Abstract
The condensation of bacteriophage phi29 genomic DNA into its preformed procapsid requires the DNA packaging motor, which is the strongest known biological motor. The packaging motor is an intricate ring-shaped protein/ RNA complex, and its function requires an RNA component called packaging RNA (pRNA). Current structural information on pRNA is limited, which hinders studies of motor function. Here, we used site-directed spin labeling to map the conformation of a pRNA three-way junction that bridges binding sites for the motor ATPase and the procapsid. The studies were carried out on a pRNA dimer, which is the simplest ring-shaped pRNA complex and serves as a functional intermediate during motor assembly. Using a nucleotide-independent labeling scheme, stable nitroxide radicals were attached to eight specific pRNA sites without perturbing RNA folding and dimer formation, and a total of 17 internitroxide distances spanning the three-way junction were measured using Double Electron–Electron Resonance spectroscopy. The measured distances, together with steric chemical constraints, were used to select 3662 viable three-way junction models from a pool of 65 billion. The results reveal a similar conformation among the viable models, with two of the helices (HT and HL) adopting an acute bend. This is in contrast to a recently reported pRNA tetramer crystal structure, in which HT and HL stack onto each other linearly. The studies establish a new method for mapping global structures of complex RNA molecules, and provide information on pRNA conformation that aids investigations of phi29 packaging motor and developments of pRNA-based nanomedicine and nanomaterial.
INTRODUCTION
RNA participates in all cellular processes associated with the maintenance and expression of genetic information, and knowledge of molecular basis of RNA function is essential for understanding basic biology.1 A key in understanding RNA function is information on its three-dimensional conformations, which can be highly complex as demonstrated by the rapidly growing number of high-resolution structures of RNA and RNA/protein complex.2,3 To advance our ability to derive RNA structural information under physiological conditions, here we present work on mapping the global structure of an RNA junction using the method of site-directed spin labeling (SDSL).4 In SDSL, chemically stable nitroxide radicals (i.e., the spin labels) are covalently attached at specific sites of a macromolecule. The behavior of the nitroxide is monitored using electron paramagnetic resonance (EPR) spectroscopy, from which local information on the macromolecule is obtained. SDSL can be applied to study structure and dynamics of large biomolecular complexes under physiological conditions, and has been demonstrated to provide unique information on proteins5–7 and nucleic acids.8–10 One of the EPR observables used in SDSL studies is the distance between a pair of nitroxides, which can be obtained by measuring electron spin dipolar coupling using either continuous-wave (cw-) EPR or more recently, pulsed EPR techniques.8,11 In particular, pulsed Double Electron–Electron Resonance (DEER or PELDOR)12–14 has been developed and successfully applied to measure distance between 20 and 80 Å in biological systems.15 In protein studies, many examples have been reported in which the DEER measured distances enable monitoring of conformational change16–22 and direct assessment of protein structure.23–30 In addition, DEER measured distances have been used to monitor RNA conformational changes upon ligand binding.31–33
Here, we report the use of multiple DEER distances for de novo mapping of the global structure (i.e., the overall shape) of a three-way junction (3-wj) in a noncoding RNA, the packaging RNA34 (pRNA, also known as prohead RNA) in the DNA packaging motor of bacteriophage phi29. Phi29 packaging motor utilizes chemical energy derived from hydrolyzing host ATP to condense its linear double-stranded DNA genome into a preformed capsid.35,36 It is reported to be the strongest bio-molecular motor, capable of generating forces that are 2- to 8-fold higher than myosin and RNA polymerase.37 Interestingly, the phi29 packaging motor is a protein/RNA complex, with the RNA component (pRNA) being essential for in vivo and in vitro motor function.34 Studies have shown that pRNA forms an oligomeric ring within the motor, with pRNA monomers interacting with each other through intermolecular base-pairing between two loop regions (i.e., the R- and L-loop, Figure 1A) in an Mg2+ dependent fashion.35,36,38 The exact composition of the pRNA ring has been a subject of debate,38–43 although it is clear that motor functions are preserved with pRNA mutants in which the R- and L-loops maintain intermolecular base-pairing.38,39 As pRNA significantly stimulates motor ATPase (i.e., gp16) activity,44,45 information on pRNA structure and function is an integral part of understanding the mechanism of the phi29 motor. Furthermore, pRNA-based constructs, such as dimers, trimers, and their variants, have been used in developing novel artificial nanostructures for material and therapeutic applications.46 Understanding of pRNA conformations will be highly beneficial for these efforts.
Figure 1.
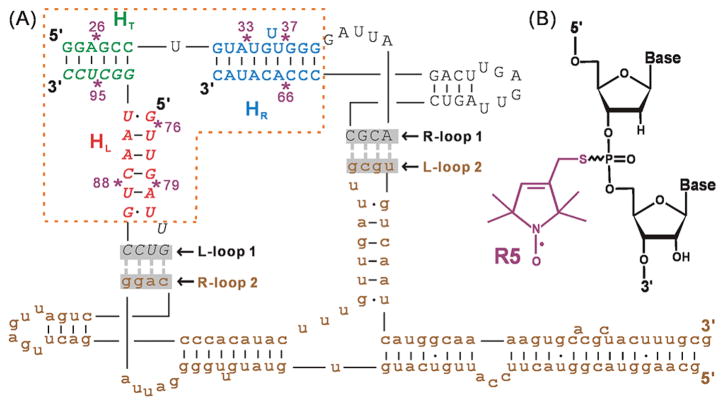
(A) Dimer construct used for SDSL mapping of pRNA 3-wj global structure (see also Supporting Information Figure S1). Upper-case letters show the two respective RNA strands constituting br_B/a′, and brown lower-case letters show the unlabeled full-length monomer A/b′. The 3-wj is indicated by the dotted box, with the HT (green), HR (blue), and HL (red) helices marked. Spin labeling sites are indicated by “*” and numbered according to the corresponding full-length pRNA sites. The two sets of interacting R- and L-loops are marked and shadowed. (B) The R5 spin label. Note that following previously validated distance measurement protocols,51,70,71 all data reported here were acquired without separating the Rp and Sp phosphorothioate diastereomers present at each attachment site.
Currently available information on the conformation of pRNA includes a model of a dimer that was initially constructed using chemical probing and photo-cross-linking data47 and then further refined using inter-pRNA distances measured by single-molecule fluorescence resonance energy transfer;48 an NMR structure of a hairpin subdomain within a pRNA monomer;49 and a 3.5 Å crystal structure of a pRNA tetramer.43 However, many questions remain unresolved. For example, the dimer model48 and the tetramer crystal structure43 show differences in the conformation of the junction defined by three pRNA helices designated as HT, HR, and HL (Figure 1A, Supporting Information Figure S1). This junction bridges binding sites for the motor ATPase (binding to the extended HT) and the procapsid (binding to HR/HL),35,36 and its conformation is of great interest in elucidating the motor mechanism. However, in the dimer studies, there is no reported distance measurement spanning this 3-wj within the monomeric unit;48 consequently, this junction conformation within a pRNA dimer remains to be determined.
In this work, using a nucleotide-independent nitroxide labeling scheme50,51 and DEER spectroscopy, 17 distances spanning the 3-wj were measured in a previously reported functional pRNA dimer. Modeling based on steric and distance constraints was carried out to reveal the spatial arrangement of the RNA helices, which defines the 3-wj global structure. The results reveal an alternative 3-wj fold in pRNA dimer as compared to that reported in the pRNA tetramer crystal structure,43 thus demonstrating versatility in pRNA conformation. These studies establish a new method for mapping global structures of complex RNA molecules, and provide information that may advance our understanding of phi29 packaging motor function as well as facilitate pRNA-based nanomedicine and nanomaterial developments.
MATERIALS AND METHODS
Synthesis of RNAs
The two pRNA constructs used in this work are designated as A/b′ and br_B/a′ (Figure 1A and Supporting Information Figure S1). The 118-nucleotide (nt) A/b′ was generated by in vitro run-off transcription using a linearized double-stranded DNA template that contains a T7 RNA polymerase promoter followed by the RNA sequence,52 with the 5′ terminus of the DNA antisense strand mutated to 5′-GCGC-3′ to allow linearization of the plasmid using the Hinp1I restriction endonuclease (cleaving 5′…G/CGC…3′, New England Biolabs, Inc.). The transcribed RNA was purified by denaturing polyacrylamide gel electrophoresis (PAGE), then quantified and stored as previously described.52
The br_B/a′ construct contains two noncovalently linked RNA strands designated as a′_23 and B_49 (Supporting Information Figure S1C), with nucleotides numbered according to the corresponding 118-nt RNA. The 23-nt a′_23 RNA was generated by solid-phase chemical synthesis. The 49-nt B_49 RNA was generated by in vitro transcription using single-stranded DNA templates52 or by solid-phase chemical synthesis. All chemically synthesized oligonucleotides were purchased from Integrated DNA Technologies, Inc. (Coralville, IA).
Spin Labeling of RNAs
One or two nitroxide spin labels, 1-oxyl-2,2,5,5-tetramethylpyrroline (R5), were attached to either a′_23 or B_49 using the phosphorothioate labeling scheme.50,51 Specifically, phosphorothioate modifications were introduced at specific sites within a′_23 or B_49 during solid-phase chemical synthesis. A R5 precursor, 1-oxyl-2,2,5,5-tetramethyl-Δ3-methane-sulfonyloxy-methylpyrroline (Toronto Research Chemicals, North York, Canada) was activated and then immediately reacted with the crude oligonucleotides.51 Each labeling site was designated by the corresponding nucleotide number. Labeled a′_23 was purified using HPLC.51 Labeled B_49 was purified by denaturing PAGE, eluted in water, and recovered by ethanol precipitation. Temperature during gel purification and elution was controlled at 4 °C to minimize label detachment from RNA. RNA concentrations were quantified by UV absorbance at 260 nm using extinction coefficients of 227 000 and 488 000 M−1 cm−1 for a′_23 and B_49, respectively. The degree of nitroxide labeling was determined using a spin-counting procedure,53 and was found to be ~100% for all samples used in pulsed EPR measurements. Labeled RNAs were resuspended in deionized water for immediate use, or stored at −80 °C.
Biochemical Characterization of Spin Labeled pRNA Dimers
The dissociation constant (Kd) between32P labeled A/b′ RNA (*A/b′) and a partner RNA (unmodified br_B/a′, or br_B/a′ with spin label(s) attached) was measured in 50 mM Tris-HCl (pH 7.6) and 3 mM MgCl2. The ratio of a′_23 and B_49 was kept at 1:1. In each measurement, proper amount of individual RNA strands (e.g., *A/b′, a′_23, and B_49 for forming a dimer of A/b′||br_B/a′) were mixed, heated at 95 °C for 1 min, then cooled down at room temperature for 2 min. Proper amount of Tris-HCl (pH 7.6) and MgCl2 were then added to achieve the desired buffer concentrations, and the mixture was incubated at 17 °C for 1 h. Monomer and dimer were resolved using a native gel, which was run at ~17 °C with 50 mM Tris-HCl (pH 7.6) and 3 mM MgCl2 present in the electrophoresis solution. Gels were then dried and quantified using a Personal Molecular Imager (Bio-Rad, Inc.). Kd values were obtained by fitting the dependence of the fraction of dimer (α) versus partner RNA concentrations ([partner RNA]) to the following equation using the program Kaleidagraph (Synergy, PA):
| (1) |
Preparation of Spin Labeled pRNA Dimers for EPR Measurements
Spin labeled dimers of A/b′||br_B/a′ were assembled using A/b′, a′_23, and B_49, with R5 attached at specific sites within br_B/a′. The ratio of A/b′, a′_23, and B_49 was kept at 1.2:1:1. For each EPR sample, A/b′, a′_23, and B_49 were mixed together and lyophilized. The sample was resuspended in a glycerol solution, incubated at 95 °C for 1 min, and then cooled down at room temperature for 2 min. Proper amount of Tris-HCl (pH 7.6) and MgCl2 was added, and each sample was then incubated at 17 °C for 1 h before transferred to an EPR capillary (see below). The final EPR sample contained 50 mM Tris-HCl (pH 7.6), 3 mM MgCl2, 60 μM R5 labeled br_B/a′, 72 μM A/b′, and 50% (v/v) glycerol. Control experiments indicated that this assembling procedure affords the best balance between the desired pRNA dimer and the undesired monomer and higher oligomers (see Results and Supporting Information Figure S2). Assembling monomeric br_B/a′ from a′_23 and B_49 prior to dimer formation has no effect on measured dimer Kd and inter-R5 distances.
EPR Spectroscopy
DEER spectroscopy was carried out to measure inter-R5 distances. The 20-μL samples were placed in a round quartz capillary (2.0 mm i.d., 2.4 mm o.d., Vitrocom, Inc., Mountain Lakes, NJ) sealed at one end, and were flash-frozen in liquid nitrogen. Measurements were carried out at 80 K on a Bruker ELEXSYS E580 X-band spectrometer with an ER4118-MS3-EN resonator. A dead-time free four-pulse scheme14 was used, with the pump pulse frequency set at the center of the nitroxide spectrum and the observer frequency being approximately 70 MHz higher. The observer π pulse was 32 ns. The pump π pulse was optimized using a nutation experiment54 and was usually set at 24 or 28 ns. The video bandwidth was fixed at 200 MHz. The shot repetition time was set at 714 μs based on a measured T1 of approximately 560 μs.55 Accumulation time in each measurement ranged from 3 to 16 h with 1024 shots per point. Interspin distance distributions were computed from the resulting dipolar evolution data using Defit 3.7.21,55,56 In the analyses, background in the dipolar evolution data was corrected by fitting an exponential decay corresponding to a homogeneous 3-dimentional distribution of electron spin to the last half of the data. The analyses yielded two key parameters that describe the interspin distance distribution: the most probable distance (r0) and the width of the distribution (characterized by the half-width at the half-maximum, wr). On the basis of repeated measurements, errors in measured r0 were less than 7% of the reported values.
Continuous-wave EPR spectroscopy was carried out on a X-band Bruker EMX spectrometer as previously reported.53
Constructing RNA Modules for 3-wj Global Structure Modeling
To model the pRNA 3-wj, RNA helices corresponding to the respective sequences of HT, HR, and HL (Figure 1A) were built using standard A-form geometry.57,58 Note that the U35 bulge in the HR helix was modeled using a homologous U-bulge structure in the 16S rRNA.59 This bulge is known to be dispensable,43,60,61 and its inclusion in the HR module minimally affects the overall helix geometry and the subsequent interhelical distance calculations. For each helix, the previously developed NASNOX program51,62 was used to obtain the ensemble of sterically allowed R5 conformers at each nucleotide. The average coordinates of the R5 nitrogen atoms for each ensemble were computed and recorded as a pseudo atom (designated as “NOX”) associated with the corresponding nucleotide in the pdb file. In addition, the helical axis was generated using the program CURVES63 and recorded within the pdb file. In subsequent transformations of each helix, all associated NOX pseudo atoms and the helical axes were subjected to the same operations as that of the other atoms. Distances between pairs of NOX pseudo atoms were used to represent the expected inter-R5 distances between the corresponding nucleotides in a given model (see below).
Grid Search for Modeling of 3-wj Global Structure
Models of 3-wj were generated using the NOX-modified helices, with HT fixed while HR and HL independently translated and rotated as rigid bodies. A translation operation was implemented by adding an offset (Δx, Δy, Δz) to the coordinate of each atom within the respective helix. Each rotation corresponded to transformations about a set of Euler angles (α,β,γ) defined with respect to the same external reference frame, and was achieved via matrix multiplication operations. In-house programs written in MATLAB were generated to systematically vary the 12 parameters corresponding to the independent transformation of HR and HL, as well as to assess each resulting model.
The grid search starts from a hand-built initial model that does not conform to the DEER measured distances but satisfies the following two sets of constraints. The first are steric constraints, which specify that the distance between any two atoms must be greater than the sum of the corresponding radii (i.e., no overlapping atoms). The second are connection constraints (affecting how far apart the helices are positioned), which are chosen based on the number of nucleotides spanning between two helices in the wild-type pRNA: HT and HL are separated by zero nucleotide, and thus, the distance from residue 91 O3′ to residue 92 C5′ was set as 0–4.5 Å (3 covalent bonds); HT and HR are separated by one nucleotide (U29), thus, the distance from residue 28 O3′ to residue 30 C5′ was set at 0–10 Å; and HR and HL are separated by three nucleotides (U72U73U74), thus, the distance from residue 71 O3′ to residue 75 C5′ was set at 0–20 Å. From the initial model, rotation parameters were individually varied from 0° to 360° with a step-size of 30°, and translation parameters were varied from −15 Å to 15 Å with a step-size of 5 Å. The resulting models that satisfy both steric and connection constraints were recorded and designated as the “sterically-allowed” ensemble.
The sterically allowable ensemble was then assessed according to the DEER measured distances. For each model, an RMSDdeer parameter was computed, which is defined as the root-mean-square-deviation (RMSDdeer) between DEER measured inter-R5 distances (rdeer) and corresponding inter-NOX distances (rmodel) (eq 2):
| (2) |
For a given model, the RMSDdeer values differ by less than 0.1 Å when rmodel was computed using the NOX pseudo atoms (see above) or calculated from the entire R5 ensembles obtained using NASNOX, with the latter being much more demanding on computation resources. Therefore, RMSDdeer calculated using NOX pseudo atoms was used throughout this work. Models with RMSDdeer ≤ 5 Å were deemed to satisfy the DEER constraints, and were designated as viable models. Upon identifying the ensemble of viable models, the model with the lowest RMSDdeer value was again used as the starting model to carry out a fine search, in which each rotation parameter was varied from the existing value in a ±15° range with a step-size of 5°, and each translation parameter was varied from the existing value in a ±1 Å range with a step-size of 1 Å. The model with the lowest RMSDdeer value from the fine search was selected as the best-fit model.
To assess the impact of the widths of DEER measured distance distributions (wr) on the outcome of 3-wj modeling, the sterically allowed ensemble described above was ranked according to a modified RMSD metric (RMSDmod) defined as:
| (3) |
Random Docking Search for Modeling the 3-wj Global Structure
With HT, HR, and HL modules described above, models of 3-wj were obtained using a conformational description similar to that presented in a previous work64 and a conformational sampling method based on Metropolis Monte Carlo algorithm. The models were assessed using steric, connection, and DEER constraints as described above.
Characterization of Structural Models
Heavy atom root-mean-square-deviations between structural models were calculated using the program VMD.65 For each model, interhelical angles between HT/HL, HT/HR, and HR/HL were computed from the dot products of corresponding helical axes.
RESULTS
Biochemical Characterization of Spin-Labeled pRNAs
Our studies were carried out on a pRNA dimer, which has been proposed to serve as an intermediate during pRNA assembly.66 A pRNA dimer also represents the simplest ring-shaped pRNA complex, as it contains two sets of intermolecular R/L loop pairing that constrain the pRNA procapsid binding domain in a closed ring topology67 (Figure 1A). Two pseudo-symmetric pRNA monomers were used, where the R- and L-loops are designed to minimize homo-oligomer formations (e.g., self-dimer, trimer, etc.) and favor heterodimer assembly.52,66 To facilitate spin labeling, one monomer was substituted by a truncated 2-piece construct (designated as br_B/a′),52 in which the HT helix noncovalently staples the R- and L-loop subdomains together (Figure 1A). This two-piece construct is fully functional in forming pRNA/pRNA complexes52 and in supporting DNA packaging.68 All results reported here were obtained in the context of this pRNA dimer.
A phosphorothioate scheme50,51 was used to efficiently attach nitroxide spin labels (designated as R5, Figure 1B) at eight br_B/a′ sites (Figure 1A) that are not involved in pRNA inter-domain interactions.69 The use of R5 for measuring nanometer distances in DNA and RNA has been experimentally validated,70,71 and a program (NASNOX) has been established for fast and accurate interpretation of measured inter-R5 distances based on the parent nucleic acid structure.51,62 These prior studies set a solid foundation for mapping the pRNA 3-wj global structure.
For each R5-labeled br_B/a′, the standard state free energy of dimer formation (ΔG0) differs by <1.0 kcal/mol from that of the unmodified construct (Table 1, Supporting Information Figure S3). As ΔG° depends on proper pRNA folding to enable simultaneous formation of the two sets of R/L loop interaction,52 the small ΔG° changes indicate R5 labeling does not significantly disrupt pRNA folding.
Table 1.
Standard State Free Energy of Dimer Formation between the 118-nt A/b′ pRNA and br_B/a′a
| label positionb | Kd (nM)c | ΔG°17 °C (kcal/mol)d | ΔΔG°17 °C (kcal/mol)e | |
|---|---|---|---|---|
| None | 175 ± 63 | −8.96 ± 0.26 | - | |
| Single labeled | (26; --) | 273 ± 99 | −8.71 ± 0.26 | 0.25 |
| (33; --) | 219 ± 94 | −8.83 ± 0.32 | 0.13 | |
| (37; --) | 196 ± 32 | −8.90 ± 0.10 | 0.06 | |
| (66; --) | 198 ± 27 | −8.89 ± 0.08 | 0.07 | |
| (76; --) | 249 ± 109 | −8.76 ± 0.33 | 0.20 | |
| (79; --) | 203 ± 101 | −8.88 ± 0.40 | 0.08 | |
| (88; --) | 601 ± 326 | −8.25 ± 0.45 | 0.71 | |
| (95; --) | 252 ± 119 | −8.75 ± 0.37 | 0.21 | |
| Double labeled | (26; 37) | 493 ± 121 | −8.36 ± 0.16 | 0.60 |
| (26; 76) | 616 ± 158 | −8.24 ± 0.17 | 0.73 | |
| (26; 79) | 657 ± 160 | −8.20 ± 0.16 | 0.76 | |
| (26; 88) | 929 ± 200 | −8.00 ± 0.14 | 0.96 | |
| (33; 76) | 449 ± 73 | −8.42 ± 0.10 | 0.52 | |
| (33; 79) | 522 ± 16 | −8.33 ± 0.02 | 0.63 | |
| (33; 95) | 411 ± 143 | −8.47 ± 0.25 | 0.49 | |
| (37; 76) | 497 ± 131 | −8.36 ± 0.18 | 0.60 | |
| (37; 79) | 320 ± 5 | −8.61 ± 0.01 | 0.35 | |
| (37; 88) | 617 ± 158 | −8.24 ± 0.17 | 0.72 | |
| (37; 95) | 313 ± 121 | −8.63 ± 0.28 | 0.33 | |
| (66; 76) | 235 ± 56 | −8.79 ± 0.16 | 0.17 | |
| (66; 79) | 319 ± 59 | −8.61 ± 0.12 | 0.35 | |
| (66; 95) | 297 ± 33 | −8.66 ± 0.07 | 0.30 | |
| (76; 95) | 228 ± 1 | −8.81 ± 0.00 | 0.15 | |
| (79; 95) | 284 ± 52 | −8.68 ± 0.12 | 0.28 | |
| (88; 95) | 402 ± 79 | −8.48 ± 0.13 | 0.48 | |
Measurements were carried out at 17 °C in 50 mM Tris-HCl, pH 7.6, 3 mM MgCl2 as described in Materials and Methods.
Designated by the sequence number(s) of the R5 attachment site(s).
Errors obtained from multiple measurements.
Errors calculated from propagating errors of Kd measurement.
ΔΔG° = ΔG°(spin labeled br_B/a′) − ΔG°(unmodified br_B/a′).
Interhelical Distances Measured Using Pulsed EPR Spectroscopy
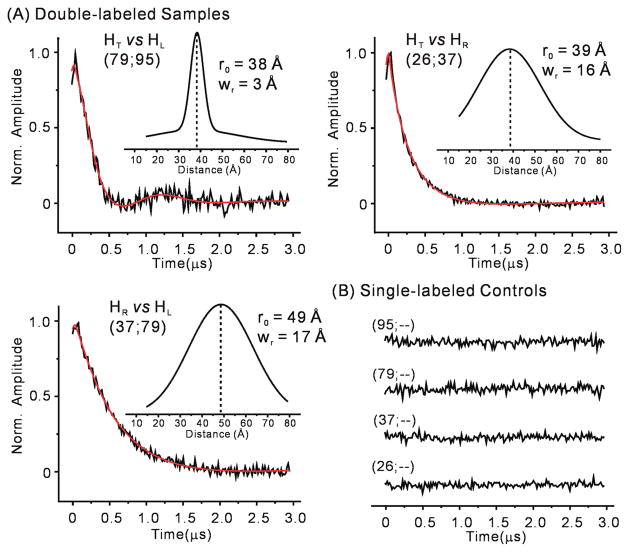
Upon confirming assembly of R5-labeled dimers under EPR conditions using native gels (Supporting Information Figure S2) and DEER (Supporting Information Figure S4), 17 sets of inter-R5 distances spanning the 3-wj were measured (Table 2). In each case, the normalized background-corrected dipolar evolution trace for the double-labeled sample showed a clear decay, while the corresponding single-labeled samples revealed flat traces without oscillation or decay pattern (Figure 2 and Supporting Information Figures S5 and S6). This ensures that distances measured using the double-labeled samples are not biased by RNA aggregation. The dipolar evolution traces were analyzed using the Defit program, in which one or more Gaussian functions are used to extract interspin distance distribution profiles.56 The analyses provide two key parameters to describe the resulting interspin distance distribution: the most probable distance (r0) and the width of distance distribution (wr) (see Materials and Methods). The measurements yielded r0 values ranging from 28 to 49 Å, with wr being 3–18 Å (Table 2, Figure 2, Supporting Information Figure S6 and Table S1). In most DEER measurements, an evolution time of 3 μs was used, which is sufficient for obtaining reliable r0 values up to 50 Å even in situations where a broad distance distribution results in a decaying DEER trace without clear oscillations.72 Consistent with previous reports,70,71 repeated measurements indicated that errors in measured r0 are <7% of the reported values. The r0 values were subsequently used as one of the main constraints in modeling (see below).
Table 2.
Interhelical Distances
| data seta | inter-R5 distance (Å)
|
|||
|---|---|---|---|---|
| DEER measuredb | EPR-based modelc | crystal structured | ||
| HT vs HL | (76; 95) | 28 | 26.4 | 27.6 |
| (79; 95) | 38 | 37.5 | 38.0 | |
| (88; 95) | 34 | 30.3 | 29.7 | |
| (26; 76) | 28e | 30.7 | 23.7 | |
| (26; 79) | 35e | 31.6 | 30.1 | |
| (26; 88) | 37 | 37.2 | 10.7 | |
| HT vs HR | (26; 37) | 39 | 38.6 | 44.8 |
| (33; 95) | 32e | 30.4 | 40.8 | |
| (37; 95) | 40 | 43.4 | 45.5 | |
| (66; 95) | 30e | 25.9 | 22.8 | |
| HR vs HL | (33; 76) | 39 | 41.0 | 23.5 |
| (33; 79) | 44 | 43.0 | 38.1 | |
| (37; 76) | 48 | 50.8 | 36.4 | |
| (37; 79) | 49 | 47.4 | 52.6 | |
| (37; 88) | 40 | 39.3 | 50.2 | |
| (66; 76) | 35 | 38.8 | 18.8 | |
| (66; 79) | 41 | 39.1 | 35.6 | |
| RMSDdeer | - | 2.43 | 10.2 | |
Each designated by corresponding labeling site numbers.
Most probable distances listed. Estimated errors are less than 7% based on repeated measurements.
Inter-NOX distances (see Materials and Methods) in the best-fit model shown in Figure 3A.
NASNOX predicted average inter-R5 distances obtained from the pRNA tetramer crystal structure.43
Major population listed.
Figure 2.
(A) Examples of DEER measured inter-R5 distances between pRNA helices. Each data set is designated by the labeling site numbers (see Figure 1A). Normalized and background-corrected experimental dipolar evolution data (black lines) were fit (red lines) using the Defit program. Insets show computed distance distribution profiles, with the most probable distance (r0) and the width of the distribution (wr) indicated. (B) Dipolar evolution data of corresponding single-labeled pRNA samples. Additional data sets are shown in Supporting Information Figures S5 and S6.
Statistical analysis built into the Defit program indicated that 13 of the 17 data sets can be adequately fit with one population of interspin distances, while the remaining 4 data sets each contains one additional population (Table 2, Supporting Information Figure S6). In these four data sets, the r0 of the longer-distance population varies substantially depending on the range of dipolar evolution data used for fitting the decay background. In addition, these r0 values fall between 47 and 60 Å, which are at the upper limit of the accurately measurable distances with the use of the corresponding dipolar evolution time (2 to 3 μs as limited by the sample phase memory time).72 These observations suggest that the longer-distance populations are likely artifacts. They were excluded from further investigation.
The width of distance distribution (wr) provides a measure of disordering in the interspin distances. The origin of disorder lies in variations in: (i) positioning of nitroxide pyrroline rings with respect to the RNA helices; and (ii) RNA conformations, including the relative spatial arrangement between RNA helices. In pRNA distance measurements, 14 of the 17 data sets give wr exceeding 5 Å (Supporting Information Table S1). These large wr values reflect distance distributions that are broader than those previously reported on DNA and RNA duplexes.70,71 They suggest disordering in the spatial arrangement between RNA helices in the 3-wj. Interestingly, interhelical distances between HT and HR, and between HL and HR, overall show larger wr than those between HT and HL (Supporting Information Table S1). This may indicate that positioning of HR is variable.
The DEER data obtained on pRNA samples were also analyzed with the DeerAnalysis program developed by Jeschke and co-workers,73 which uses a model free Tikhonov regularization method to extract interspin distance distributions. Data sets with oscillating echo evolution traces (and therefore narrow distance distributions, e.g., (79; 95)) show consistent distance distribution profiles when analyzed using either Defit or DeerAnalysis (Supporting Information Figure S7). For those data sets without oscillations, such as (33; 76), the optimized regularization parameter used in Tikhonov fits is undetermined, and consequently, the shape of distance distribution profiles cannot be adequately determined (Supporting Information Figure S8). This hampers further efforts on dissecting subpopulations in distance distribution profiles using a previously reported procedure.22 None the less, Defit and DeerAnalysis render similar average distances (i.e., r0) and comparable distribution widths when the entire distance distribution profiles are considered (Supporting Information Figure S8). As Defit has been successfully applied in studies where disordering in protein structures results in DEER traces without oscillations,21,56 it was used in this work to describe inter-R5 distance distribution profiles.
Modeling the Three-Way Junction Global Structure Using DEER Measured Distances
Using a set of in-house programs, a grid search approach was employed to evaluate over 65 billion unique models where the spatial arrangement was varied between three A-form helices corresponding to HT, HR, and HL (see Materials and Methods). The search yielded ~480 000 sterically allowed models that conform to steric and chemical bonding constraints. An RMSDdeer metric was then computed, which corresponds to the root-mean-square-deviation of inter-R5 distances between DEER-measured values (i.e., the r0 values) and those derived on each model. With a DEER constraint of RMSDdeer ≤ 5 Å, which slightly exceeds errors of the measured r0 (≤7% of measured values, see above), 3662 viable models were found to satisfy the DEER constraints, which is <0.8% of the sterically allowed population. Expanding the range of translation parameters from ±15 to ±20 Å resulted in no significant increase of viable models, indicating sufficient coverage of the parameter space.
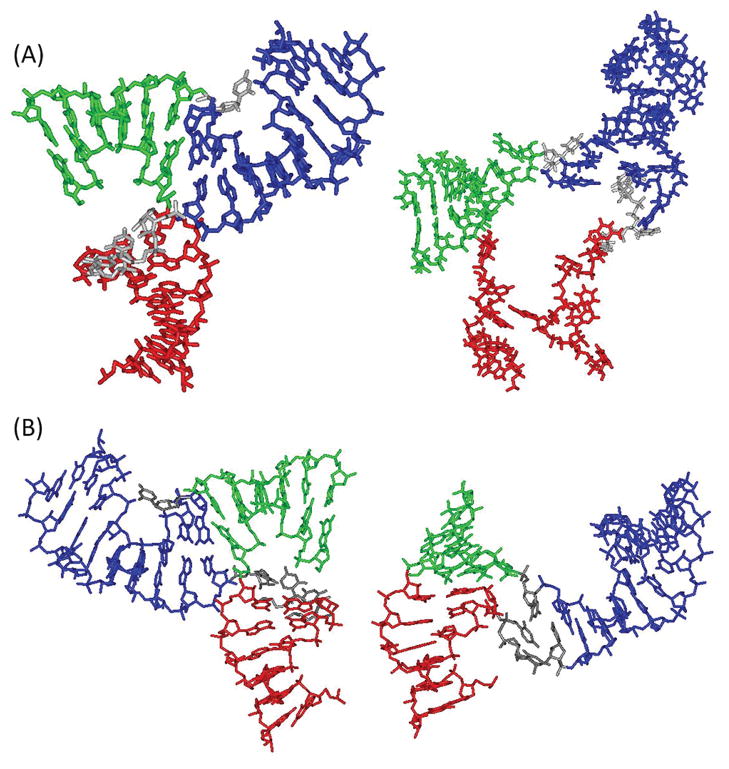
The grid search yielded a best-fit model with an RMSDdeer of 2.43 Å (Figure 3A, Table 2). It shows a T-shaped 3-wj, with an acute bent between HT and HL (interhelical angel θT,L = 93°) and an approximately linear arrangement between HR and HL (θR,L = 142°) (Figure 3A). Further analyses revealed that, when compared to the best-fit model, the ensemble of 3662 viable models shows a heavy atom root-mean-square-deviation distribution of 5.0 ± 1.4 Å (Supporting Information Figure S9). Distribution of each interhelical angle shows predominately one population, with angles between HT/HL, HT/HR, and HL/HR being 99° ± 19°, 57° ± 19°, and 147° ± 15°, respectively (Figure 3B). Overall, EPR data reveal that, in pRNA dimer, the 3-wj adopts one family of conformation with an acute kink between HT and HL.
Figure 3.
Models of the pRNA 3-wj derived based on DEER distances measured in pRNA dimer. (A) Best-fit model obtained from grid search using the RMSDdeer criterion. HT, HL, and HR are shown in green, red, and blue, respectively. Angles between the respective helical axes are shown in the inset. To demonstrate that the best-fit model does satisfy the connection constraint, connections between HT/HR (U29), HR/HL (U72U73U74), and HT/HL (no nucleotide present) were manually built-in using conformations observed in the pRNA tetramer crystal structure43 (pdb no. 3R4F) with slight adjustment of backbone torsion angles. (B) Interhelical angle distributions from the 3662 viable models obtained using the RMSDdeer criterion. (C) Best-fit model obtained from grid search using the Rmsdmod criterion. The interhelical angles between HT/HL, HT/HR, and HR/HL are 90°, 46°, and 134°, respectively. (D) Best-fit model obtained from random docking search.
To assess the impact of the widths of distance distribution (wr) on the outcome of 3-wj modeling, the sterically allowed ensemble identified in the grid search was ranked according to a modified RMSD metric (RMSDmod, see Materials and Methods). The top-ranked model obtained using the RMSDmod criterion shows minimal difference from the best-fit model identified by the RMSDdeer criterion (compare panels A and C in Figure 3), with the root-mean-square-deviation between corresponding heavy atoms being 1.7 Å. Furthermore, an allowable ensemble was constructed by selecting 3662 lowest RMSDmod models, which matches the number of models satisfying the RMSDdeer ≤ 5 Å criterion. This ensemble also shows one population of RNA conformation, and interhelical angles between HT/HL, HT/HR, and HL/HR are 77° ± 16°, 59° ± 26°, and 120° ± 26°, respectively. Importantly, no allowable model has θT,L < 45°, which suggests HT and HL are kinked rather than linearly stacked. All these characteristics match those observed in the RMSDdeer ≤5 Å ensemble. We note that there are alternative means to incorporate wr into the search criterion. In addition, for DEER traces that show decay without oscillation due to intrinsic flexibility of pRNA, our ability to measure wr accurately is limited. Nonetheless, RMSDmod analyses suggest that the uncertainty arisen from the measured distance distribution width is unlikely to alter the conclusion that the EPR-based 3-wj models adopt a kinked configuration between HT and HL.
Models of 3-wj were also obtained using a random docking search (Figure 3D), which uses a fundamentally different algorithm as compared to the grid search (see Materials and Methods). From this search, the model that fits best to the DEER measured distance has an RMSDdeer of 2.86 Å. The structural differences between the best models from grid search and random docking search are minimal, with heavy atom root-mean-square-deviation being approximately 2.4 Å between these two models (compare panels A and D in Figure 3).
We note that in the pRNA dimer construct used in this study, U72U73U74 is present in the full length A/b′ monomer, but absent in br_B/a′ (Figure 1A). Considering the symmetry of the pRNA dimer, enforcing the connection constraint from residue 71 O3′ to residue 75 C5′ (i.e., accounting for U72U73U74) is justified. Furthermore, the distance between residue 71 O3′ and residue 75 C5′ is 16.5 Å in the EPR-based model (Figure 3A), which is very similar to the value measured in the crystal structure (16.7 Å).43 As controls, modeling without enforcing the connection constraint from residue 71 O3′ to residue 75 C5′ was carried out. Such a search yielded the same top-ranked model and a similar interhelical angle distribution pattern as compared to the search with the constraint, although the resulting viable model ensemble is larger as expected.
Assessing Previously Reported Three-Way Junction Conformations Using DEER Measured Distances
The DEER measured distances allow direct assessments of two pRNA 3-wj conformations that were reported during the course of this SDSL work. The Guo group has reported a model of pRNA dimer, which was obtained based on biochemical data and distances measured using single-molecule fluorescence resonance energy transfer (smFRET).47,48 In this FRET-based model, the pRNA 3-wj shows a kinked HT/HL conformation, which is characteristically similar to the EPR-based model described above (Figure 4A). However, in the FRET study, there is no reported FRET measurement spanning the pRNA 3-wj,48 and clear differences are present between the FRET-model and the EPR-model (Figure 4A).
Figure 4.
Comparison of 3-wj models obtained from different studies. In each panel, the HT, HL, and HR helices are shown in green, red, and blue, respectively. (A) Comparison between the EPR-derived model (left) and the FRET-based model48 (right). Inter-R5 distances were not predicted for the FRET model, which shows largely irregular helical conformations around the 3-wj. Nevertheless, both models show a kinked conformation between HT and HL, and a more linear conformation between HL and HR. (B) Comparison between the EPR-derived model (left) and that observed in the tetramer crystal structure43 (right). The two models were aligned according to HL. The EPR model shows a kinked conformation between HT and HL, while the crystal structure shows a linearly stacked HT and HL.
Very recently, a 3.5 Å resolution crystal structure of a pRNA tetramer was reported (PDB no. 3R4F).43 When R5 was modeled at the corresponding sites of this tetramer crystal structure, the resulting inter-R5 distances deviate from the DEER measured distances with an RMSDdeer of 10.2 Å, with five of the data sets showing deviation >10 Å (Table 2, bolded). Particularly, for data set (26; 88), which measures distance between HT and HL, r0 predicted using the tetramer crystal structure differs from the DEER measured value by 26 Å (10.7 vs 37 Å, Table 2), which significantly exceeds the measured distance distribution half-width (wr = 17 Å, Supporting Information Table S1). Modeling using the crystal structure also shows that at these two sites, R5 can be adequately accommodated simultaneously without distorting the RNA. Furthermore, cw-EPR spectra of all double-labeled samples, including that of (26; 88), show no line-broadening compared to single-labeled controls (Supporting Information Figure S10). This indicates that inter-R5 distance at (26; 88) is >20 Å,74–76 which is not compatible with the 10.7 Å value predicted based on the tetramer crystal structure. Overall, even though many of the DEER measured distances reported here show broad distributions, the significant deviations between the measured r0 and those predicted based on the crystal structure suggest that the majority of the 3-wj conformation in the pRNA dimer in solution deviates from the conformation reported in the tetramer crystal structure.
Examination of the crystal structure43 reveals that HT, HL, and HR each adopts an A-form conformation as assumed in our model search. However, in the crystal structure, HT and HL are stacked nearly linearly, while HR and HL adopt a relatively kinked conformation (θT,L = 15°, θT,R = 78°, and θL,R = 93°) (Figure 4B). This is characteristically different from the kinked HT/HL conformation in the EPR-derived model. As discussed above, the kinked HT/HL conformation persists in all EPR-based models, including those obtained using the RMSDmod criterion that takes into account the widths of measured distance distributions. The analyses therefore support the conclusion that the 3-wj conformations are different between the pRNA tetramer crystal structure and the pRNA dimer in solution state.
DISCUSSION
Global Conformation of the pRNA Three-Way Junction
EPR measured distances have been widely used to assess the viability of existing models or to distinguish competing models.6,7,21,77 Results reported here show that the 3-wj conformation in the pRNA tetramer crystal structure does not conform to the DEER measured interhelical distances in the dimer in solution state (see Results). Consequently, the EPR-based 3-wj model displays clear differences in the relative orientations between HT, HL, and HR as compared to the tetramer crystal structure (Figure 4B). Furthermore, the EPR-derived model shows similar characteristics to the model constructed based on biochemical and smFRET data (Figure 4A). Together the results suggest that the 3-wj conformation in the dimer in solution is different from that in the tetramer crystal structure. We do note that, in addition to variations in salt and buffer conditions, EPR and crystallography studies used different monomeric pRNA constructs, although both contructs represent a truncated pRNA resembling the procapsid binding domain and have been shown to be functional.43,52,68 For example, in the EPR study, the U72U73U74 linker was deleted in one of the pRNA monomer (Figure 1A). This may affect relative positioning of HR with respect to HL and HT, and indeed, there are indications that positioning of HR is more variable than that of HL and HT (see Results). However, deleting U72U73U74 should have a much less drastic effect on the relative spatial arrangement between HT and HL, which shows the biggest deviation between the crystal structure and the EPR-based model (Figure 4B).
In a pRNA oligomer, HR and HL are constrained by the intermolecular R/L loop base-pairing and ultimately interact to form the ring-shaped pRNA/pRNA interface (Figure 1A).38,39,66 Between pRNA dimer and tetramer, changes in relative HR/HL configurations are likely needed to accommodate the increased ring size. Interestingly, results presented here indicate that relative positioning of HT with respect to HL and HR is different between the dimer and the tetramer. If one considers that HR and HL define the pRNA ring, this will imply that the relative positioning of HT with respect to the pRNA ring changes in different pRNA oligomeric states. Note that the (extended) HT contains the binding site for the phi29 motor ATPase (i.e., gp16), while HL and HR form the pRNA ring that binds to the procapsid.35,36 Global structural changes in 3-wj, which alter the relative spatial arrangement between HT, HR and HL, will change the spatial relationship between the motor ATPase and the procapsid. This is consistent with a recent proposal that pRNA serves as a communicator to bridge different parts of the motor during packaging.43 In addition, the linearly stacked HT/HL conformation observed in the tetramer crystal structure has been used directly to model pRNA pentamer and hexamer.43 The variability of junction conformation between the dimer and the tetramer reported here suggests more complexity in modeling pRNA oligomers.
Information on the pRNA 3-wj also impacts efforts on developing pRNA-based artificial nanostructures for material and therapeutic applications.46 For example, taking advantage of the R/L loop interactions, pRNA variants have been engineered to assemble oligomeric RNA arrays and superstructures.78 The 3-wj strongly influences the shape of the monomeric pRNA unit and, therefore, impacts the morphology of the arrays and superstructures. As such, information obtained here on 3-wj conformation, as well as how it may change in different oligomeric states, should aid the rational design of these superstructures. In addition, pRNA 3-wj has been used as a scaffold for assembling therapeutic modules such as interfering RNA, ribozyme, and small molecule agents (e.g., folate), and the resulting multifunctional nanoparticles seem to be able to function in vivo.79 As the 3-wj controls the related positioning of the therapeutic modules, information on its conformation reported here should benefit these developments.
Modeling RNA Global Structure Using Multiple DEER Measured Distances
While there are now a growing number of reports on SDSL mapping of protein conformations,23–30 SDSL mapping of nucleic acids conformation is limited.31–33 In this study, we demonstrated a general strategy to map the global structure of nucleic acids. Multiple distances in the nanometer range were measured using an advanced pulsed EPR methodology (i.e., DEER) and a nucleotide independent nitroxide label (i.e., R5) that can be efficiently attached to multiple RNA sites. In parallel, a de novo pool of RNA models were constructed, which allows explicit computation of internitroxide distances in each model. The DEER measured distances were then used as constraints to select viable RNA conformations. When all 17 sets of distances were applied, less than 0.8% of sterically allowed models were deemed viable, clearly demonstrating the power of DEER measured long-range distances on mapping RNA global structure.
However, there remain a number of unanswered questions. For example, how does the amount of DEER distances affect modeling? As a first step in addressing this question, searches were carried out using only 12 of the 17 DEER distances while omitting data sets (26; 76), (26; 79), (26; 88), (26; 37), and (37; 88) (see Table 2). This yielded 26 390 viable models with RMSDdeer < 5 Å, which is approximately 7 times larger than that obtained with all 17 distances. Key characteristics between these two pools of model are very similar: the top-ranked models deviate with a heavy atom root-mean-square-deviation of 3.5 Å; and the average interhelical angles are almost identical (Supporting Information Figure S11). This further strengthens the confidence on the 3-wj conformation reported above (Figure 3). The pool obtained using 12 DEER distances does show broader interhelical angle distributions (Supporting Information Figure S11) and, more interestingly, includes a small fraction of models (3.6%) with θT,L < 45°, which is approaching a more linear HT/HL configuration. Therefore, the additional 5 distances, including data set (26; 88) that shows a large deviation between the DEER measurement and that predicted from the tetramer crystal structure, do push the models further away from the linearly stacked HT/HL configuration. We note that a prior study in protein indicates both the amount of distance constraints and the location of the labeling sites are important for optimal structural determination.80 Further studies are needed in order to achieve rational selection of optimal labeling sites for mapping RNA global structure.
SDSL uses a small nitroxide probe that is less intrusive as compared to most fluorophores, and avoids a number of issues faced by crystallography (e.g., crystalline sample preparation, interference from lattice packing) and NMR (e.g., limitation on molecule size). The nanometer distances measured by SDSL/EPR provide a unique set of long-range constraints. While work reported here demonstrates de novo mapping of RNA global structure, combining SDSL/EPR with other experimental and computational approaches should be particularly powerful in mapping tertiary structure of complex nucleic acid and protein/nucleic acid systems.
Supplementary Material
Acknowledgments
We thank NIH (GM069557 and 1S10RR028992) and NSF (MCB 054652) for financial support, the Environmental Molecular Sciences Laboratory of the Pacific Northwest National Laboratory for a pulsed EPR instrumentation grant, Drs. M. Bowman, B. Hegde, E. Walter for assistance on pulsed EPR, and Y. Fang for assistance on pRNA preparation and characterization. Part of the computation work was supported by the University of Southern California Center for High-Performance Computing and Communications.
Footnotes
ASSOCIATED CONTENT
Supplemental data and discussions. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Gesteland RF, Atkins JF, Cech TR, editors. RNA World. 3. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. [Google Scholar]
- 2.Holbrook SR. Annu Rev Biophys. 2008;37:445–464. doi: 10.1146/annurev.biophys.36.040306.132755. [DOI] [PubMed] [Google Scholar]
- 3.Butcher SE, Pyle AM. Acc Chem Res. 2011;44:1302–1311. doi: 10.1021/ar200098t. [DOI] [PubMed] [Google Scholar]
- 4.Altenbach C, Flitsch SL, Khorana HG, Hubbell WL. Biochemistry. 1989;28:7806–7812. doi: 10.1021/bi00445a042. [DOI] [PubMed] [Google Scholar]
- 5.Hubbell WL, Altenbach C. Curr Opin Struct Biol. 1994;4:566–573. [Google Scholar]
- 6.Hubbell WL, Cafiso DS, Altenbach C. Nat Struct Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 7.Fanucci GE, Cafiso DS. Curr Opin Struct Biol. 2006;16:644–653. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 8.Sowa GZ, Qin PZ. Prog Nucleic Acid Res Mol Biol. 2008;82:147–197. doi: 10.1016/S0079-6603(08)00005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen P, Qin PZ. Wiley Interdiscip Rev: RNA. 2012;3:62–72. doi: 10.1002/wrna.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, Qin PZ. Biophysics of RNA Folding. Springer; New York: 2012. [Google Scholar]
- 11.Schiemann O, Prisner TF. Q Rev Biophys. 2007;40:1–53. doi: 10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- 12.Milov A, Maryasov A, Tsvetkov Y. Appl Magn Reson. 1998;15:107–143. [Google Scholar]
- 13.Martin RE, Pannier M, Diederich F, Gramlich V, Hubrich M, Spiess HW. Angew Chem, Int Ed. 1998;37:2833–2837. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2833::AID-ANIE2833>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 14.Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. J Magn Reson. 2000;142:331–40. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 15.Jeschke G, Bender A, Paulsen H, Zimmermann H, Godt A. J Magn Reson. 2004;169:1–12. doi: 10.1016/j.jmr.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 16.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. Proc Natl Acad Sci USA. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim M, Xu Q, Murray D, Cafiso DS. Biochemistry. 2008;47:670–679. doi: 10.1021/bi7016415. [DOI] [PubMed] [Google Scholar]
- 18.Klein JC, Burr AR, Svensson B, Kennedy DJ, Allingham J, Titus MA, Rayment I, Thomas DD. Proc Natl Acad Sci USA. 2008;105:12867–12872. doi: 10.1073/pnas.0802286105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grote M, Polyhach Y, Jeschke G, Steinhoff HJ, Schneider E, Bordignon E. J Biol Chem. 2009;284:17521–17526. doi: 10.1074/jbc.M109.006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou P, Bortolus M, McHaourab HS. J Mol Biol. 2009;393:586–597. doi: 10.1016/j.jmb.2009.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vileno B, Chamoun J, Liang H, Brewer P, Haldeman BD, Facemyer KC, Salzameda B, Song L, Li H-C, Cremo CR, Fajer PG. Proc Natl Acad Sci USA. 2011;108:8218–8223. doi: 10.1073/pnas.1014137108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kear JL, Blackburn ME, Veloro AM, Dunn BM, Fanucci GE. J Am Chem Soc. 2009;131:14650–14651. doi: 10.1021/ja907088a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown LJ, Sale KL, Hills R, Rouviere C, Song L, Zhang X, Fajer PG. Proc Natl Acad Sci USA. 2002;99:12765–12770. doi: 10.1073/pnas.202477399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park SY, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, Bilwes AM, Crane BR. Nat Struct Mol Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- 25.Hilger D, Polyhach Y, Padan E, Jung H, Jeschke G. Biophys J. 2007;93:3675–3683. doi: 10.1529/biophysj.107.109769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jao CC, Hegde BG, Chen J, Haworth IS, Langen R. Proc Natl Acad Sci USA. 2008;105:19666–19671. doi: 10.1073/pnas.0807826105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Endeward B, Butterwick JA, MacKinnon R, Prisner TF. J Am Chem Soc. 2009;131:15246–15250. doi: 10.1021/ja904808n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowman A, Ward R, El-Mkami H, Owen-Hughes T, Norman DG. Nucleic Acids Res. 2010;38:695–707. doi: 10.1093/nar/gkp1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai AL, Huang H, Herrick DZ, Epp N, Cafiso DS. J Mol Biol. 2011;405:696–706. doi: 10.1016/j.jmb.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim S, Brandon S, Zhou Z, Cobb CE, Edwards SJ, Moth CW, Parry CS, Smith JA, Lybrand TP, Hustedt EJ, Beth AH. J Biol Chem. 2011;286:20746–20757. doi: 10.1074/jbc.M111.230326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim NK, Bowman MK, DeRose VJ. J Am Chem Soc. 2010;132:8882–8884. doi: 10.1021/ja101317g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krstic I, Frolow O, Sezer D, Endeward B, Weigand JE, Suess B, Engels JW, Prisner TF. J Am Chem Soc. 2010;132:1454–1455. doi: 10.1021/ja9077914. [DOI] [PubMed] [Google Scholar]
- 33.Wunnicke D, Strohbach D, Weigand JE, Appel B, Feresin E, Suess B, Muller S, Steinhoff HJ. RNA. 2011;17:182–188. doi: 10.1261/rna.2222811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo PX, Erickson S, Anderson D. Science. 1987;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 35.Grimes S, Jardine PJ, Anderson D. Adv Virus Res. 2002;58:255–294. doi: 10.1016/s0065-3527(02)58007-6. [DOI] [PubMed] [Google Scholar]
- 36.Guo P, Lee TJ. Mol Microbiol. 2007;64:886–903. doi: 10.1111/j.1365-2958.2007.05706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 38.Guo P, Zhang C, Chen C, Garver K, Trottier M. Mol Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 39.Zhang F, Lemieux S, Wu X, St-Arnaud D, McMurray CT, Major F, Anderson D. Mol Cell. 1998;2:141–147. doi: 10.1016/s1097-2765(00)80123-9. [DOI] [PubMed] [Google Scholar]
- 40.Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shu D, Zhang H, Jin J, Guo P. EMBO J. 2007;26:527–537. doi: 10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao F, Zhang H, Guo P. Nucleic Acids Res. 2008;36:6620–6632. doi: 10.1093/nar/gkn669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding F, Lu C, Zhao W, Rajashankar KR, Anderson DL, Jardine PJ, Grimes S, Ke A. Proc Natl Acad Sci USA. 2011;108:7357–7362. doi: 10.1073/pnas.1016690108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grimes S, Anderson D. J Mol Biol. 1990;215:559–566. doi: 10.1016/s0022-2836(05)80168-8. [DOI] [PubMed] [Google Scholar]
- 45.Ibarra B, Valpuesta JMa, Carrascosa JL. Nucleic Acids Res. 2001;29:4264–4273. doi: 10.1093/nar/29.21.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo P. Nat Nanotechnol. 2010;5:833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoeprich S, Guo P. J Biol Chem. 2002;277:20794–20803. doi: 10.1074/jbc.M112061200. [DOI] [PubMed] [Google Scholar]
- 48.Shu D, Zhang H, Petrenko R, Meller J, Guo P. ACS Nano. 2010;4:6843–6853. doi: 10.1021/nn1014853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris S, Schroeder SJ. Biochemistry. 2010;49:5989–5997. doi: 10.1021/bi100393r. [DOI] [PubMed] [Google Scholar]
- 50.Qin PZ, Butcher SE, Feigon J, Hubbell WL. Biochemistry. 2001;40:6929–6936. doi: 10.1021/bi010294g. [DOI] [PubMed] [Google Scholar]
- 51.Qin PZ, Haworth IS, Cai Q, Kusnetzow AK, Grant GP, Price EA, Sowa GZ, Popova A, Herreros B, He H. Nat Protoc. 2007;2:2354–2365. doi: 10.1038/nprot.2007.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fang Y, Cai Q, Qin PZ. Biochemistry. 2005;44:9348–9358. doi: 10.1021/bi0475020. [DOI] [PubMed] [Google Scholar]
- 53.Zhang X, Cekan P, Sigurdsson ST, Qin PZ. Methods Enzymol. 2009;469:303–328. doi: 10.1016/S0076-6879(09)69015-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weber RT. Pulsed ELDOR Option User’s Manual. Bruker BioSpin Corporation; Bellerica, MA: 2006. [Google Scholar]
- 55.Fajer PG, Brown L, Song L. In: ESR Spectroscopy in Membrane Biophysics. Hemminga MA, Berliner LJ, editors. Vol. 27. Springer; New York: 2007. pp. 95–128. [Google Scholar]
- 56.Sen KI, Logan TM, Fajer PG. Biochemistry. 2007;46:11639–11649. doi: 10.1021/bi700859p. [DOI] [PubMed] [Google Scholar]
- 57.Tung CS, Carter ESII. Comput Appl Biosci. 1994;10:427–433. doi: 10.1093/bioinformatics/10.4.427. [DOI] [PubMed] [Google Scholar]
- 58.Tung CS, Soumpasis DM, Hummer G. J Biomol Struct Dyn. 1994;11:1327–1344. doi: 10.1080/07391102.1994.10508071. [DOI] [PubMed] [Google Scholar]
- 59.Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 60.Reid RJ, Bodley JW, Anderson D. J Biol Chem. 1994;269:5157–5162. [PubMed] [Google Scholar]
- 61.Chen C, Zhang C, Guo P. RNA. 1999;5:805–818. doi: 10.1017/s1355838299990350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Price EA, Sutch BT, Cai Q, Qin PZ, Haworth IS. Biopolymers. 2007;87:40–50. doi: 10.1002/bip.20769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lavery R, Sklenar H. J Biomol Struct Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 64.Tung CS, Walsh DA, Trewhella J. J Biol Chem. 2002;277:12423–12431. doi: 10.1074/jbc.M110298200. [DOI] [PubMed] [Google Scholar]
- 65.Humphrey W, Dalke A, Schulten K. J Mol Graphics. 1996;14:33–8. 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 66.Chen C, Sheng S, Shao Z, Guo P. J Biol Chem. 2000;275:17510–17516. doi: 10.1074/jbc.M909662199. [DOI] [PubMed] [Google Scholar]
- 67.Moll WD, Guo P. J Nanosci Nanotechnol. 2007;7:3257–3267. doi: 10.1166/jnn.2007.914. [DOI] [PubMed] [Google Scholar]
- 68.Fang Y, Shu D, Xiao F, Guo P, Qin PZ. Biochem Biophys Res Commun. 2008;372:589–594. doi: 10.1016/j.bbrc.2008.05.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang C, Trottier M, Chen C, Guo P. Virology. 2001;281:281–293. doi: 10.1006/viro.2000.0771. [DOI] [PubMed] [Google Scholar]
- 70.Cai Q, Kusnetzow AK, Hubbell WL, Haworth IS, Gacho GP, Van Eps N, Hideg K, Chambers EJ, Qin PZ. Nucleic Acids Res. 2006;34:4722–4730. doi: 10.1093/nar/gkl546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cai Q, Kusnetzow AK, Hideg K, Price EA, Haworth IS, Qin PZ. Biophys J. 2007;93:2110–2117. doi: 10.1529/biophysj.107.109439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jeschke G, Polyhach Y. Phys Chem Chem Phys. 2007;9:1895–1910. doi: 10.1039/b614920k. [DOI] [PubMed] [Google Scholar]
- 73.Jeschke G, Chechik V, Ionita P, Godt A, Zimmermann H, Banham J, Timmel C, Hilger D, Jung H. Appl Magn Reson. 2006;30:473–498. [Google Scholar]
- 74.Eaton GR, Eaton SS, Berliner LJ, editors. Distance Measurements in Biological Systems by EPR. Vol. 19 Kluwer; New York, NY: 2000. [Google Scholar]
- 75.Fajer PG. In: Encyclopedia of Analytical Chemistry. Meyers R, editor. John Wiley & Sons; Chichester: 2000. pp. 5725–5761. [Google Scholar]
- 76.Rabenstein MD, Shin YK. Proc Natl Acad Sci USA. 1995;92:8239–8243. doi: 10.1073/pnas.92.18.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Herrick DZ, Kuo W, Huang H, Schwieters CD, Ellena JF, Cafiso DS. J Mol Biol. 2009;390:913–923. doi: 10.1016/j.jmb.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shu D, Moll WD, Deng Z, Mao C, Guo P. Nano Lett. 2004;4:1717–1723. doi: 10.1021/nl0494497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. Nat Nano. 2011;6:658–667. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kazmier K, Alexander NS, Meiler J, McHaourab HS. J Struct Biol. 2011;173:549–557. doi: 10.1016/j.jsb.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.