 87, 97). An edited transcript mRNA has important effects, and sometimes half of the nucleotides are altered (Fig. 8). It should be noted that although the changes may be small, its effect is important. For example, replacement of a C nucleotide by U in the human apolipoprotein B transcript (Fig. 9) leads to conversion of a glutamine codon to a stop codon. This edited transcript produces a truncated protein (85).
Fig. 8.
Model RNA editing in the kinetoplast: addition of U (left), removal of U (center) or formation of a chimera (right) in an mRNA transcript are performed by TUTase (http://dna.kdna.ucla.edu/trypanosome/images/kablea.JPG)
Fig. 9.
RNA editing in human apolipoprotein B
The TGA codon (stop codon) of the Leishmania spp. maxicircle encodes tryptophan (98). When the parasite glycosomal cycle is reduced, the mitochondrial (kinetoplast) volume is increased, and vice versa (99). The characteristics of kinetoplast DNA has led to its choice as a target for drug therapy (99, 100).
Discussion
Leishmania is a protozoan parasite with some similarities and differences as compared to other eukaryotic cells. It shares some characteristics with prokaryotic cells, such as polycistronic transcription (31, 38). Researchers have been attracted to its unique characteristics. In recent years, Leishmania spp. has been used as a host for production of recombinant proteins. An appropriate host is an important factor in production of recombinant proteins (drugs). Prokaryotes such as Escherichia coli need simple and inexpensive culture media and have a short proliferation time. This provides high yields in the production of recombinant proteins. However, prokaryotes do not generate posttranslational modifications such as gly-cosylation, phosphorylation, and car-boxylation. Some eukaryotic proteins are non-functional after translation in E. coli, and some of them become aggregated as inclusion bodies in the host cell cytoplasm. These proteins cannot fold appropriately if they are expressed in a prokaryotic host.
Replication of yeasts such as Pichia pastoris, Saccharomyces cerevisiae, and Schizosaccharomyces pombe also requires significant culture time, and posttranslational modifications are not perfect processes. Other types of eukaryotic cell cultures tend to be expensive and require specialized culture conditions and laboratory equipment. Because Leishmania spp. is maintained easily in NNN culture medium at low cost and can multiply quickly, it is preferred over other species. Eukaryotic Leishmania spp. can perform posttranslational modifications (21). This makes Leishmania spp. a suitable host for production of recombinant protein drugs (101–104). Efforts undertaken thus far have allowed the production of some therapeutic proteins (105–108). However, more research is needed before it can be used extensively as a host for the production of recombinant proteins. Researchers in biochemistry, pharmacology, and immunology, who are engaged in new drug development as well as production and testing vaccines, need appropriate cell models. Leishmania spp. is expected to be an appropriate candidate (109,110). Anti-folates are used to treat malaria, bacterial infections, and cancer. Leishmania spp. is an appropriate model for testing such enzyme inhibitors to investigate the progression of anti-parasitic and anti-cancer drugs (111, 112). Posttranslational modifications play important roles in various cellular processes. Small ubiquitin-like modifier protein (SUMO) is a fusion protein that can be added as a reversible tag to N terminal recombinant proteins (eukaryotes and prokaryotes) to provide stability and solubility to proteins (113, 114). SUMO is produced by Leishmania spp. and can be used as a stable and soluble factor for the production of recombinant proteins in Leishmania promastigote (115). One disadvantage is that non-coding RNAs act as mRNA stability factors are not transcribed in the Leishmania promastigote. Because mRNA is not stable in promastigotes (50), this form of Leishmania spp. is not appropriate for use as a host in the preparation of recombinant proteins.
Glossary
Biosynthesis de novo: The de novo purine biosynthetic pathway produces purines which represent the building blocks for DNA and RNA synthesis
Cistron: A segment DNA equivalent to gene for function (protein or enzymes)
Diploid; An organism with sexual cycle is diploid and has one chromosome set from each of its parents
Haploid: The haploid means usual number of chromosomes set in somatic cells of common organisms. Organisms that have not sexual cycle are haploid
Inversion: Chromosome break of the two areas separated pieces is back by reversal from chromosome breakage
Inverted repeats: Is a sequence of nucleotides that is the reversed complement of another sequence further downstream
Poly cistronic: There are some cistrons on one mRNA
Polyploidy: Increase in chromosome set number
Pseudo-genes: Are copy of original gene sequence, but lacked the necessary sequences for function. These genes from genetically similar to functional genes, but they have containing multiple mutations
snRNP: Small nuclear ribonucleoproteins, are RNA-protein complexes, they will combined with unmodified pre-mRNA and various other proteins to spliceosome formation.
Sub telomeric: Sub telomeric is a region near the end of chromosomes composed of polymorphic repetitive DNA. Damage to this area in humans lead to mental retardation
Acknowledgements
The author thanks the authorities of the Cellular and Molecular Biology Research Center of Shahid Beheshti University of Medical Sciences for their collaborations and declares that there is no conflict of interests.
References
- 1.Handman E. Cell biology of Leishmania . Adv Parasitol. 1999;44:1–39. doi: 10.1016/s0065-308x(08)60229-8. [DOI] [PubMed] [Google Scholar]
- 2.Bard E. Molecular biology of Lei-shmania . Biochem Cell Biol. 1989;67(9):516–24. doi: 10.1139/o89-083. [DOI] [PubMed] [Google Scholar]
- 3.Panton LJ, Tesh RB, Nadeau KC, Beverley SM. A test for genetic exchange in mixed infections of Leishmania major in the sand fly Phlebotomus papatasi. J Protozool. 1991;38(3):224–8. doi: 10.1111/j.1550-7408.1991.tb04433.x. [DOI] [PubMed] [Google Scholar]
- 4.Cruz AK, Titus R, Beverley SM. Plasticity in chromosome number and testing of essential genes in Leishmania by targeting. Proc Nati Acad Sci USA. 1993;90:1599–603. doi: 10.1073/pnas.90.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Victoir k, Dujardin JC. How to succeed in parasitic life without sex? Asking leishmania . Trends Parasitol. 2002;18(2):81–5. doi: 10.1016/s1471-4922(01)02199-7. [DOI] [PubMed] [Google Scholar]
- 6.6- Tibayrenc M, Kjellberg F F, Arnaud J, Oury B, Brenipre F, Dardi ML, Ayala FJ. Are eukaryotic microorganisms clonal or sexual? A population genetics vantage. Proc Natl Acad Sci USA. 1991;88:5129–33. doi: 10.1073/pnas.88.12.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Youssef MY, Eissa MM, el Mansoury ST. Evidence of sexual reproduction in the protozoan parasite Leishmania of the Old World. J Egypt Soc Parasitol. 1997;27(3):651–7. [PubMed] [Google Scholar]
- 8.Kreutzer RD, Yemma JJ, Grogl M, Tesh RB, Martin TI. Evidence of sexual reproduction in the protozoan parasite Leishmania (Kinetoplastida: Trypanosomatidae) Am J Trop Med Hyg. 1994;51(3):301–7. doi: 10.4269/ajtmh.1994.51.301. [DOI] [PubMed] [Google Scholar]
- 9.Tait A. Sexual processes in the kinetoplastida. Parasitology. 1983;86(Pt 4):29–57. doi: 10.1017/s0031182000050836. [DOI] [PubMed] [Google Scholar]
- 10.Jenni L. Sexual stages in trypanosomes and implications. Ann Parasitol Hum Comp. 1990;65(Suppl 1):19–21. doi: 10.1051/parasite/1990651019. [DOI] [PubMed] [Google Scholar]
- 11.Gibson W, Stevens J. Genetic exchange in the trypanosomatidae. Adv Parasitol. 1999;43:1–46. doi: 10.1016/s0065-308x(08)60240-7. [DOI] [PubMed] [Google Scholar]
- 12.Turner CM, Sternberg J, Buchanan N, Smith E, Hide G, Tait A. Evidence that the mechanism of gene exchange in Trypanosoma brucei involves meiosis and syngamy. Parasitology. 1990;101(Pt 3):377–86. doi: 10.1017/s0031182000060571. [DOI] [PubMed] [Google Scholar]
- 13.Chang KP, Reed SG, McGwire BS, Soong L. Leishmania model for microbial virulence: the relevance of parasite multiplication and pathoantigenicity. Acta Tropica. 2003;85(3):375–90. doi: 10.1016/s0001-706x(02)00238-3. [DOI] [PubMed] [Google Scholar]
- 14.Mattner J, Wandersee-Steinhäuser A, Pahl A, Röllinghoff M, Majeau G R, . Hochman PS, Bogdan C. Protection against Progressive Leishmaniasis by IFN-β. J Immunol. 2004;172:7574–82. doi: 10.4049/jimmunol.172.12.7574. [DOI] [PubMed] [Google Scholar]
- 15.Scott P. IFN-gamma modulates the early development of Th1 and Th2 responses in a murine model of cutaneous leishmaniasis. J Immunol. 1991;147(9):3149–55. [PubMed] [Google Scholar]
- 16.Almeida R, Norrish A, Levick M, Vetrie D, Freeman T, Vilo J, Ivens A, Lange U, Stober C, McCann S, Blackwell JM. From genomes to vaccines: Leishmania as a model. Philos Trans R Soc Lond B Biol Sci. 2002;357(1417):5–11. doi: 10.1098/rstb.2001.0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das BB, Sen N, Dasgupta SB, Ganguly A, Das R, Majumder HK. Topoisomerase research of kin-etoplastid parasite Leishmania, with special reference to development of therapeutics. Indian J Med Res. 2006;123:221–32. [PubMed] [Google Scholar]
- 18.Dutta S, Kolli BK, Tang A, Sassa S, Chang KP. Transgenic Leishmania model for delta-aminolevulinate-inducible monospecific uroporphyria: cytolytic phototoxicity initiated by singlet oxygen-mediated inactivation of proteins and its ablation by endosomal mobilization of cytosolic uroporphyrin. Eukaryot Cell. 2008;7(7):1146–57. doi: 10.1128/EC.00365-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Louis J, Gumy A, Voigt H, Röcken M, Launois P. Experimental cutaneous Leishmaniasis: a powerful model to study in vivo the mechanisms underlying genetic differences in Th subset differentiation. Euro J Dermatol. 2002;12(4):316–8. [PubMed] [Google Scholar]
- 20.El Fadili K, Imbeault M, Messier N, Roy G, Gourbal B, Bergeron M, Tremblay MJ, Légaré D, Ouellette M. Modulation of gene expression in human macrophages treated with the anti-leishmania pentavalent antimonial drug sodium stibogluconate. Ant-imicrob Agents Chemother. 2008;52(2):526–33. doi: 10.1128/AAC.01183-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, et al. The genome of the kinetoplastid parasite, Leishmania major . Science. 2005;309(5733):436–42. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, Quail MA. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet. 2007;39(7):839–47. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Britto C, Ravel C, Bastien P, Blaineau C, Pagès M, Dedet JP, Wincker P. Conserved linkage groups associated with large-scale chromosomal rea-rrangements between Old World and New World Leishmania genomes. Gene. 1998;222(1):107–17. doi: 10.1016/s0378-1119(98)00472-7. [DOI] [PubMed] [Google Scholar]
- 24.Wincker P, Ravel C, Blaineau C, Pages M, Jauffret Y, Dedet JP, Bastien P. The Leishmania genome comprises 36 chromosomes conserved across widely divergent human pathogenic species. Nucleic Acids Res. 1996;24(9):1688–94. doi: 10.1093/nar/24.9.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravel C, Macari F, Bastien P, Pagès M, Blaineau C. Conservation among Old World Leishmania species of six physical linkage groups defined in Leishmania infantum small chromosomes. Mol Biochem Parasitol. 1995;69(1):1–8. doi: 10.1016/0166-6851(94)00166-k. [DOI] [PubMed] [Google Scholar]
- 26.Ravel C, Dubessay P, Britto C, Blaineau C, Bastien P, Pagès M. High conservation of the fine-scale org-anisation of chromosome 5 between two pathogenic Leishmania species. Nucleic Acids Res. 1999;27(12):2473–7. doi: 10.1093/nar/27.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDonagh PD, Peter J., Myler PJ, Stuart K. The unusual gene organization of Leishmania major chromosome 1 may reflect novel transcription processes. Nucleic Acids Res. 2000;28(14):2800–3. doi: 10.1093/nar/28.14.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myler PJ, Audleman L, deVos T, Hixson G, Kiser P, Lemley C, Magness C, Rickel E, Sisk E, Sunkin S, Swartzell S, Westlake T, Bastien P, Fu G, Ivens A, Stuart K. Leishmania major Friedlin chromosome 1 has an unusual distribution of protein-coding genes. Proc Natl Acad Sci U S A. 1999;96(6):2902–6. doi: 10.1073/pnas.96.6.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myler PJ, Sisk E, . McDonagh PD, Martınez-Calvillo S, Schnaufer A, Sunkin SM, Yan S, Madhubala R, Ivens A, Stuart K. Genomic org-anization and gene function in leishmania . Bioche Soc Trans. 2000;28(5):527–31. doi: 10.1042/bst0280527. [DOI] [PubMed] [Google Scholar]
- 30.Myler PJ, Beverley SM, Cruz AK, Dobson DE, Ivens AC, McDonagh PD, Madhubala R, Martınez-Calvillo S, Ruiz JC, Saxena A, Sisk E, Sunkin SM, Worthey E, Yan S, Stuart KD. The Leishmania genome project: new insights into gene organization and function. Med Microbiol Immunol. 2001;190(1-2):9–12. doi: 10.1007/s004300100070. [DOI] [PubMed] [Google Scholar]
- 31.Worthey EA, Martınez-Calvillo S, Schnaufer A, et al. Leishmania major chromosome 3 contains two long convergent polycistronic gene clusters separated by a tRNA gene. Nucleic Acids Res. 2003;31(14):4201–10. doi: 10.1093/nar/gkg469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen-Freue G, Holzer TR, Forney JD, McMaster WR. Global gene expression in Leishmania . Int J Parasitol. 2007;37(10):1077–86. doi: 10.1016/j.ijpara.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 33.Lighthall GK, Giannini SH. The chromosomes of Leishmania . Parasitol Today. 1992;8(6):192–9. doi: 10.1016/0169-4758(92)90263-2. [DOI] [PubMed] [Google Scholar]
- 34.Ouellette M, Papadopoulou B. Mechanisms of drug resistance in Leishmania . Parasitol Today. 1993;9(5):150–3. doi: 10.1016/0169-4758(93)90135-3. [DOI] [PubMed] [Google Scholar]
- 35.Segovia M. Leishmania gene amplification: a mechanism of drug resistance. Ann Trop Med Parasitol. 1994;88(2):123–30. doi: 10.1080/00034983.1994.11812849. [DOI] [PubMed] [Google Scholar]
- 36.Singh AK, Papadopoulou B, Ouellette M. Gene amplification in amp-hotericin B-resistant Leishmania tarentolae . Exp Parasitol. 2001;99(3):141–7. doi: 10.1006/expr.2001.4663. [DOI] [PubMed] [Google Scholar]
- 37.Haimeur A, Ouellette M. Gene Amplification in Leishmania tarentolae Selected for Resistance to Sodium Stibogluconate. Antimicrob Agents Chemother. 1998;24(7):1689–94. doi: 10.1128/aac.42.7.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flinn HM, Smith DF. Genomic organization and expression of a differentially regulated gene family from Leishmania major . Nucleic Acids Res. 1992;20(4):755–62. doi: 10.1093/nar/20.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gentil LG, Lasakosvitsch F, Silveira JF, Santos MR, Barbiéri CL. Analysis and chromosomal mapping of Leishmania (Leishmania) amazonensis amastigote expressed sequence tags. Mem Inst Oswaldo Cruz. 2007;102(6):707–11. doi: 10.1590/s0074-02762007005000065. [DOI] [PubMed] [Google Scholar]
- 40.Fong D, Lee B. Beta tubulin gene of the parasitic protozoan Leishmania Mexicana . Mol Biochem Parasitol. 1983;31(1):97–106. doi: 10.1016/0166-6851(88)90149-1. [DOI] [PubMed] [Google Scholar]
- 41.Hejazi SH, Zia-Jahromi N, Bandehpour M, Eslami G, Salehi R, Khamesipour A, Kazemi B. Gene Cloning of Iranian Leishmania major Mannose-1-Phosphate Guanyltransferase. Iran J Parasitol. 2009;4(3):1–9. [Google Scholar]
- 42.Kheirandish F, Bandehpour M, Haghighi A, Mohebali M, Mahboudi F, Kazemi B. Molecular cloning, Expression and Enzymatic assay of Iranian Leishmania major pteridine reductase 1. Iran j Parasitol. 2008;3(2):1–9. [Google Scholar]
- 43.Rasouli M, Zavaran Hoseini A, Kazemi B, Alborzi A, Kiany S. Expression of heat shock protein 70 of MCAN/IT/96/LON-49, a tool for diagnosis and future vaccine research. Iran J Immunol. 2009;6(2):75–86. [PubMed] [Google Scholar]
- 44.Kazemi B, Tohidi F, Bandehpour M, Yarian F. Molecular Cloning, Expression, and Enzymatic Assay of Pteridine Reductase 1 from Iranian Lizard Leishmania . Iran Biomed J. 2010;14(3):97–102. [PMC free article] [PubMed] [Google Scholar]
- 45.Shaddel M, Oormazdi H, Akhlaghi L, Kazemi B, Bandehpour M. Cloning of Leishmania Major P4 Gene. Yahkhteh Medical Journal. 2008;10(3):201–4. [Google Scholar]
- 46.Lee MG, Van der Ploeg LH. Transcription of protein- coding genes in trypanosomes by RNA polymerase I. Ann Rev Microbiol. 1997;51:463–89. doi: 10.1146/annurev.micro.51.1.463. [DOI] [PubMed] [Google Scholar]
- 47.Borst P. Discontinuous transcription and antigenic variation in try-panosomes. Annu Rev Biochem. 1986;55:701–32. doi: 10.1146/annurev.bi.55.070186.003413. [DOI] [PubMed] [Google Scholar]
- 48.MartI′nez-Calvillo S, Yan S, Nguyen D, Fox M, Stuart K, Myler PJ. Transcription of Leishmania major Friedlin Chromosome 1 Initiates in Both Directions within a Single Region. Mol Cell. 2003;11(5):1291–9. doi: 10.1016/s1097-2765(03)00143-6. [DOI] [PubMed] [Google Scholar]
- 49.Monnerat S, Martınez-Calvillo S, Worthey E, Myler PJ, Stuart KD, Fasel N. Genomic organization and gene expression in a chromosomal region of Leishmania major . Mol Biochem Parasitol. 2004;134(2):233–43. doi: 10.1016/j.molbiopara.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 50.Dumas C, Chow C, Müller M, Papadopoulou B. A Novel Class of Developmentally Regulated Non-coding RNAs in Leishmania . Eukaryot Cell. 2006;5(12):2033–2046. doi: 10.1128/EC.00147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller SI, Wirth DF. Trans splicing in Leishmania enriettii and identification of ribonucleoprotein complexes containing the spliced leader and U2 equivalent RNAs. Mol Cell Biol. 1988;8(6):2597–603. doi: 10.1128/mcb.8.6.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang Y, Walsh CE. Spliceosome-Mediated RNA Trans-splicing. Mol Ther. 2005;12(6):1006–12. doi: 10.1016/j.ymthe.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 53.Nilsen TW. Trans-splicing: an update. Mol Biochem Parasitol. 1995;73(1-2):1–6. doi: 10.1016/0166-6851(94)00107-x. [DOI] [PubMed] [Google Scholar]
- 54.Nilsen TW. Evolutionary origin of SL-addition trans-splicing: still an enigma. Trends in Genet. 2001;17(12):678–80. doi: 10.1016/s0168-9525(01)02499-4. [DOI] [PubMed] [Google Scholar]
- 55.Pouchkina-Stantcheva NN, Tunnacliffe A. Spliced leader RNA-mediated trans-splicing in phylum Rotifera. Mol Biol Evol. 2005;22(6):1482–9. doi: 10.1093/molbev/msi139. [DOI] [PubMed] [Google Scholar]
- 56.Zhang H, Hou Y, Miranda L, Campbell DA, Sturm NR, Gaasterland T, Lin S. Spliced leader RNA trans-splicing in dinoflagellates. Proc Natl Acad Sci USA. 2007;104(11):4618–23. doi: 10.1073/pnas.0700258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nilsen TW. Trans-splicing in protozoa and helmiths. Infect Agents Dis. 1992;1(4):212–8. [PubMed] [Google Scholar]
- 58.Nilsen TW. Trans-splicing of nematode premessenger RNA. Annu Rev Microbiol. 1993;47:413–40. doi: 10.1146/annurev.mi.47.100193.002213. [DOI] [PubMed] [Google Scholar]
- 59.Brehm K, Jensen K, Frosch M. mRNA Trans-splicing in the Human Parasitic Cestode Echinococcus multilocularis . J Biol Chem. 2000;275(49):38311–8. doi: 10.1074/jbc.M006091200. [DOI] [PubMed] [Google Scholar]
- 60.Davis RE. Spliced leader RNA trans-splicing in metazoa. Parasitol Today. 1996;12(1):33–40. doi: 10.1016/0169-4758(96)80643-0. [DOI] [PubMed] [Google Scholar]
- 61.Requena JM, Quijada L, Soto M, Alonso C. Conserved nucleotides surrounding the trans-splicing acceptor site and the translation initiation codon in leishmania genes. Exp Parasitol. 2003;103(1-2):78–81. doi: 10.1016/s0014-4894(03)00061-4. [DOI] [PubMed] [Google Scholar]
- 62.Gopal S, Awadalla S, Gaasterland T, Cross GA. A computational inv-estigation of kinetoplastid trans-splicing. Genome Biol. 2005;6(11):R95. doi: 10.1186/gb-2005-6-11-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carter NS, Yates P, Arendt CS, Boitz JM, Ullman B. Purine and pyrimidine metabolism in Leishmania . Adv Exp Med Biol. 2008;625:141–54. doi: 10.1007/978-0-387-77570-8_12. [DOI] [PubMed] [Google Scholar]
- 64.Marr JJ, Berens RL, Nelson DJ. Purine metabolism in Leishmania donovani and Leishmania braziliensis . Biochim Biophys Acta. 1978;544(2):360–71. doi: 10.1016/0304-4165(78)90104-6. [DOI] [PubMed] [Google Scholar]
- 65.LaFon SW, Nelson DJ, Berens RL, Marr JJ. Purine and pyrimidine salvage pathways in Leishmania donovani . Biochem Pharmacol. 1982;31(2):231–8. doi: 10.1016/0006-2952(82)90216-7. [DOI] [PubMed] [Google Scholar]
- 66.Datta AK, Datta R, Sen B. Antiparasitic chemotherapy: tinkering with the purine salvage pathway. Adv Exp Med Biol. 2008;625:116–32. doi: 10.1007/978-0-387-77570-8_10. [DOI] [PubMed] [Google Scholar]
- 67.Cunningham ML, Beverley SM. Pteridine salvage throughout the Leishmania infectious cycle: imp-lications for antifolate chemotherapy. Mol Biochem Parasitol. 2001;113:199–213. doi: 10.1016/s0166-6851(01)00213-4. [DOI] [PubMed] [Google Scholar]
- 68.Morales J, Mogi T, Mineki S, Takashima E, Mineki R, Hirawake H, Sakamoto K, Omura S, Kita K. Novel mitochondrial complex II isolated from Trypanosoma cruzi is composed of 12 peptides including a heterodimeric Ip subunit. J Biol Chem. 2009;284(11):7255–63. doi: 10.1074/jbc.M806623200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kita K, Nihei C, Omitsuka E. Parasite mitochondria as drug target: diversity and dynamic changes during the life cycle. Curr Med Chem. 2003;10(23):2535–48. doi: 10.2174/0929867033456549. [DOI] [PubMed] [Google Scholar]
- 70.Biscardi AM, Lopez LM, de Pahn EM, Pellegrino de Iraldi A, Stoppani AO. Effect of dyskinetoplastic agents on ultrastructure and oxidative pho-sphorylation in Crithidia fasciculate . Biocell. 2001;25(1):43–51. [PubMed] [Google Scholar]
- 71.Yatawara L, Le TH, Wickramasinghe S, Agatsuma T. Maxicircle (mitochondrial) genome sequence (partial) of Leishmania major: gene content, arrangement and composition compared with Leishmania tarentolae . Gene. 2008;424(1-2):80–6. doi: 10.1016/j.gene.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 72.Chen J, Rauch CA, White JH, Englund PT, Cozzarelli NR. The topology of the kinetoplast DNA network. Cell. 1995;80(1):61–9. doi: 10.1016/0092-8674(95)90451-4. [DOI] [PubMed] [Google Scholar]
- 73.Englund PT. Free minicircles of kinetoplast DNA in Crithidia fasciculata . J Biol Chem. 1979;254(11):4895–900. [PubMed] [Google Scholar]
- 74.Martynkina LP, Novikova EG, Kolesnikov AA, Strel'tsov SA, Semenov TE, Vengerov IuIu. Structural organization of kinetoplast DNA and its compactization in a model system in vitro. Mol Biol (Mosk) 1989;23(6):1645–57. [PubMed] [Google Scholar]
- 75.Ntambi JM, Englund PT. A gap at a unique location in newly replicated kinetoplast DNA minicircles from Trypanosoma equiperdum . J Biol Chem. 1985;260(9):5574–9. [PubMed] [Google Scholar]
- 76.Ntambi JM, Shapiro TA, Ryan KA, Englund PT. Ribonucleotides associated with a gap in newly replicated kinetoplast DNA minicircles from Trypanosoma equiperdum . J Biol Chem. 1986;261(25):11890–5. [PubMed] [Google Scholar]
- 77.Pérez-Morga DL, Englund PT. The attachment of minicircles to kinetoplast DNA networks during replication. Cell. 1993;74(4):703–11. doi: 10.1016/0092-8674(93)90517-t. [DOI] [PubMed] [Google Scholar]
- 78.Chen J, Englund PT, Cozzarelli NR. Changes in network topology during the replication of kinetoplast DNA. EMBO J. 1995;14(24):6339–47. doi: 10.1002/j.1460-2075.1995.tb00325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lindsay ME, Gluenz E, Gull K, Englund PT. A new function of Trypanosoma brucei mitochondrial topoisomerase II is to maintain kinetoplast DNA network topology. Mol Microbiol. 2008;70(6):1465–76. doi: 10.1111/j.1365-2958.2008.06493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sela D, Yaffe N, Shlomai J. Enzymatic mechanism controls redox-mediated protein-DNA interactions at the replication origin of kinetoplast DNA minicircles. J Biol Chem. 2008;283(46):32034–44. doi: 10.1074/jbc.M804417200. [DOI] [PubMed] [Google Scholar]
- 81.Onn I, Milman-Shtepel N, Shlomai J. Redox potential regulates binding of universal minicircle sequence binding protein at the kinetoplast DNA replication origin. Eukaryot Cell. 2004;3(2):277–87. doi: 10.1128/EC.3.2.277-287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Milman N, Motyka SA, Englund PT, Robinson D, Shlomai J. Mitochondrial origin-binding protein UMSBP mediates DNA replication and segregation in trypanosomes. Proc Natl Acad Sci USA. 2007;104(49):19250–5. doi: 10.1073/pnas.0706858104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sela D, Shlomai J. Regulation of UMSBP activities through redox-sensitive protein domains. Nucleic Acids Res. 2009;37(1):279–88. doi: 10.1093/nar/gkn927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ferguson ML, Torri AF, Pérez-Morga D, Ward DC, Englund PT. Kinetoplast DNA replication: mechanistic differences between Trypanosoma brucei and Crithidia fasciculata . J Cell Biol. 1994;126(3):631–9. doi: 10.1083/jcb.126.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Horton TL, Landweber LF. Rewriting the information in DNA: RNA editing in kinetoplastids and myxomycetes. Curr Opin Microbiol. 2002;5(6):620–6. doi: 10.1016/s1369-5274(02)00379-x. [DOI] [PubMed] [Google Scholar]
- 86.Estévez AM, Simpson L. Uridine insertion/deletion RNA editing in trypanosome mitochondria–a review. Gene. 1990;240(2):247–60. doi: 10.1016/s0378-1119(99)00437-0. [DOI] [PubMed] [Google Scholar]
- 87.Benne R. RNA editing in mitochondria of Leishmania tarentolae and Crithidia fasciculata . Semin Cell Biol. 1993;4(4):241–9. doi: 10.1006/scel.1993.1029. [DOI] [PubMed] [Google Scholar]
- 88.Sturm NR, Simpson L. Leishmania tarentolae minicircles of different sequence classes encode single guide RNAs located in the variable region approximately 150 bp from the conserved region. Nucleic Acids Res. 1991;19(22):6277–81. doi: 10.1093/nar/19.22.6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Weng J, Aphasizheva I, Etheridge RD, Huang L, Wang X, Falick AM, Aphasizhev R. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol Cell. 2008;32(2):198–209. doi: 10.1016/j.molcel.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60(2):189–98. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- 91.de la Cruz VF, Lake JA, Simpson AM, Simpson L. A minimal ribosomal RNA: sequence and secondary structure of the 9S kinetoplast ribosomal RNA from Leishmania tarentolae . Proc Natl Acad Sci USA. 1985;82(5):1401–5. doi: 10.1073/pnas.82.5.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Simpson L, Simpson AM, Kidane G, Livingston L, Spithill W. The kinetoplast DNA of the hemoflagellate protozoa. Am J Trop Med Hyg. 1980;29(5 Suppl):1053–63. doi: 10.4269/ajtmh.1980.29.1053. [DOI] [PubMed] [Google Scholar]
- 93.Masuda H, Simpson L, Rosenblatt H, Simpson AM. Restriction map, partial cloning and localization of 9S and 12S kinetoplast RNA genes on the maxicircle component of the kinetoplast DNA of Leishmania tarentolae . Gene. 1979;6(1):51–73. doi: 10.1016/0378-1119(79)90085-4. [DOI] [PubMed] [Google Scholar]
- 94.Simpson AM, Neckelmann N, de la Cruz VF, Muhich ML, Simpson L. Mapping and 5' end determination of kinetoplast maxicircle gene transcripts from Leishmania tarentolae . Nucleic Acids Res. 1985;13(16):5977–93. doi: 10.1093/nar/13.16.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.de la Cruz VF, Simpson AM, Lake JA, Simpson L. Primary sequence and partial secondary structure of the 12S kinetoplast (mitochondrial) ribosomal RNA from Leishmania tarentolae: conservation of peptidyl-transferase structural elements. Nucleic Acids Res. 1985;13(7):2337–56. doi: 10.1093/nar/13.7.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rauch CA, Perez-Morga D, Cozzarelli NR, Englund PT. The absence of supercoiling in kinetoplast DNA minicircles. EMBO J. 1993;12(2):403–11. doi: 10.1002/j.1460-2075.1993.tb05672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Golas MM, Böhm C, Sander B, Effenberger K, Brecht M, Stark H, Ulrich Göringer HU. Snapshots of the RNA editing machine in trypanosomes captured at different assembly stages in vivo. EMBO J. 2009;28:766–78. doi: 10.1038/emboj.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.de la Cruz VF, Neckelmann N, Simpson L. Sequences of six genes and several open reading frames in the kinetoplast maxicircle DNA of Leishmania tarentolae . J Biol Chem. 1984;259(24):15136–47. [PubMed] [Google Scholar]
- 99.de Souza W, Attias M, Rodrigues JC. Particularities of mitochondrial structure in parasitic protists (Apicomplexa and Kinetoplastida) Int J Biochem Cell Biol. 2009;41(10):2069–80. doi: 10.1016/j.biocel.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 100.Motta MC. Kinetoplast as a potential chemotherapeutic target of trypanosomatids. Curr Pharm Des. 2008;14(9):847–54. doi: 10.2174/138161208784041051. [DOI] [PubMed] [Google Scholar]
- 101.Rosenzweig D, Smith D, Myler PJ, Olafson RW, Zilberstein D. Post-translational modification of cellular proteins during Leishmania donovani differentiation. Proteomics. 2008;8(9):1843–50. doi: 10.1002/pmic.200701043. [DOI] [PubMed] [Google Scholar]
- 102.Hem S, Gherardini PF, Osorio y Fortéa J, Hourdel V, Morales MA, Watanabe R, Pescher P, Kuzyk MA, Smith D, Borchers CH, Zilberstein D, Helmer-Citterich M, Namane A, Späth GF. Identification of Leishmania-specific protein phosphorylation sites by LC-ESI-MS/MS and comparative genomics analyses. Proteomics. 2010;10(21):3868–83. doi: 10.1002/pmic.201000305. [DOI] [PubMed] [Google Scholar]
- 103.Kaur J, Sundar S, Singh N. Molecular docking, structure-activity relationship and biological evaluation of the anticancer drug monastrol as a pteridine reductase inhibitor in a clinical isolate of Leishmania donovani . J Antimicrob Chemother. 2010;65(8):1742–8. doi: 10.1093/jac/dkq189. [DOI] [PubMed] [Google Scholar]
- 104.Kumar P, Kumar A, Verma SS, Dwivedi N, Singh N, Siddiqi MI, Tripathi RP, Dube A, Singh N. Leishmania donovani pteridine reductase 1: biochemical properties and structure-modeling studies. Exp Parasitol. 2008;120(1):73–9. doi: 10.1016/j.exppara.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 105.Basile G, Peticca M. Recombinant protein expression in Leishmania tarentolae . Mol Biotechnol. 2009;43(3):273–8. doi: 10.1007/s12033-009-9213-5. [DOI] [PubMed] [Google Scholar]
- 106.Soleimani M, Mahboudi F, Davoudi N, Amanzadeh A, Azizi M, Adeli A, Rastegar H, Barkhordari F, Mohajer-Maghari B. Expression of human tissue plasminogen activator in the trypanosomatid protozoan Leishmania tarentolae . Biotechnol Appl Biochem. 2007;48(Pt 1):55–61. doi: 10.1042/BA20060217. [DOI] [PubMed] [Google Scholar]
- 107.Hemayatkar M, Mahboudi F, Majidzadeh-A K, Davami F, Vaziri B, Barkhordari F, Adeli A, Mahdian R, Davoudi N. Increased expression of recombinant human tissue pla-sminogen activator in Leishmania tarentolae . Biotechnol J. 2010;5(11):1198–206. doi: 10.1002/biot.201000233. [DOI] [PubMed] [Google Scholar]
- 108.Mirzaahmadi S, Asaadi Tehrani G, Bandehpour M, Davoudi N, Tahmasbi L, Mirzahoseini H, Parivar K, Kazemi B. Expression of recombinant human coagulation factor VII by the Lizard Leishmania expression system. JBB. 2011 doi: 10.1155/2011/873874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sereno D, Lemesre JL. Axenically Cultured Amastigote Forms as an In Vitro Model for Investigation of Antileishmanial Agents. Antimicrob Agen Chemothe. 1997;41(5):972–6. doi: 10.1128/aac.41.5.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gupta S, Nishi Visceral leishmaniasis: Experimental models for drug discovery. Indian J Med Res. 2011;133:27–39. [PMC free article] [PubMed] [Google Scholar]
- 111.Tulloch LB, Martini VP, Iulek J, Huggan JK, Lee JH, Gibson CL, Smith TK, Suckling CJ, Hunter WN. Structure-based design of pteridine reductase inhibitors targeting African sleeping sickness and the leishmaniases. J Med Chem. 2010;53(1):221–229. doi: 10.1021/jm901059x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cavazzuti A, Paglietti G, Hunter WN, Gamarro F, Piras S, Loriga M, Allecca S, Corona P, McLuskey K, Tulloch L, Gibellini F, Ferrari S, Costi MP. Discovery of potent pteridine reductase inhibitors to guide antiparasite drug development. Proc Natl Acad Sci U S A. 2008;105(5):1448–1453. doi: 10.1073/pnas.0704384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Panavas T, Sanders C, Butt TR. SUMO fusion technology for enhanced protein production in prokaryotic and eukaryotic expression systems. Methods Mol Biol. 2009;497:303–317. doi: 10.1007/978-1-59745-566-4_20. [DOI] [PubMed] [Google Scholar]
- 114.Peroutka Iii RJ, Orcutt SJ, Strickler JE, Butt TR. SUMO fusion technology for enhanced protein expression and purification in prokaryotes and eukaryotes. Methods Mol Biol. 2011;705:15–30. doi: 10.1007/978-1-61737-967-3_2. [DOI] [PubMed] [Google Scholar]
- 115.García-Estrada C, Reguera RM, Villa H, Requena JM, Müller S, Pérez-Pertejo Y, Balaña-Fouce R, Ordóñez D. Identification of a gene in Leishmania infantum encoding a protein that contains a SP-RING/MIZ zinc finger domain. Biochim Biophys Acta. 2003;1629(1-3):44–52. doi: 10.1016/j.bbaexp.2003.07.001. [DOI] [PubMed] [Google Scholar]