Abstract
Objective:
In families with autosomal dominant partial epilepsy with auditory features (ADPEAF) with mutations in the LGI1 gene, we evaluated clustering of mutations within the gene and associations of penetrance and phenotypic features with mutation location and predicted effect (truncation or missense).
Methods:
We abstracted clinical and molecular information from the literature for all 36 previously published ADPEAF families with LGI1 mutations. We used a sliding window approach to analyze mutation clustering within the gene. Each mutation was mapped to one of the gene's 2 major functional domains, N-terminal leucine-rich repeats (LRRs) and C-terminal epitempin (EPTP) repeats, and classified according to predicted effect on the encoded protein (truncation vs missense). Analyses of phenotypic features (age at onset and occurrence of auditory symptoms) in relation to mutation site and predicted effect included 160 patients with idiopathic focal unprovoked seizures from the 36 families.
Results:
ADPEAF-causing mutations clustered significantly in the LRR domain (exons 3–5) of LGI1 (p = 0.026). Auditory symptoms were less frequent in individuals with truncation mutations in the EPTP domain than in those with other mutation type/domain combinations (58% vs 80%, p = 0.018).
Conclusion:
The LRR region of the LGI1 gene is likely to play a major role in pathogenesis of ADPEAF.
Autosomal dominant partial epilepsy with auditory features (ADPEAF) (OMIM 600512) is an idiopathic focal epilepsy syndrome with auditory symptoms or receptive aphasia as a prominent ictal manifestation.1–3 These symptoms strongly suggest localization to the lateral temporal lobe; hence, the syndrome is also called autosomal dominant lateral temporal lobe epilepsy. A substantial proportion (approximately 50%) of affected families have mutations in the leucine-rich, glioma inactivated 1 (LGI1) gene,4–6 with an average penetrance of 67%.7
LGI1 encodes a secreted protein, Lgi1, with 2 major domains: an N-terminal leucine-rich repeat (LRR) domain containing 4 LRRs flanked by 2 conserved cysteine-rich regions, and a C-terminal epitempin (EPTP) domain containing 7 EPTP repeats.8,9 To date, 33 unique LGI1 mutations have been reported in ADPEAF families (n = 36) and sporadic patients with idiopathic focal epilepsy with auditory symptoms (n = 2).7,8,10–15 Missense and truncation mutations have been found in both LRR and EPTP domains. Although previous studies have reported that pathogenic LGI1 mutations are uniformly distributed across the gene,8 none has used a quantitative approach to assess mutation clustering or investigated genotype-phenotype correlations in detail. Establishment of genotype-phenotype associations has the potential to elucidate the biologic pathways involving LGI1, including the mechanisms leading to ADPEAF symptoms.
In this study, we tested 2 hypotheses: 1) the distribution of ADPEAF-causing mutations is not uniform across the LGI1 gene and 2) penetrance and phenotypic features in ADPEAF-affected individuals differ, depending on the predicted effect (truncation or missense) or domain location (LRR or EPTP) of mutations.
METHODS
ADPEAF families.
Molecular and clinical information from all 36 previously reported ADPEAF families with LGI1 mutations (11 reported by our group and 25 reported by others) was collected and assembled into a database. For our previously reported families,2–4,13,16 we obtained this information directly from our existing database. For families reported by others, we abstracted the clinical information from published reports.6,10–12,14,15,17–26
Mutation density and clustering analysis.
To visualize the distribution of previously reported mutations in the gene, we first computed mutation density by counting the number of mutations in each exon and dividing the count by the nucleotide length of the exon (excluding intronic regions). For statistical analysis of mutation clustering across the coding sequence of LGI1, we used a sliding window approach. We considered all overlapping windows of a fixed length L, and for each such window W we calculated a statistic SW defined as the maximum number of mutations over all windows of length L, i.e., SL = maxWSW. Statistical significance was assessed using a classic approach from scan statistic theory that efficiently corrects for the multiple correlated windows that are considered.27 SW is sensitive to L and the total length of the nucleotide sequence scanned.
Genotype-phenotype correlation analysis.
We estimated penetrance through a method similar to that in our previous article, which was based on the proportion affected among obligate carriers (i.e., parents of affected individuals who were not married into the family).7 To increase precision, we modified our previous approach by including offspring of affected individuals in addition to obligate carriers. In each family, we estimated the number of mutation carriers (denominator of the penetrance estimate) from the published pedigree, assuming that all obligate carriers and half of the offspring of affected individuals were carriers. The number of these individuals who were affected served as the numerator of the penetrance estimate. To increase the likelihood that the offspring included in these calculations were old enough to have passed through the risk period for ADPEAF, we included only offspring of affected individuals aged >40 years at the time of the study or death, as reported in the original articles. (Age of the affected individuals served as a proxy for age of their offspring, because age of the offspring was seldom reported.) Then we divided the families into 2 groups based on the median penetrance estimate among all families and used χ2 tests to assess differences in the proportion with high vs low penetrance in relation to the mutations' domain localization (LRR or EPTP) and predicted effect (missense or truncation).
For genotype-phenotype association analyses, we examined age at onset of unprovoked seizures and occurrence of auditory symptoms in 160 patients from 32 families with idiopathic focal unprovoked seizures in LGI1 mutation-positive ADPEAF families. (For the remaining 4 families, information from published reports was not sufficiently detailed for analysis.) These 2 features were selected because they were the only ones systematically documented in the families reported in the literature. As described above, analyses were performed by stratifying the mutations by their domain location (LRR or EPTP) and predicted effect (missense or truncation). Genotype-phenotype association was assessed by linear regression (for age at onset) and logistic regression (for auditory symptoms) using generalized estimating equations to account for the clustering of individuals within families. All analyses were performed using SPSS (IBM, Chicago IL).
Standard protocol approvals, registrations, and patient consents.
This study was approved by the Columbia University Medical Center Institutional Review Board, and all participants from our research group gave written informed consent. Data from individuals studied by other research groups were taken from published reports and were anonymous.
RESULTS
Mutation density and clustering analyses.
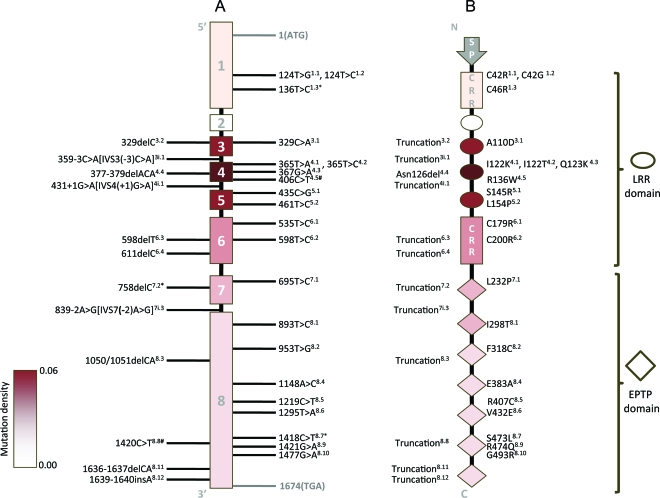
Figure 1A summarizes the genomic structure of LGI1 with all 33 reported pathogenic mutations labeled. As shown in the figure, 5 mutations have been observed more than once, either in 2 families (n = 3) or in a family and a sporadic case (n = 2). Mutation density within each exon is represented by the color intensity of the exon; higher color intensity represents higher density. Exons 1–6 encode the protein's LRR domain, and exons 7–8 encode the EPTP domain. Figure 1B is a schematic diagram of the Lgi1 protein with predicted effects of the mutations. With the exception of one in-frame deletion mutation (377–379delACA) resulting in the deletion of asparagine in the encoded protein,11 all other mutations are predicted to produce either amino acid substitution (missense mutations, n = 21, 66% of all mutations) or truncation of the Lgi1 protein (n = 11, 34% of all mutations).
Figure 1. LGI1 mutations cluster in leucine-rich repeat (LRR) domain.
(A) LGI1 gene structure with the reported mutation sites indicated. For superscripts next to mutations, the first digit indicates the exon localization of the mutation, and the second digit (after the decimal point) indicates the order of the mutation within the exon (mutations at the same nucleotide numbered arbitrarily). i = intronic mutation; ∗ = mutation found in 2 families (n = 3); # = mutation found in a family and a sporadic case (n = 2). (B) Lgi1 protein with effect of mutations on protein summarized. The 4 LRRs in the LRR domain are represented by ovals and the 7 epitempin (EPTP) repeats in the EPTP domain are represented by diamonds. Different intensities of the red shade correspond to the density of mutations in each exon. The superscripts next to protein variants are cross-indexed to the nucleotide changes listed in the gene structure (A). CRR = cysteine-rich region; SP = signal peptide.
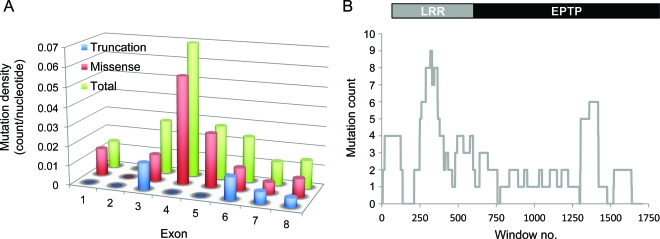
Figure 2A shows that mutation density is the highest in exon 4, followed by exon 5 and exon 3. Stratification of mutations by their predicted effects on the protein revealed that the high density of mutations in exon 4 is primarily attributed to missense mutations, which are more common than truncation mutations.
Figure 2. Significant clustering of pathogenic LGI1 mutations in the leucine-rich repeat (LRR) domain.
(A) Mutation density per nucleotide within each exon. Blue bars represent truncation mutations, red bars represent missense mutations, and green bars represent total mutations. (B) A sliding window analysis was performed on the sequence length of LGI1 mRNA amplified by the reported sequencing primers (1,823 nucleotides). A window size of 118 bp (median size of exons in LGI1) was used and the maximum number of mutations observed per window was 9 (p = 0.026). EPTP = epitempin.
To assess the significance of mutation clustering by the sliding window analysis, we chose a window length L = 118 bp, the median size of the LGI1 exons, to scan the mRNA length of the gene. Because the scanning statistic is sensitive to the length of mRNA analyzed, we chose 2 lengths (1,774 and 1,823 nucleotides [nt]) to capture the sequencing range covered in the literature.28,29 Among the total of 38 mutational events, 35 were included in this scanning analysis. The remaining 3 mutations were intronic and were excluded because intronic sequences have not been systematically sequenced. As shown in figure 2B, the maximum number of mutational events observed in any window is 9, i.e., SL = 9. The resulting p values for 2 scanning lengths are 0.030 and 0.026 for nt = 1,774 and nt = 1,823, respectively. Moreover, 2 of the 3 previously reported intronic mutations are located within the genomic sequence spanning exons 3–5 of LGI1, corresponding to the region where the exonic mutations cluster. The number of mutations was too small for formal statistical analysis of missense and truncation mutations separately.
To confirm that the clustering of LGI1 variants observed in the ADPEAF families is specific to pathogenic mutations, we searched the UCSC Genome Browser (db build 131) for LGI1 variants.30 The search returned 266 variants in the LGI1 gene, of which 170 were validated by HapMap (270 individuals) or the 1000 Genomes Project (629 individuals). Only 2 of 170 validated variants were in the coding region, one encoding a synonymous change in exon 6 and the other encoding a change in the 3′ untranslated region of exon 8. Neither of these has been associated with ADPEAF. The relative invariability of the protein coding sequence of LGI1 in reference genomes indicates that the gene is highly conserved, and the clustering of ADPEAF-causing mutations in the LRR region is probably specific to pathogenic mutations.
Genotype-phenotype correlation analyses.
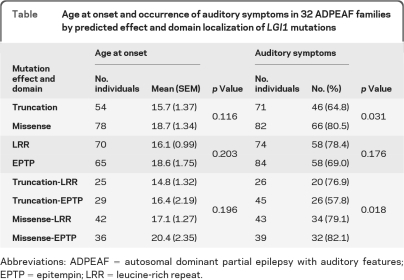
The overall penetrance estimate was 66% (SE 3.6%), with a median of 75% among the 36 families. The proportion of families with penetrance ≥75% was similar among those with missense (52%) vs truncation (58%) mutations. The proportion with penetrance ≥75% was higher among families with mutations in the LRR (58%) vs EPTP (47%) domain, but the difference was not significant.
The prevalence of auditory symptoms was significantly lower among individuals with truncation mutations than among those with missense mutations (65% vs 81%, p = 0.031) but did not differ according to domain location of mutations (table). Further stratification by both mutation effect and domain showed that the lower prevalence of auditory symptoms in individuals with truncation mutations was attributed to a lower frequency in those with truncation mutations in the EPTP domain (58% vs 80% in those with other mutation types/localizations, p = 0.018). Age at onset was not associated with either domain location or predicted effect of mutations (table). These results were not changed after adjustment for the current age of the patients.
Table.
Age at onset and occurrence of auditory symptoms in 32 ADPEAF families by predicted effect and domain localization of LGI1 mutations
Abbreviations: ADPEAF = autosomal dominant partial epilepsy with auditory features; EPTP = epitempin; LRR = leucine-rich repeat.
DISCUSSION
Establishment of genotype-phenotype relationships is fundamental to elucidating pathogenic effects of human mutations. In addition to influencing susceptibility to ADPEAF, recent studies have also shown that LGI1 plays an important role in limbic encephalitis, a disorder in which autoimmunity against the Lgi1 protein is the underlying cause.31 These observations support the importance of LGI1 in a spectrum of neurologic functions.
We found that ADPEAF-causing mutations cluster significantly within the LRR domain of LGI1. This finding was primarily attributed to missense mutations. The most likely explanation for this finding is that mutations in the LRR domain confer a greater susceptibility to ADPEAF than those in the EPTP domain, although mutations in both domains are clearly pathogenic. In addition, given the very strong conservation of this gene, an alternative explanation is that mutations in the LRR domain are more compatible with life and hence are observed more frequently than those in the EPTP domain. Either or both possibilities could underlie the mutation clustering in the LRR domain.
We observed a significantly lower frequency of auditory symptoms among individuals with truncation mutations in the EPTP domain than among those with other mutation types/locations. This finding is difficult to interpret with our current state of understanding of the biological function of the LGI1 gene products. Analysis of disease severity (e.g., medication response or seizure frequency) might have been more revealing but was not possible with the data abstracted from the literature. One possible explanation for the lower frequency of auditory symptoms among patients with truncation mutations in the EPTP domain is that they had a milder form of epilepsy, with fewer seizures overall and less recognition of associated symptoms.
Further elucidation of the function of LGI1 and its products is essential for better understanding of the possible phenotypic consequences of particular mutations. The pathogenicity of LGI1 mutations may involve both haploinsufficiency and dominant-negative mechanisms. In Lgi1 mouse knockout models, homozygous ablation of the gene leads to spontaneous seizures and early death,32,33 whereas heterozygous Lgi1+/− mice showed a lowered threshold for auditory stimuli-induced seizures.32 These findings are consistent with a mechanism involving haploinsufficiency, because the phenotype is more extreme in homozygotes than in heterozygotes. Although in many genes, truncation mutations lead to nonsense-mediated decay of the abnormal mRNA,34 evidence from animal and human studies indicates that this process does not always occur with truncation mutations in LGI1.35,36 For example, in transgenic mice with an extra copy of the LGI1 gene containing a truncation mutation in exon 6, maturation of glutamatergic synapses was arrested, clearly indicating a dominant-negative effect of the mutation.36 These results imply that the abnormal mRNA transcript resulting from the truncation mutation was not simply degraded by nonsense-mediated decay.
Protein-protein interactions involving Lgi1's EPTP domain have been studied more extensively than those involving the LRR domain. Several studies have used EPTP truncation mutations to investigate interactions of Lgi1 with Kv1 channels in presynaptic neurons35 or with ADAM proteins at transsynaptic sites.37,38 However, a recent model proposed that the formation of Lgi1 dimers through the LRR domain may influence pre- and postsynaptic transport or communication.39 These interactions may underlie the maturation and activity of glutamatergic synapses and thus modify the manifestation of ADPEAF.36,37 Given our finding of mutation clustering in the LRR domain, the reported dominant-negative effects of the EPTP truncation mutants, and the observation of naturally occurring Lgi1 isoforms that resemble EPTP truncation mutants,40 further investigation of protein-protein interactions or other molecular mechanisms involving the LRR domain that might influence epileptogenesis is warranted.
Our inclusion of all known LGI1 mutation-positive ADPEAF families in these genotype-phenotype association analyses provided an opportunity to gain insights into the structure-function relationships of LGI1 in a physiologic context. The sliding window analysis used here may be applicable to the analysis of mutation clustering in other disease genes.
Conversely, the lack of consistent reporting of phenotypic characteristics in the literature restricted our genotype-phenotype association analysis to penetrance, age at onset, and auditory symptoms. Statistical power was also limited by the relatively few previously reported ADPEAF families with LGI1 mutations. We are in the process of developing a more structured and standardized platform that can be used across different research centers for collection of clinical information on ADPEAF. Future analyses using a larger collection of ADPEAF families and more comprehensive phenotypic information will facilitate further elucidation of genotype-phenotype relationships of LGI1 mutations.
ACKNOWLEDGMENT
The authors thank the Division of Statistical Genetics, Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, NY, for their support and are also grateful to Matthew P. Anderson, MD, PhD, for critical comments.
Footnotes
- ADPEAF
- autosomal dominant partial epilepsy with auditory features
- EPTP
- epitempin
- LRR
- leucine-rich repeat
- nt
- nucleotides.
AUTHOR CONTRIBUTIONS
Dr. Ho drafted and helped to revise the manuscript and contributed to the study concept and design, data collection, statistical analysis, and interpretation. Dr. Ionita-Laza helped to revise the manuscript and contributed to the study design and statistical analysis. Dr. Ottman helped to revise the manuscript, contributed to the study concept and design and to data analysis and interpretation, and was responsible for data acquisition, study supervision, and obtaining funding.
DISCLOSURE
Dr. Ho has received postdoctoral research fellowship support from the NIH/NIMH. Dr. Ionita-Laza has received research support from the National Science Foundation and the NIH. Dr. Ottman serves on the scientific advisory board for and holds stock options in Trigeminal Solutions, Inc.; has received funding for travel from the International League Against Epilepsy, Ortho McNeil-Janssen Scientific Affairs, LLC, the NIH, Fallon Medica, LLC, and EDJ Associates, Inc.; has received speaker honoraria for non–industry-sponsored lectures; serves as a consultant to Ortho McNeil-Janssen Scientific Affairs, LLC; and has received research support from the Italian Ministry of Health and the NIH.
REFERENCES
- 1. Michelucci R, Pasini E, Nobile C. Lateral temporal lobe epilepsies: clinical and genetic features. Epilepsia 2009; 50 (suppl 5): 52–54 [DOI] [PubMed] [Google Scholar]
- 2. Ottman R, Risch N, Hauser WA, et al. Localization of a gene for partial epilepsy to chromosome 10q. Nat Genet 1995; 10: 56–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Winawer MR, Ottman R, Hauser WA, Pedley TA. Autosomal dominant partial epilepsy with auditory features: defining the phenotype. Neurology 2000; 54: 2173–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ottman R, Winawer MR, Kalachikov S, et al. LGI1 mutations in autosomal dominant partial epilepsy with auditory features. Neurology 2004; 62: 1120–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Michelucci R, Poza JJ, Sofia V, et al. Autosomal dominant lateral temporal epilepsy: clinical spectrum, new epitempin mutations, and genetic heterogeneity in seven European families. Epilepsia 2003; 44: 1289–1297 [DOI] [PubMed] [Google Scholar]
- 6. Berkovic SF, Izzillo P, McMahon JM, et al. LGI1 mutations in temporal lobe epilepsies. Neurology 2004; 62: 1115–1119 [DOI] [PubMed] [Google Scholar]
- 7. Rosanoff MJ, Ottman R. Penetrance of LGI1 mutations in autosomal dominant partial epilepsy with auditory features. Neurology 2008; 71: 567–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nobile C, Michelucci R, Andreazza S, Pasini E, Tosatto SC, Striano P. LGI1 mutations in autosomal dominant and sporadic lateral temporal epilepsy. Hum Mutat 2009; 30: 530–536 [DOI] [PubMed] [Google Scholar]
- 9. Staub E, Perez-Tur J, Siebert R, et al. The novel EPTP repeat defines a superfamily of proteins implicated in epileptic disorders. Trends Biochem Sci 2002; 27: 441–444 [DOI] [PubMed] [Google Scholar]
- 10. Di Bonaventura C, Carni M, Diani E, et al. Drug resistant ADLTE and recurrent partial status epilepticus with dysphasic features in a family with a novel LGI1 mutation: electroclinical, genetic, and EEG/fMRI findings. Epilepsia 2009; 50: 2481–2486 [DOI] [PubMed] [Google Scholar]
- 11. de Bellescize J, Boutry N, Chabrol E, et al. A novel three base-pair LGI1 deletion leading to loss of function in a family with autosomal dominant lateral temporal epilepsy and migraine-like episodes. Epilepsy Res 2009; 85: 118–122 [DOI] [PubMed] [Google Scholar]
- 12. Kawamata J, Ikeda A, Fujita Y, Usui K, Shimohama S, Takahashi R. Mutations in LGI1 gene in Japanese families with autosomal dominant lateral temporal lobe epilepsy: the first report from Asian families. Epilepsia 2010; 51: 690–693 [DOI] [PubMed] [Google Scholar]
- 13. Heiman GA, Kamberakis K, Gill R, et al. Evaluation of depression risk in LGI1 mutation carriers. Epilepsia 2010; 51: 1685–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Di Bonaventura C, Operto FF, Busolin G, et al. Low penetrance and effect on protein secretion of LGI1 mutations causing autosomal dominant lateral temporal epilepsy. Epilepsia 2011; 52: 1258–1264 [DOI] [PubMed] [Google Scholar]
- 15. Striano P, Busolin G, Santulli L, et al. Familial temporal lobe epilepsy with psychic auras associated with a novel LGI1 mutation. Neurology 2011; 76: 1173–1176 [DOI] [PubMed] [Google Scholar]
- 16. Winawer MR, Boneschi FM, Barker-Cummings C, et al. Four new families with autosomal dominant partial epilepsy with auditory features: clinical description and linkage to chromosome 10q24. Epilepsia 2002; 43: 60–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brodtkorb E, Gu W, Nakken KO, Fischer C, Steinlein OK. Familial temporal lobe epilepsy with aphasic seizures and linkage to chromosome 10q22–q24. Epilepsia 2002; 43: 228–235 [DOI] [PubMed] [Google Scholar]
- 18. Chabrol E, Popescu C, Gourfinkel-An I, et al. Two novel epilepsy-linked mutations leading to a loss of function of LGI1. Arch Neurol 2007; 64: 217–222 [DOI] [PubMed] [Google Scholar]
- 19. Fertig E, Lincoln A, Martinuzzi A, Mattson RH, Hisama FM. Novel LGI1 mutation in a family with autosomal dominant partial epilepsy with auditory features. Neurology 2003; 60: 1687–1690 [DOI] [PubMed] [Google Scholar]
- 20. Hedera P, Abou-Khalil B, Crunk AE, Taylor KA, Haines JL, Sutcliffe JS. Autosomal dominant lateral temporal epilepsy: two families with novel mutations in the LGI1 gene. Epilepsia 2004; 45: 218–222 [DOI] [PubMed] [Google Scholar]
- 21. Ikeda A, Kunieda T, Miyamoto S, Fukuyama H, Shibasaki H. Autosomal dominant temporal lobe epilepsy in a Japanese family. J Neurol Sci 2000; 176: 162–165 [DOI] [PubMed] [Google Scholar]
- 22. Kobayashi E, Santos NF, Torres FR, et al. Magnetic resonance imaging abnormalities in familial temporal lobe epilepsy with auditory auras. Arch Neurol 2003; 60: 1546–1551 [DOI] [PubMed] [Google Scholar]
- 23. Pisano T, Marini C, Brovedani P, et al. Abnormal phonologic processing in familial lateral temporal lobe epilepsy due to a new LGI1 mutation. Epilepsia 2005; 46: 118–123 [DOI] [PubMed] [Google Scholar]
- 24. Pizzuti A, Flex E, Di Bonaventura C, et al. Epilepsy with auditory features: a LGI1 gene mutation suggests a loss-of-function mechanism. Ann Neurol 2003; 53: 396–399 [DOI] [PubMed] [Google Scholar]
- 25. Poza JJ, Saenz A, Martinez-Gil A, et al. Autosomal dominant lateral temporal epilepsy: clinical and genetic study of a large Basque pedigree linked to chromosome 10q. Ann Neurol 1999; 45: 182–188 [DOI] [PubMed] [Google Scholar]
- 26. Striano P, de Falco A, Diani E, et al. A novel loss-of-function LGI1 mutation linked to autosomal dominant lateral temporal epilepsy. Arch Neurol 2008; 65: 939–942 [DOI] [PubMed] [Google Scholar]
- 27. Naus J. The distribution of the size of the maximum cluster of points on a line. J Am Stat Assoc 1965; 60: 532–538 [Google Scholar]
- 28. Kalachikov S, Evgrafov O, Ross B, et al. Mutations in LGI1 cause autosomal-dominant partial epilepsy with auditory features. Nat Genet 2002; 30: 335–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morante-Redolat JM, Gorostidi-Pagola A, Piquer-Sirerol S, et al. Mutations in the LGI1/epitempin gene on 10q24 cause autosomal dominant lateral temporal epilepsy. Hum Mol Genet 2002; 11: 1119–1128 [DOI] [PubMed] [Google Scholar]
- 30. Fujita PA, Rhead B, Zweig AS, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res 2011; 39: D876–D882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Irani SR, Alexander S, Waters P, et al. Antibodies to Kv1 potassium channel-complex proteins leucine-rich, glioma inactivated 1 protein and contactin-associated protein-2 in limbic encephalitis, Morvan's syndrome and acquired neuromyotonia. Brain 2010; 133: 2734–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chabrol E, Navarro V, Provenzano G, et al. Electroclinical characterization of epileptic seizures in leucine-rich, glioma-inactivated 1-deficient mice. Brain 2010; 133: 2749–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu YE, Wen L, Silva J, et al. Lgi1 null mutant mice exhibit myoclonic seizures and CA1 neuronal hyperexcitability. Hum Mol Genet 2010; 19: 1702–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bhuvanagiri M, Schlitter AM, Hentze MW, Kulozik AE. NMD: RNA biology meets human genetic medicine. Biochem J 2010; 430: 365–377 [DOI] [PubMed] [Google Scholar]
- 35. Schulte U, Thumfart JO, Klocker N, et al. The epilepsy-linked Lgi1 protein assembles into presynaptic Kv1 channels and inhibits inactivation by Kvβ1. Neuron 2006; 49: 697–706 [DOI] [PubMed] [Google Scholar]
- 36. Zhou YD, Lee S, Jin Z, Wright M, Smith SE, Anderson MP. Arrested maturation of excitatory synapses in autosomal dominant lateral temporal lobe epilepsy. Nat Med 2009; 15: 1208–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fukata Y, Adesnik H, Iwanaga T, Bredt DS, Nicoll RA, Fukata M. Epilepsy-related ligand/receptor complex LGI1 and ADAM22 regulate synaptic transmission. Science 2006; 313: 1792– 1795 [DOI] [PubMed] [Google Scholar]
- 38. Sagane K, Ishihama Y, Sugimoto H. LGI1 and LGI4 bind to ADAM22, ADAM23 and ADAM11. Int J Biol Sci 2008; 4: 387– 396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leonardi E, Andreazza S, Vanin S, Busolin G, Nobile C, Tosatto SC. A computational model of the LGI1 protein suggests a common binding site for ADAM proteins. PLoS One 2011; 6: e18142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sirerol-Piquer MS, Ayerdi-Izquierdo A, Morante-Redolat JM, et al. The epilepsy gene LGI1 encodes a secreted glycoprotein that binds to the cell surface. Hum Mol Genet 2006; 15: 3436–3445 [DOI] [PubMed] [Google Scholar]