Abstract
Objective:
To identify copy number variant (CNV) causes of periventricular nodular heterotopia (PNH) in patients for whom FLNA sequencing is negative.
Methods:
Screening of 35 patients from 33 pedigrees on an Affymetrix 6.0 microarray led to the identification of one individual bearing a CNV that disrupted FLNA. FLNA-disrupting CNVs were also isolated in 2 other individuals by multiplex ligation probe amplification. These 3 cases were further characterized by high-resolution oligo array comparative genomic hybridization (CGH), and the precise junctional breakpoints of the rearrangements were identified by PCR amplification and sequencing.
Results:
We report 3 cases of PNH caused by nonrecurrent genomic rearrangements that disrupt one copy of FLNA. The first individual carried a 113-kb deletion that removes all but the first exon of FLNA. A second patient harbored a complex rearrangement including a deletion of the 3′ end of FLNA accompanied by a partial duplication event. A third patient bore a 39-kb deletion encompassing all of FLNA and the neighboring gene EMD. High-resolution oligo array CGH of the FLNA locus suggests distinct molecular mechanisms for each of these rearrangements, and implicates nearby low copy repeats in their pathogenesis.
Conclusions:
These results demonstrate that FLNA is prone to pathogenic rearrangements, and highlight the importance of screening for CNVs in individuals with PNH lacking FLNA point mutations. Neurology® 2012;78:269–278
Congenital disorders of human brain development represent a diverse group of conditions, clinically and genetically. Loss-of-function mutations in FLNA, encoding the actin cross-linking protein Filamin A, cause one of the most prevalent brain malformations encountered clinically: X-linked periventricular nodular heterotopia (PNH).1–3 In PNH, the neurons accumulate as nodules along the surface of the lateral ventricles,1 resulting in seizures by early adulthood, and sometimes impeding normal psychomotor development.1 Since loss-of-function mutations in FLNA are most often lethal prenatally in males, X-linked PNH is clinically encountered primarily in females.1,4
FLNA point mutations are found in most, but not all, patients with X-linked familial PNH,5,6 and in only 26% of sporadic patients with bilateral PNH.5 Some of the missing heritability may be due to genetic heterogeneity.7–10 Alternatively, lesions in Filamin A undetectable by traditional sequencing may also cause disease. One category of such lesions is copy number variants (CNVs), which are an increasingly recognized cause of disease11,12 and trait variation.13–15 FLNA resides in a rearrangement-prone region of the X chromosome at Xq28.16,17 We hypothesized that CNVs in this region might represent a previously unrecognized cause of PNH.
Here, we describe 3 patients with pathogenic FLNA CNVs as a cause of otherwise unexplained PNH. Custom tiling oligo comparative genomic hybridization (CGH) revealed distinct CNV boundaries consistent with rare, nonrecurrent events, likely stimulated by nearby genomic low copy repeats (LCRs). These results demonstrate that genomic rearrangements are an important cause of FLNA mutation, and establish the clinical importance of screening for such rearrangements in PNH.
METHODS
Diagnosis.
Clinically suspected PNH in patients was confirmed by brain MRI.
Affymetrix 6.0 microarray analysis.
DNA samples were analyzed on an Affymetrix genome-wide SNP Array 6.0. Data were processed using Birdsuite18 to identify candidate copy number changes. Birdsuite calls were loaded into a MySQL database and filtered by score (lod >6) to generate a list of candidate CNVs. Separately, SNP calls were analyzed using PennCNV19 with the software's recommended default settings.
Multiple ligation dependent probe amplification analysis.
Multiple ligation dependent probe amplification (MLPA) analysis was performed in individuals from 3 families using the Lissencephaly probe kit (SALSA P061-B2, MRC Holland, Amsterdam, the Netherlands), including 7 paired probes specific for the FLNA gene (exons 4, 11, 22, 25, 29, 39, and 46). The resulting products were separated and sized on an ABI 3130XL sequencer (Applied Biosystems). Analysis was performed according to the manufacturer protocol.
Quantitative PCR validation.
Taqman copy number assays from Applied Biosystems were used to validate CNVs (table e-1 on the Neurology® Web site at www.neurology.org) with RNAseP as the internal control. DNA samples from neurologically normal Caucasian controls were used to calibrate the data. The data were analyzed, plotted, and 95% confidence intervals calculated using CopyCaller software from Applied Biosystems.
Oligo microarray CGH.
A custom CGH array (Nimblegen) with 137,408 probes was used to detect copy number of genomic DNA in areas of interest. The probes were designed to span the entire FLNA genomic locus, including neighboring genes (chrX:152,943,207–153,559,506, hg18), with an average spacing of 30 bp. The data were plotted using the statistical package R. Copy number values were calculated by log2 (Cy3/Cy5), where Cy3 is the probe signal of the proband and Cy5 is the probe signal from a reference female control. To reduce noise, values were normalized to CGH results of a second reference female control, previously established by quantitative PCR (qPCR) to lack copy number variation at the locus. Exon numbering is provided relative to UCSC gene ID uc004fkk.2.
Junctional breakpoint sequencing.
PCR amplification (Promega GoTaq) was employed to sequence the breakpoints of the FLNA locus rearrangements. Primer sequences for family 1: 5′-CTCTGTGAGCCGCAAAGTGT and 5′-CCCCCACAGCTGTTTAGAGA; family 2: 5′-ATGCAGTGCGAGATGTGGAC and 5′-CAGCCTGAAAATCCCTGGTA. Sequencing results were mapped against the human hg18 genomic reference sequence.
Standard protocol approvals, registrations, and patient consents.
Clinical details and biological samples were obtained with written informed consent from the participants or their parents/guardians at Children's Hospital Boston. The study was approved by the Human Research Ethics Committee of all participating institutions and conducted as part of the National Institute of Neurological Disorders and Stroke Trial, Human Epilepsy Genetics–Neuronal Migration Disorders Study #NCT00041600.
RESULTS
Clinical information.
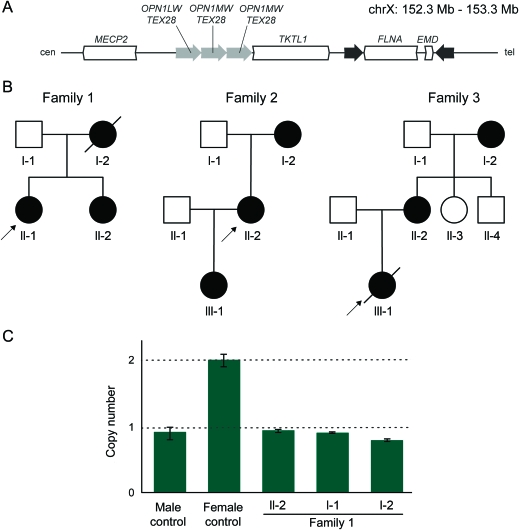
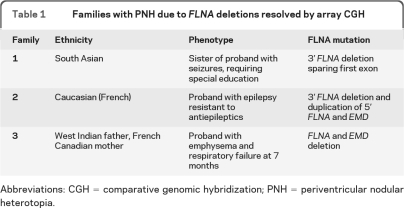
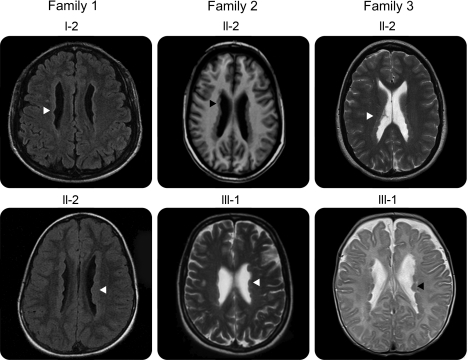
Family 1 is a nonconsanguineous family of South Asian ancestry with multiple women affected by PNH (figure 1B, table 1). Screening ultrasound of female II-1 revealed irregular linings of the ventricles bilaterally, and brain MRI established a diagnosis of PNH. Developmental milestones were achieved appropriately with the exception of walking, which was mildly delayed at 18 months. She is currently pursuing a university degree. Her older sister, II-2, had normal early development but developed frequent seizures in childhood requiring antiepileptics. She had slight right-sided weakness. She required special education and is currently in a certificate program. MRI revealed bilateral nodular heterotopia (figure 2). She remains on antiepileptic medications. Their mother, I-2, was diagnosed with PNH by MRI following her daughters' diagnoses. She died at age 49 of undetermined causes.
Figure 1. The FLNA locus is prone to rearrangement.
(A) The FLNA and EMD genes are flanked by 11.3-kb inverted repeats at Xq28. FLNA encodes for the protein Filamin A, an actin cross-linking protein. EMD encodes for Emerin, a protein disrupted in Emery-Dreifuss muscular dystrophy. Recombination at this locus results in benign inversions. FLNA also lies telomeric to 3 low copy repeats of the color vision gene OPN1 and TEX28. Nonallelic homologous recombination–mediated deletion of OPN1-TEX28 repeats is a known cause of human color blindness. (B) Pedigrees of families 1, 2, and 3. Females are represented by circles, affected individuals by dark circles, and deceased individuals by an angled line. Arrows indicate the proband in each family. (C) Quantitative PCR (qPCR) confirmation of FLNA copy number loss in II-2 and I-2 of family 1. qPCR results targeting sequence centered on chrX:153,240,806. When normalized to a female control, it is apparent that the affected mother and daughter have one copy of the allele at that location, whereas the female control has 2 copies. The father, I-1, has one copy, which is expected for an unaffected male. The 95% confidence intervals were calculated using CopyCaller software by Applied Biosystems.
Table 1.
Families with PNH due to FLNA deletions resolved by array CGH
Abbreviations: CGH = comparative genomic hybridization; PNH = periventricular nodular heterotopia.
Figure 2. Brain MRI in affected individuals from families 1–3.
Representative axial brain MRI images from affected individuals in this study. Bilateral gray matter nodules (arrowheads) line and project into the lateral ventricles, the classic appearance of periventricular nodular heterotopia. Family 1, I-2 and II-2, family 2, II-2: T1-weighted. Family 2, III-1, family 3, II-2 and III-1: T2-weighted.
Family 2 is a multigeneration family affected by PNH (figure 1B, table 1). The proband (II-2) experienced seizures resistant to multiple antiepileptics beginning in childhood. Her cognitive development and capabilities were within normal limits. MRI demonstrated classic, bilateral PNH (figure 2). The proband's mother, I-2, and the proband's daughter, III-1, were both asymptomatic and their diagnoses were ascertained after PNH was diagnosed in the proband, II-2.
Three generations of women in family 3 are affected by bilateral PNH (figure 1B, table 1), with some additional pathogenic features. The proband III-1 was born at 39 weeks to a 40-year-old mother, II-2. The pregnancy was significant for slowed intrauterine growth from 28 to 29 weeks. At 2 months of age, she became irritable, with decreased feeding, poor sleep, stridor, and weak cry. She was admitted to the hospital where investigations revealed right ventricular hypertrophy, a patent foramen ovale, and pulmonary emphysema affecting multiple lobes. She was seen by the Genetics team because of possible dysmorphic features (blue sclerae, joint hypermobility, and loose skin); routine chromosome analysis and array CGH studies were normal. Her pulmonary function deteriorated progressively, and she died of respiratory insufficiency at 7 months. Autopsy showed bilateral, symmetric PNH, pulmonary panacinar emphysema, and tricuspid and mitral valve dysplasia. Her mother and grandmother were subsequently also diagnosed with PNH. Her mother also had cardiac valvular abnormalities and apical bullae of the lungs; her grandmother had lobar emphysema. No seizures were evident in any of the 3 affected individuals.
Molecular investigations in family 1.
The proband of family 1 was identified in a screen for FLNA CNVs in a cohort of 35 PNH patients (from 33 pedigrees) genotyped with Affymetrix 6.0 microarrays. The resulting data were used to screen for CNVs using 2 algorithms, Birdsuite18 and PennCNV.19 Applying Birdsuite, we found 3 patients with evidence suggestive of copy number loss overlapping FLNA. Only one of these events was predicted with PennCNV, and qPCR confirmed only the single true positive. In this female individual (figure 1B, family 1, individual II-1), Birdsuite predicted a single copy deletion spanning 19.8 kb (hg18, chrX:153,228,832–153,248,612), disrupting the 3′ end of FLNA. qPCR in the proband using multiple probes spanning the FLNA locus, as well as MLPA, confirmed a copy number of 1 over this interval (data not shown); this copy number change was also found in 2 other affected family members (figure 1C, family 1, sister II-2, and mother I-2), both of whom had MRI-confirmed PNH (figure 2).
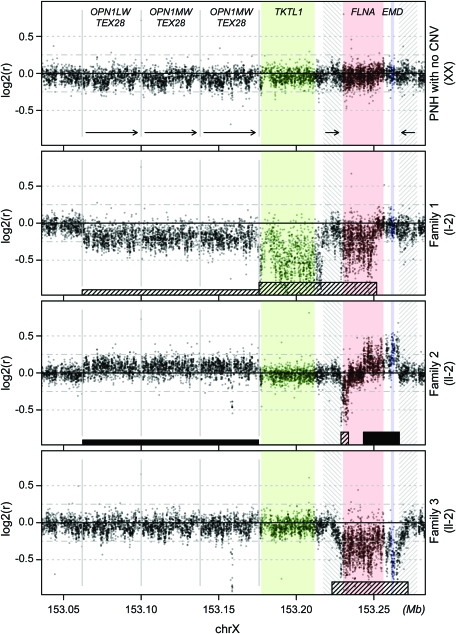
To understand the rearrangement that led to this copy number change, we generated a custom microarray to fine-tile the FLNA locus with overlapping 40–60 bp probes at a spacing of 15–30 bp, and performed CGH. CGH of individual I-2 from family 1 confirmed the presence of a FLNA 3′ deletion, which was significantly larger than originally predicted by the Affymetrix 6.0 data. Decreased signal intensities were observed over the entire TKTL1 gene and nearly all of FLNA, consistent with loss of a single copy (figure 3, second panel). This lesion spares only FLNA exon 1 (noncoding) and FLNA exon 2 (the first coding exon), and is therefore predicted to abrogate FLNA function. The remainder of the locus demonstrated partially depressed signal intensities over the ∼120 kb region centromeric to TKTL1, as well as over both inverted 11.3 kb LCRs flanking FLNA (figure 3, second panel). The former region contains 3 39-kb direct LCRs that include the opsin genes responsible for human color vision.20 Each repeating subunit contains one copy of the opsin gene OPN1 and one copy of TEX28 (figure 3, first panel). Since the CGH probes in the OPN1-TEX28 locus cannot distinguish one direct repeat from another, a deletion of one of the 3 LCRs on one chromosome would be predicted to cause a one-sixth reduction in observed copy number relative to a diploid control [expected log2ratio of log2(5/6) = −0.26]. We therefore interpret the slight but uniform decrease in log2ratio across the OPN1-TEX28 region to indicate deletion of one of the 3 LCRs on one chromosome.
Figure 3. High-resolution array comparative genomic hybridization (CGH) of the FLNA locus delineates distinct genomic rearrangements in each family with periventricular nodular heterotopia (PNH).
Top panel: A female individual with PNH, but without FLNA copy number change detected by quantitative PCR (qPCR), demonstrates signal intensities consistent with normal copy number across the locus. Second panel: Affected individual I-2 of family 1 demonstrates single-copy loss of all but the 5′ end of FLNA, as well as the adjacent TKTL1 gene. Third panel: Affected individual II-2 of family 2 demonstrates a single copy deletion of the 3′ end of FLNA, and duplication of the 5′ end of FLNA and EMD. Bottom panel: Affected individual II-2 of family 3 demonstrates deletion of both FLNA and EMD. The y-axis represents the log2 ratio of the signal intensity from the patient compared to a reference female control. Tall striped rectangles = inferred deletions; tall filled rectangle = inferred duplications; short striped and solid rectangles = regions of slightly decreased and increased signal intensity, respectively, across entire OPN1-TEX28 locus; green, red, blue shading = TKTL1, FLNA, EMD gene loci, respectively; gray hashes and short arrows = boundaries and direction of the inverted repeats flanking FLNA and EMD; gray vertical lines and long arrows = boundaries and direction of the OPN1-TEX28 tandem repeats. CNV = copy number variant.
Molecular investigations in families 2 and 3.
Screening additional cohorts of PNH families using MLPA revealed 2 more pathogenic FLNA deletions. Family 2 is a 3-generation family with PNH (figure 1B). DNA was obtained from the proband, II-2 (DNA samples were not available from I-2 and III-1). MLPA revealed the proband to be missing one copy of FLNA (data not shown). Fine-tiling array CGH delineated a 4.8-kb deletion removing exons 42–48 of FLNA (figure 3, third panel), accompanied by a 23-kb duplication spanning the 5′ end of FLNA and all of EMD (figure 3, third panel; see Discussion). This mutation is likely to be deleterious, since distal truncating mutations as late as exon 47 in FLNA cause PNH.21 There is also a slight but uniform increase in signal intensity in the OPN1-TEX28 tandem repeat region, consistent with a duplication of one repeat on one chromosome [expected log2ratio, log2(7/6) = 0.22].
Family 3 is another 3-generation family exhibiting X-linked PNH (figures 1B and 2). MLPA screening of individual III-1 and her mother II-2 revealed a FLNA copy number loss (data not shown), and fine-tiling array CGH in II-2 demonstrated a 39-kb deletion with breakpoints mapping to the 11.3-kb inverted LCRs (figure 3, fourth panel). The identical deletion was detected by array CGH in individual III-1 (data not shown). This deletion removes one copy of the entire FLNA gene, providing an explanation for PNH in this pedigree. The deletion also removes one copy of the neighboring EMD gene. Mutations in EMD cause X-linked recessive Emery-Dreifuss muscular dystrophy, a degenerative myopathy without involvement of the CNS.22 Female carriers of EMD mutations are at risk for cardiac abnormalities, including cardiomyopathy and conduction defects.23,24
Junctional breakpoint analysis.
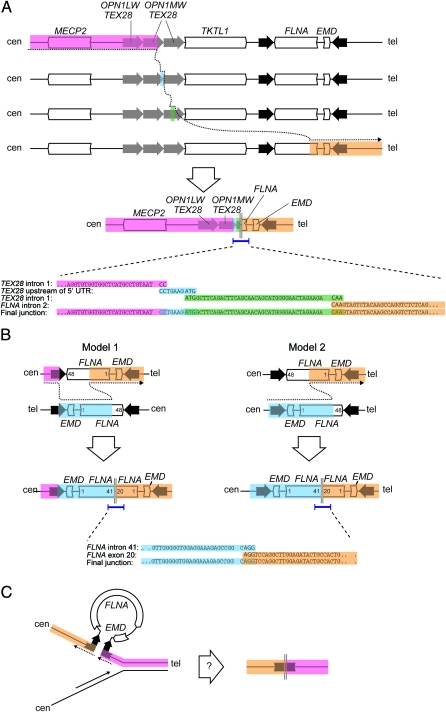
A series of PCR assays were designed to interrogate the rearrangement breakpoint junctions. We successfully identified breakpoint PCR products in 2 of the 3 families. In family 1, PCR was attempted under the assumption of a simple, contiguous 113-kb deletion extending from TKTL1 to FLNA (see Discussion). A junctional amplification product was obtained of the approximate size predicted (data not shown), but sequence analysis revealed evidence of a more complex mechanism. Instead of a contiguous 113-kb deletion, the junctional product contained sequence from TEX28 intron 1, followed by an apparent 1,700-bp deletion, followed by 10 base pairs of TEX28 promoter, followed by a 33,403-bp deletion, followed by 43 base pairs of TEX28 intron 1, followed by a 77,942-bp deletion, followed by sequence from FLNA intron 2 (figure 4A). Closer inspection revealed 2 to 3 base pairs of microhomology between adjacent fragments (figure 4A).
Figure 4. Copy number changes at the FLNA locus in 3 families with periventricular nodular heterotopia implicate distinct molecular mechanisms.
(A) Breakpoint sequencing supports a mechanism for family 1 rearrangement involving replication fork stalling and template switching/microhomology-mediated break-induced replication with template switches occurring between the second OPN1-TEX28 tandem repeat, the junction between the first and second OPN1-TEX28 tandem repeats, the third OPN1-TEX28 repeat, and FLNA intron 2. The result is loss of 1 OPN1-TEX28 tandem repeat and a deletion of all but the 5′ end of FLNA. (B) Breakpoint sequencing in family 2 supports 2 alternative models. In model 1, nonallelic homologous recombination between mispaired inverted repeats is followed by nonhomologous end joining (NHEJ) between 2 inverted copies of FLNA on opposite chromatids, leading to deletion of the 3′ end of FLNA and duplication of the 5′ end of FLNA and EMD. Model 2 presupposes a heterozygous background for the benign FLNA-EMD inversion, in which case a single NHEJ event between FLNA intron 41 on one chromatid and FLNA exon 20 on the second chromatid is sufficient to generate the observed rearrangement. (C) A speculative, replication-based model for FLNA rearrangement in family 3, in which the inverted repeats flanking FLNA and EMD form a single-strand secondary structure that results in contiguous deletion of both genes and the inner half of both repeats.
In family 2, array CGH data suggested a head-to-tail partial duplication of FLNA (see Discussion). We designed PCR primers extending in the centromeric direction from exon 20 and from exon 41 and successfully obtained an amplification product, analysis of which revealed a junction between FLNA intron 41 and exon 20 (figure 4B). Three base pairs of microhomology were shared between the 2 fragments.
DISCUSSION
Point mutations in FLNA are found in most but not all patients with X-linked familial PNH, and only 26% of sporadic cases,5,6 suggesting the existence of other causative genetic lesions. Here, we applied a variety of methods to identify CNVs overlapping FLNA in patients with developmental brain disorders. Three families harboring pathogenic FLNA deletions are described: family 1, carrying a deletion of the majority of the 3′ end of FLNA; family 2, with a deletion of the 3′ end of FLNA and partial duplication of the 5′ end and EMD; and family 3, with a deletion of the entire FLNA gene and EMD. Deletions of EMD have been previously described,16,17 but to our knowledge, this is the first report of deletion involving both EMD and FLNA.
The FLNA-EMD locus has been reported to be prone to genomic rearrangements, likely stimulated by multiple LCRs in the region.16 Two 11.3-kb inverted repeats flank FLNA and EMD. Inverted repeats promote genomic instability25 by a variety of mechanisms,26,27 one of which is nonallelic homologous recombination (NAHR). NAHR between the inverted repeats results in benign inversion of FLNA-EMD in up to 18% of patients of European descent.16 Mispairing between the highly homologous 11.3-kb inverted repeats, followed by nonhomologous end joining (NHEJ) between an intervening Alu element and FLNA intron 28, was shown to result in deletion of EMD (and partial duplication of FLNA) in a patient with Emery-Dreifuss muscular dystrophy.16 Inspection of the copy number pattern observed in family 2 (figure 3) suggests a similar mechanism of rearrangement (figure 4B): NAHR between 2 misaligned inverted repeats, followed by NHEJ between exon 20 of FLNA and intron 41 of FLNA on the opposite chromatid, resulting in deletion of the 3′ end of FLNA and head-to-tail duplication of the 5′ end of FLNA and EMD. Alternatively, a second model is possible in which this rearrangement first occurred in a maternal ancestor heterozygous for one copy of the benign FLNA-EMD inversion (figure 4B). NAHR between properly aligned LCRs, followed by NHEJ between FLNA exon 20 and intron 41, would result in the same final product.
The boundaries of the FLNA deletion observed in family 1 suggest a distinct, more complicated mechanism. While array CGH results (figure 3) initially suggested a single 113-kb deletion, breakpoint sequencing revealed that the expected contiguous deletion was actually interrupted by 2 sequences, 10 and 43 base pairs long, corresponding to sequences upstream of the TEX28 5′ UTR and sequences from TEX28 intron 1, respectively, with 2–3 base pairs of microhomology at each sequence junction (figure 4A). This pattern, in which the resulting product appears as if the polymerase “skipped” several times across the template with evidence of microhomology at each junction, suggests that the family 1 rearrangement arose via a replication Fork Stalling and Template Switching (FoSTeS)/microhomology-mediated break-induced replication (MMBIR).28 FoSTeS/MMBIR has been observed to underlie other CNV-associated disorders such as Pelizaeus-Merzbacher disease (PMD),29 Potocki-Lupski microduplication syndrome (PTLS)/Smith-Magenis microdeletion syndrome, and Charcot-Marie-Tooth disease type 1A (CMT1A).30 The resulting product in family 1 is an allele deleting one OPN1-TEX28 unit, all of TKTL1 and the centromeric 11.3-kb inverted repeat, and the 3′ end of FLNA (figure 4A). We believe the PNH phenotype in this family is explained entirely by the FLNA truncation; to our knowledge, no clinical diseases have been ascribed to TKTL1 haploinsufficiency, although there are hypothesized links to susceptibility to Wernicke-Korsakoff syndrome.31–34
In family 3, array CGH predicts a clean excision of FLNA and EMD, as well as the inner half of each flanking repeat. This is despite the fact that direct homologous recombination between 2 inverted repeats is expected to cause inversion, not deletion. Examination of the Alu-rich inverted repeats for direct repeats that could lead to deletion revealed no such sequence (data not shown). We speculate that a mechanism for the family 3 rearrangement might be one akin to replication slippage,28 in which the inverted DNA repeats—exposed during replication as single-stranded DNA—form a secondary structure that leads to deletion of the intervening DNA (figure 4C). This explanation is not wholly satisfactory, however, as the size of this deletion (39 kb) is considerably larger than those typically observed in replication slippage. Since we were unable to amplify the breakpoint sequence, we cannot rule out the possibility of FoSTeS/MMBIR in this family as well.
The reported spectrum of clinically relevant FLNA variations includes not only loss-of-function mutations (resulting in PNH) but also gain-of-function mutations (resulting in otopalatodigital spectrum disorders) and partial loss-of-function mutations (associated with milder PNH and male survival).35 The clinical presentations of the patients in this study are generally indistinguishable from those of other loss-of-function alleles. This is consistent with these lesions acting as molecular nulls.
Affected individuals in family 3 exhibited additional pulmonary abnormalities, a finding that has been recently reported in 2 cases, in the setting of a missense mutation (p.G74R) and a truncating mutation (p.K331X).36,37 Our finding that deletion can phenocopy these changes indicates that the pulmonary phenotype is the result of FLNA loss-of-function, rather than a novel gain-of-function effect specific to the 2 other alleles. Although the true incidence of pulmonary disease in individuals with FLNA mutation remains to be established, it is unclear why pulmonary abnormalities are more severe in some individuals. In this respect, while affected individuals in family 3 are unique in that they harbor a concomitant EMD deletion, this is probably insufficient to account for their pulmonary presentation: EMD mutations are not known to affect the lung, and the previously reported pulmonary cases36,37 are not known to have additional mutations in EMD.
Here we have described 3 cases of FLNA genomic rearrangements leading to PNH, each with a unique pattern of copy number loss or gain that implies distinct, nonrecurrent mutational mechanisms. These results add to the rich variety of human variation documented at the Xq28 locus—opsin gene deletion causing color blindness, benign FLNA-EMD inversion, EMD deletion causing muscular dystrophy, and now FLNA disruption causing PNH—and underscore the importance of molecular screening for such rearrangements.
Supplementary Material
GLOSSARY
- CGH
comparative genomic hybridization
- CMT1A
Charcot-Marie-Tooth disease type 1A
- CNV
copy number variant
- FoSTeS
Fork Stalling and Template Switching
- LCR
low copy repeat
- MLPA
multiple ligation dependent probe amplification
- MMBIR
microhomology-mediated break-induced replication
- NAHR
nonallelic homologous recombination
- NHEJ
nonhomologous end joining
- PMD
Pelizaeus-Merzbacher disease
- PNH
periventricular nodular heterotopia
- PTLS
Potocki-Lupski microduplication syndrome
- qPCR
quantitative PCR.
Footnotes
Supplemental data at www.neurology.org
AUTHOR CONTRIBUTIONS
K.R.C. performed Birdsuite copy number analyses, QPCR experiments, and wrote the manuscript. T.W.Y. designed and performed Birdsuite copy number analyses, QPCR and breakpoint localization experiments, and wrote the manuscript. V.S.G. helped perform copy number analyses, helped design and perform breakpoint localization experiments, and helped write the manuscript. B.B. organized clinical information and patient samples. Y.C. performed PennCNV copy number analyses. D.M., B.F., and E.P. identified and clinically characterized family 2, and D.M. and E.P. performed MLPA analysis of family 2. L.D. and M.N. identified and characterized family 1. C.S. and C.K. identified and clinically characterized family 3. R.G. directed the overall research and wrote the manuscript. C.A.W. directed the overall research and wrote the manuscript.
STUDY FUNDING
K.R.C. and T.W.Y. were supported by the Nancy Lurie Marks family foundation. T.W.Y. was additionally supported by the Clinical Investigator Training Program at Harvard-MIT Health Sciences and Technology and Beth Israel Deaconess Medical Center, in collaboration with Pfizer, Inc. and Merck and Co., Inc. This work was also supported by grants from the NINDS (RO1 NS35129) and the Manton Center for Orphan Disease Research (to CAW), ISS grant PRE 178/07 COR-F (to R.G.), and a SPARC grant from the Broad Institute of Harvard and Massachusetts Institute of Technology, and by the Sixth Framework Programme of the EU project grant LSH-2005-2.1.3-2 (to R.G.). C.A.W. is an Investigator of the Howard Hughes Medical Institute.
DISCLOSURE
K.R. Clapham reports no disclosures. Dr. Yu has received research support from the Nancy Lurie Marks Foundation and from the Clinical Investigator Training Program at Harvard-MIT Health Sciences and Technology and Beth Israel Deaconess Medical Center, in collaboration with Pfizer Inc and Merck and Co., Inc. V.S. Ganesh, B. Barry, and Y.R. Chan report no disclosures. Dr. Mei serves on the editorial board of Epilepsia. Dr. Parrini, Dr. Funalot, and L. Dupuis report no disclosures. Dr. Nezarati and her spouse hold stock in Johnson & Johnson. C. du Souich reports no disclosures. Dr. van Karnebeek receives research support from the BC Children's Hospital Foundation, Vancouver, Canada. Dr. Guerrini has served on scientific advisory boards for Biocodex, UCB, Eisai Inc., ValueBox, and EMA (European Medicine Agency); has received funding for travel and/or speaker honoraria from Biocodex, Eisai Inc., Japanese Epilepsy Society, and Weill Cornell Medical College in Qatar; serves as Associate Editor of Epilepsia and on the international advisory board of Progress in Epileptic Disorders; serves/has served on the editorial boards of Neuropediatrics, the Journal of Child Neurology, Seizure, Epileptic Disorders, the European Neurological Journal, BMC Medical Genetics, Topics in Epilepsy, and Journal of Pediatric Epilepsy; receives royalties from the publication of Epilepsy and Movement Disorders (Cambridge University Press, 2002), Aicardi's Epilepsy in Children (Lippincott Williams & Wilkins, 2004), Progress in Epileptic Spasms on West Syndrome (John Libbey, Eurotext, 2007), and The Causes of Epilepsy (Cambridge University Press, 2011); has received honoraria from sanofi-aventis and Eisai Inc.; and receives research support from the Italian Ministry of Health, the European Community Sixth Framework Thematic Priority Life Sciences, Genomics and Biotechnology for Health, the Italian Ministry of Education, University and Research, the Tuscany Region, the Telethon Foundation, and the Mariani Foundation. Dr. Walsh serves on scientific advisory boards for Autism Consortium, Merck Foundation, McKnight Foundation, and Generation Health; has received funding for travel or speaker honoraria from Pfizer Inc; is an employee of Howard Hughes Medical Institute; serves as a consultant for Anne and Paul Marcus Foundation and MPM Capital; and receives research support from the NIH (NINDS/NIMH), NLM Family Foundation, and Simons Foundation.
REFERENCES
- 1. Ekşioğlu YZ, Scheffer IE, Cardenas P, et al. Periventricular heterotopia: an X-linked dominant epilepsy locus causing aberrant cerebral cortical development. Neuron 1996;16:77–87 [DOI] [PubMed] [Google Scholar]
- 2. Fox JW, Lamperti ED, Ekşioğlu YZ, et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron 1998;21:1315–1325 [DOI] [PubMed] [Google Scholar]
- 3. Zenker M, Rauch A, Winterpacht A, et al. A dual phenotype of periventricular nodular heterotopia and frontometaphyseal dysplasia in one patient caused by a single FLNA mutation leading to two functionally different aberrant transcripts. Am J Hum Genet 2004;74:731–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guerrini R, Mei D, Sisodiya S, et al. Germline and mosaic mutations of FLN1 in men with periventricular heterotopia. Neurology 2004;63:51–56 [DOI] [PubMed] [Google Scholar]
- 5. Parrini E, Ramazzotti A, Dobyns WB, et al. Periventricular heterotopia: phenotypic heterogeneity and correlation with Filamin A mutations. Brain 2006;129:1892–1906 [DOI] [PubMed] [Google Scholar]
- 6. Sheen VL, Dixon PH, Fox JW, et al. Mutations in the X-linked filamin 1 gene cause periventricular nodular heterotopia in males as well as in females. Hum Mol Genet 2001;10:1775–1783 [DOI] [PubMed] [Google Scholar]
- 7. Sheen VL, Topçu M, Berkovic S, et al. Autosomal recessive form of periventricular heterotopia. Neurology 2003;60:1108–1112 [DOI] [PubMed] [Google Scholar]
- 8. Sheen VL, Wheless JW, Bodell A, et al. Periventricular heterotopia associated with chromosome 5p anomalies. Neurology 2003b;60:1033–1036 [DOI] [PubMed] [Google Scholar]
- 9. Sheen VL, Basel-Vanagaite L, Goodman JR, et al. Etiological heterogeneity of familial periventricular heterotopia and hydrocephalus. Brain Dev 2004;26:326–334 [DOI] [PubMed] [Google Scholar]
- 10. Sheen VL, Ganesh VS, Topcu M, Sebire G, et al. Mutations in ARFGEF2 implicate vesicle trafficking in neural progenitor proliferation and migration in the human cerebral cortex. Nat Genet 2004;36:69–76 [DOI] [PubMed] [Google Scholar]
- 11. Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet 2009;10:451–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat Genet 2007;39(7 suppl):S37–S42 [DOI] [PubMed] [Google Scholar]
- 13. Carvalho CMB, Zhang F, Lupski JR. Evolution in health and medicine: Sackler colloquium: genomic disorders: a window into human gene and genome evolution. Proc Natl Acad Sci USA 2010;107(suppl 1):1765–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee JA, Lupski JR. Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders. Neuron 2006;52:103–121 [DOI] [PubMed] [Google Scholar]
- 15. Walsh CA, Engle EC. Allelic diversity in human developmental neurogenetics: insights into biology and disease. Neuron 2010;68:245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Small K, Iber J, Warren ST. Emerin deletion reveals a common X-chromosome inversion mediated by inverted repeats. Nat Genet 1997;16:96–99 [DOI] [PubMed] [Google Scholar]
- 17. Small K, Warren ST. Emerin deletions occurring on both Xq28 inversion backgrounds. Hum Mol Genet 1998;7:135–139 [DOI] [PubMed] [Google Scholar]
- 18. Korn JM, Kuruvilla FG, Mccarroll SA, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet 2008;40:1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang K, Li M, Hadley D, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 2007;17:1665–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nathans J, Thomas D, Hogness DS. Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science 1986;232:193–202 [DOI] [PubMed] [Google Scholar]
- 21. Moro F, Carrozzo R, Veggiotti P, et al. Familial periventricular heterotopia: missense and distal truncating mutations of the FLN1 gene. Neurology 2002;58:916–921 [DOI] [PubMed] [Google Scholar]
- 22. Bione S, Maestrini E, Rivella S, et al. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat Genet 1994;8:323–327 [DOI] [PubMed] [Google Scholar]
- 23. Funakoshi M, Tsuchiya Y, Arahata K. Emerin and cardiomyopathy in Emery-Dreifuss muscular dystrophy. Neuromuscul Disord 1999;9:108–114 [DOI] [PubMed] [Google Scholar]
- 24. Karst ML, Herron KJ, Olson TM. X-linked nonsyndromic sinus node dysfunction and atrial fibrillation caused by emerin mutation. J Cardiovasc Electrophysiol 2008;19:510–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gordenin DA, Lobachev KS, Degtyareva NP, Malkova AL, Perkins E, Resnick MA. Inverted DNA repeats: a source of eukaryotic genomic instability. Mol Cell Biol 1993;13:5315–5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lobachev KS, Shor BM, Tran HT, et al. Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics 1998;148:1507–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Voineagu I, Narayanan V, Lobachev KS, Mirkin SM. Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proc Natl Acad Sci USA 2008;105:9936–9941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nature Reviews Genetics 2009;10:551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee JA, Carvalho CMB, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 2007;131:1235–1247 [DOI] [PubMed] [Google Scholar]
- 30. Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, Lupski JR. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet 2009;41:849–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blass JP, Gibson GE. Abnormality of a thiamine-requiring enzyme in patients with Wernicke-Korsakoff syndrome. N Engl J Med 1977;297:1367–1370 [DOI] [PubMed] [Google Scholar]
- 32. Mukherjee AB, Svoronos S, Ghazanfari A, et al. Transketolase abnormality in cultured fibroblasts from familial chronic alcoholic men and their male offspring. J Clin Invest 1987;79:1039–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leigh D, McBurney A, McIlwain H. Erythrocyte transketolase activity in the Wernicke-Korsakoff syndrome. Br J Psychiatry 1981;139:153–156 [DOI] [PubMed] [Google Scholar]
- 34. Leigh D, McBurney A, McIlwain H. Wernicke-Korsakoff syndrome in monozygotic twins: a biochemical peculiarity. Br J Psychiatry 1981;139:156–159 [DOI] [PubMed] [Google Scholar]
- 35. Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol 2004;6:1034–1038 [DOI] [PubMed] [Google Scholar]
- 36. Masurel-Paulet A, Haan E, Thompson EM, et al. Lung disease associated with periventricular nodular heterotopia and an FLNA mutation. Eur J Med Genet 2010;54:25–28 [DOI] [PubMed] [Google Scholar]
- 37. de Wit MCY, Tiddens HAWM, de Coo IFM, Mancini GMS. Lung disease in FLNA mutation: confirmatory report. Eur J Med Genet 2011;54:299–300 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.