Abstract
Background
What characterizes individuals whose natural killer (NK) cells are able to respond to HIV-1 peptides is not known.
Methods
The association between NK cell responses and KIR gene profiles and HLA-B and HLA-C alleles was investigated among 76 HIV-1-infected women in South Africa previously categorized as responders (n = 39) or nonresponders (n = 37) to HIV-1 peptide pools in a whole blood intracellular cytokine assay. Viral load was significantly lower and CD4 T-cell counts higher among responders compared with nonresponders (P = 0.023 and P = 0.030, respectively).
Results
Possession of one HLA-C1 allele associated with increased magnitude of NK cell responses to Env (P = 0.031) and significantly decreased viral load (P = 0.027) compared with its absence. There was a trend to increased possession of KIR2DL3+HLA-C1 in responders (71.8% vs 51.4%, P = 0.098) and decreased possession of KIR2DL3/2DL3+C2C2 (2.6% vs 16.2%, P = 0.053). A total of 64.1% of responders versus 32.4% of nonresponders had 13 or more KIR genes (P = 0.0067). Notably, the 13-KIR gene containing the Bx21 genotype (has eight inhibitory and three activating genes KIR2DS2, 2DS4, 2DS5) showed substantially higher representation among the responders (28.2% vs 2.6%, P = 0.001). A significantly higher proportion of responders had both KIR2DS2 and KIR2DS5 compared with either gene alone (72.4% vs 37%; P = 0.015). At least one HLA-C1 allele together with 13 or more KIR genes was associated with NK cell responsiveness (48.7% vs 13.5%; P = 0.001).
Conclusion
NK cell responses to HIV-1 peptides are more likely to occur among individuals with a genotype supporting a more activating NK cell phenotype and who possess at least one HLA-C1 allele.
Keywords: HIV-specific NK cell responses, KIR, HLA-B, HLA-C
INTRODUCTION
Natural killer (NK) cells are highly versatile cells that contribute to both innate and adaptive immunity. NK cells under normal, noninflammatory conditions are strictly dominated by inhibitory signals, a mechanism that ensures that healthy cells are not inadvertently destroyed. Under altered conditions, through recognition of “missing self” (loss or downregulation of HLA Class I molecules), “induced self” through stress (upregulated host molecules), or through foreign recognition (allogeneic cells or pathogen infection), NK cells overcome inhibitory signals and become activated culminating in killing of target cells. These interactions between NK cells and target cells are all mediated by various activating and inhibitory receptors on NK cells and their corresponding ligands on target cells.1,2 Among these receptors are killer-cell immunoglobulin-like receptors (KIRs) that bind specific human leukocyte antigen (HLA) Class I molecules that have, in several genetic studies, shown importance in relation to control of HIV-1 infection.3–5
HIV-1-infected individuals who are viremic display altered NK receptor ligand expression,6 upregulation of inhibitory NK cell receptors,7,8 and reduced expression of activating NK cell receptors.7,9–11 These alterations would be expected to collectively predispose NK cells to a more inhibitory type of phenotype, raising the threshold that would need to be overcome for NK cells to contribute to control of HIV-1 infection through activation and subsequent elimination of virus-infected cells.
Our recent findings have described NK cell (non-T-cell/CD3-negative) responses to HIV-1 peptides among HIV-1-infected mothers and their infants that were associated with reduced maternal–infant HIV-1 transmission and associated with significantly lower viral loads and higher CD4 T-cell counts in the mothers.12,13 Because exposure to HIV-1 peptides in the assay results in activation of NK cells (measured by intracellular detection of interferon-γ) in some individuals, we postulated that these “responders” would possess a more activating KIR gene profile or particular KIR–HLA combinations, that would explain the ability of their NK cells to overcome inhibitory signals and so be able to mount NK cell responses in the presence of HIV-1 peptides. In this study, we describe the KIR and HLA-B and HLA-C genes in relation to the detection of NK cell responses to HIV-1 peptides of the HIV-1-infected women from the mother–child cohort.12,13 We show that the greater the KIR gene number, the Bx21 KIR genotype and KIR gene number in combination with an HLA-C1 allele characterize those individuals with NK responses to HIV-1 peptides.
MATERIALS AND METHODS
Study Samples
Genomic DNA was extracted from whole blood of a total of 76 HIV-1-infected women using the QIAamp DNA Mini Kit (Qiagen, Dusseldorf, Germany) according to the manufacturer’s instructions. These 76 were when sufficient sample was available of 79 HIV-1-infected women whose HIV-specific NK cell responses (CD3-negative) were previously described.12,13 All were recruited at one of two sites in Johannesburg, South Africa, during the postpartum period as part of a study of maternal–infant HIV transmission.12,13 Of these women, 39 had positive NK cell responses (forthwith termed “responders”) to at least one HIV-1 peptide pool (Gag, Pol, Nef, Reg, and Env peptide pools tested) and 37 had no detectable NK cell responses (termed “nonresponders”); three of the original nonresponder group of 40 had no DNA sample available. Magnitudes of each individual’s peptide pool(s) response are reported in Table 2 of reference 12 and graphically shown with corresponding magnitudes of patient CD4 and CD8 T-cell responses in the study by Tiemessen et al.13 HIV-1 RNA levels (expressed as log10 units) were quantitated using the Roche Amplicor RNA Monitor assay (Roche Diagnostic Systems, Inc, Branchburg, NJ) with a lower detection limit of 400 HIV-1 RNA copies/mL. CD4 T-cell counts were determined using the commercially available FACSCount System from Becton Dickinson (San Jose, CA). The median viral load for the total group was 4.10 log10 (range, 2.6–5.69 log) (n = 76), and the median CD4 T-cell count was 436 cells/μL (range, 40–1655 cells/μL) (n = 57). Only one woman who was in the NK responder group received triple-drug HIV treatment. None of the others had received HIV treatment, although most had received single-dose nevirapine for the prevention of maternal–infant HIV transmission. For comparisons involving CD4 T-cell counts or viral load, exclusion of this sample did not alter any outcomes so was therefore included throughout.
This study was approved by the University of Wit-watersrand Committee for Research on Human Subjects and the Institutional Review Board of Columbia University and signed informed consent was obtained from all participants.
KIR GENOTYPING
KIR genotyping was performed using sequence-specific primer polymerase chain reaction (Olerup SSP KIR Genotyping kit; Olerup SSP AB, Stockholm, Sweden). Genomic DNA was genotyped for the presence or absence of the following KIR genes: KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL4, KIR2DL5, KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5, KIR3DL1, KIR3DL2, KIR3DL3, KIR3DS1, KIR2DP1, and KIR3DP1. Group B haplotypes possess one or more of the following genes: KIR2DL5, KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS5, and KIR3DS1. Group A haplotypes were defined by the absence of all Group B genes and the presence of nine genes: KIR2DL1, KIR2DL3, KIR2DL4, KIR2DS4, KIR2DP1, KIR3DL1, KIR3DL2, KIR3DL3, and KIR3DP1 (14th International HLA and Immunogenetics Workshop, 2005). The Group B haplotypes were collectively termed Bx, because they constitute a mixture of AB and BB haplotypes. KIR genotype profiles were assigned to the AA and Bx haplotype groups using the New Allele Frequency Database: http://www.allelefrequencies.net.14
HLA CLASS I GENOTYPING
HLA-B and HLA-C high-resolution genotyping was performed using a sequence-based typing strategy and the protocol previously described by Cereb et al15 for heterozygous amplification of exon 2, intron 2, and exon 3 of the HLA loci. Nucleotide sequencing was performed on an ABI 3730 Genetic Analyzer using Big Dye Terminator Version 1.1 chemistry (Applied Biosystems, Foster City, CA). Allele assignment was performed using SeqScape Version 2.5 software (Applied Biosystems) and a library compiled from the 2.17.0 release of the IMGT/HLA Database.
Statistical Analysis
Fisher exact tests were performed using SISA: Simple Interactive Statistical Analysis16 and exact 95% confidence intervals of odds ratios of genotype frequency differences calculated. Two-sided tests were used and statistical significance was considered at P < 0.05. No adjustment was made for multiple comparisons. Mann-Whitney U tests were performed using SPSS Version 15.0 software (SPSS Inc, Chicago, IL).
RESULTS
KIR Genes and HLA-B and HLA-C Allotype Representation Among Natural Killer Responders and Nonresponders
KIR gene profiles and HLA-B and HLA-C alleles were determined for 76 HIV-1-infected women (39 responders and 37 nonresponders) and are described in Table 1 together with their viral loads, CD4 T-cell counts, and the HIV-1 peptide pool specificities originally determined for the positive NK cell responses.12,13 Viral load was significantly lower and CD4 T-cell counts higher among the responders when compared with nonresponders (P = 0.023 and P = 0.030, respectively).
TABLE 1.
KIR and HLA-B and HLA-C Genotypes and Clinical Parameters of Patients Stratified According to the Presence or Absence of Natural Killer Cell Responses to HIV-1 Peptides

|
nd: not determined; Env: envelope peptide pool, Reg: regulatory regions (Tat, Rev, Vif, Vpu, and Vpr) peptides combined
C1C1: two group-1 HLA-C alleles, C2C2: two group-2 HLA-C alleles, C1C2: heterozygote; Bw4/4: two HLA-Bw6 alleles, Bw6/6: two HLA-Bw6 alleles, Bw4/6: heterozygote 2DL, 3DL: inhibitory KIR; 2DS, 3DS: activating KIR; 2DP1, 3DP1: pseudogenes
To date 14 distinct KIR genes and two pseudogenes have been described (Carrington and Norman, http://www.ncbi.nlm.nih.gov/books/bookres.fcgi/mono_003/ch1d1.pdf and http://www.ebi.ac.uk/ipd/kir/). The extracellular region of KIR receptors binds with particular ligands (HLA Class I molecules); the cytoplasmic tail, which can be either short-tailed (S) or long-tailed (L), transduces the receptor-mediated signals. The L forms are usually inhibitory in function, whereas the S forms are stimulatory in function. Fourteen KIR genes and two pseudogenes were determined for each individual (Table 1). The most notable difference when comparing possession of individual KIR genes between responders and nonresponders was for KIR2DS5 (56.4% vs 37.8%, respectively; P = 0.11). Overall, more individuals who were responders possessed activating KIR genes (2DS1, 2DS2, 2DS5, 3DS1, although all independently; P > 0.05).
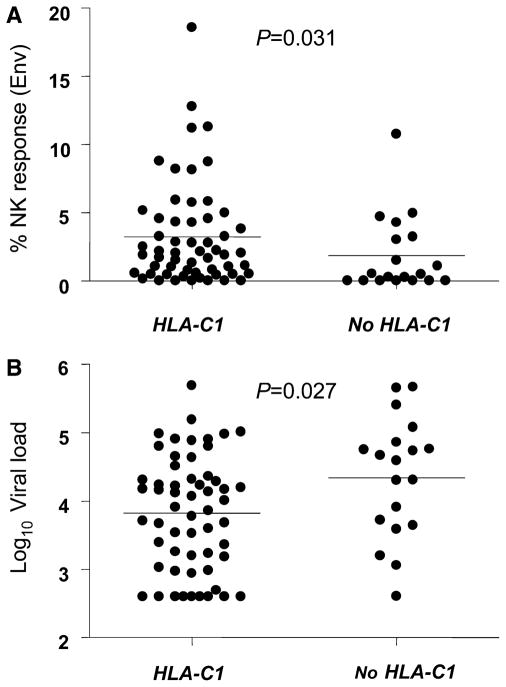
HLA-B alleles were further grouped as HLA-Bw4 (Bw4) and Bw6 allotypes based on five variable amino acids spanning residues 77–83 at the carboxyl-terminal end of the α1 helix.17,18 The Bw4 allotype subset Bw4-80Ile contains an isoleucine at position 80 as opposed to the Bw4-80Thr subset that contains a threonine at the same position and serves as the better ligand for KIR3DL119–21 and is a putative ligand for KIR3DS1 based on epidemiologic data.4HLA-C alleles were grouped as Group 1 or C1 allotypes (asparagine at residue 80 and are known ligands for KIR2DL2, KIR2DL3, and KIR2DS2) or Group 2 or C2 allotypes (lysine at residue 80, known ligands for KIR2DL1 and KIR2DS1).22 Data were analyzed between groups as presence of at least one allele of a particular allotype (C1 or C2, Bw4 or Bw6) and total allelic representation or allelic dose (C1C1, C2C2, C1C2 and Bw4/4, Bw6/6, Bw4/6). There was no significant difference in representation of HLA-Bw4 and Bw6 allotypes or of the Bw4-80Ile allotype subset among Bw4-possessing individuals, or of HLA-C1 and HLA-C2, between responders and nonresponders (P > 0.05). However, among women who had at least one HLA-C1 allele (n = 57), the magnitude of their NK cell responses to Env was significantly increased (P = 0.031) and viral load was significantly decreased (P = 0.027) when compared with those without a HLA-C1 allele (ie, C2C2 homozygotes, n = 19) (Fig. 1).
FIGURE 1.
Magnitudes of natural killer (NK) cell responses to Env (A) and viral loads (B) of individuals in the total group stratified according to the presence or absence of an HLA-C1 allele. Responses represent the percent of CD3-negative (NK) cells that produce interferon-γ in response to the Env peptide pool after subtraction of background (described in the study by Tiemessen et al13). Possession of one HLA-C1 allele groups together individuals who are C1C1 homozygotes or C1C2 heterozygotes (n = 57) in the total group; no HLA-C1 indicates C2C2 homozygotes (n = 19). Log10 viral load: HIV-1 RNA copies/mL.
Influence of KIR Genes and Corresponding HLA-B/C Ligands
KIR2DL2 and KIR2DL3 segregate as alleles of the same locus and so allelic dose of these genes and their respective HLA-C allotypes as well as the compound effects of KIR–HLA combinations was analyzed in the context of the ability to develop HIV-specific NK cell responses (Table 2). The most notable difference between responders and nonresponders was the possession of KIR2DL3+HLA-C1 (71.8% vs 51.4%, P = 0.098) and KIR2DL3/2DL3+C2C2 (2.6% vs 16.2%, P = 0.053). These trends highlight the potential importance of possession of at least one HLA-C1 allele and the presence of KIR2DL3 in likelihood of NK cell responsiveness to HIV-1 peptides.
TABLE 2.
Comparison of Frequencies of KIR2DL2, KIR2DL3, and HLA-C Allotypes and Combinations of KIR-HLA-C Between Responders and Nonresponders
| Responders (n = 39) | Nonresponders (n = 37) | Responders vs Nonresponders | |||
|---|---|---|---|---|---|
| Percent Representation | Odds Ratio | 95% Confidence Interval | P | ||
| KIR alleles | |||||
| 2DL2/2DL2 | 20.5 | 27 | 0.70 | 0.24–2.02 | 0.594 |
| 2DL2/2DL3 | 53.8 | 43.2 | 1.53 | 0.62–3.79 | 0.370 |
| 2DL3/2DL3 | 25.6 | 29.7 | 0.82 | 0.30–2.23 | 0.799 |
| HLA-C alleles | |||||
| C1/C1 | 20.5 | 24.3 | 0.93 | 0.32–2.69 | 1.000 |
| C1/C2 | 59 | 45.9 | 1.52 | 0.61–3.76 | 0.491 |
| C2/C2 | 20.5 | 29.7 | 0.61 | 0.21–1.74 | 0.431 |
| KIR-HLA combinations | |||||
| 2DL1+C2 | 76.9 | 75.7 | 1.07 | 0.372–3.09 | 1.000 |
| 2DL2+C1 | 56.4 | 56.8 | 0.99 | 0.40–2.44 | 1.000 |
| 2DL3+C1 | 71.8 | 51.4 | 2.41 | 0.93–6.23 | 0.098 |
| 2DS1+C2 | 12.8 | 5.4 | 2.57 | 0.47–14.18 | 0.432 |
| 2DS2+C1 | 53.8 | 45.9 | 1.37 | 0.56–3.38 | 0.646 |
| 2DL1+C2C2 | 20.5 | 29.7 | 0.61 | 0.21–1.74 | 0.431 |
| 2DL2+C1C1 | 7.7 | 16.2 | 0.43 | 0.10–1.87 | 0.303 |
| 2DL3+C1C1 | 25.6 | 21.6 | 1.25 | 0.43–3.62 | 0.790 |
| 2DS1+C2C2 | 5.1 | 2.7 | 1.95 | 0.17–22.4 | 1.000 |
| 2DS2+C1C1 | 7.7 | 13.5 | 0.53 | 0.12–2.41 | 0.475 |
| 2DL2/2DL2+C1C1 | 0 | 2.7 | 0.487 | ||
| 2DL2/2DL2+C2C2 | 12.8 | 8.1 | 1.67 | 0.37–7.53 | 0.712 |
| 2DL2/2DL2+C1C2 | 7.7 | 16.2 | 0.43 | 0.10–1.87 | 0.303 |
| 2DL3/2DL3+C1C1 | 12.8 | 8.1 | 1.67 | 0.37–7.53 | 0.712 |
| 2DL3/2DL3+C2C2 | 2.6 | 16.2 | 0.14 | 0.02–1.19 | 0.053 |
| 2DL3/2DL3+C1C2 | 10.3 | 5.4 | 2.00 | 0.34–11.64 | 0.675 |
| 2DL2/2DL3+C1C1 | 10.3 | 13.5 | 0.73 | 0.18–2.96 | 0.733 |
| 2DL2/2DL3+C2C2 | 5.1 | 5.4 | 0.95 | 0.13–7.09 | 1.000 |
| 2DL2/2DL3+C1C2 | 38.5 | 24.3 | 0.70 | 0.72–5.23 | 0.222 |
Bold P values indicate trends (0.05 < P < 0.1).
As for KIR2DL2 and KIR2DL3, KIR3DS1 and KIR3DL1 segregate as alleles of the same locus,23,24 but given that KIR3DS1 is present in only a few individuals (5.2% in the total group) and as only one copy (KIR3DS1/3DL1 heterozygotes), the majority of individuals are KIR3DL1/3DL1 homozygotes and so effects of dose of either KIR gene could not be determined. In addition, combinations of KIR3DL1+Bw4, KIR3DL1+Bw4-80Ile, and KIR3DL1-80Thr yield the same findings as if the HLA-B allotype groupings are tested independently.
KIR Gene Numbers and Genotypes
As can be seen from the KIR gene profiles (Table 1), there exists variation in numbers of KIR genes (nine to 16) and in combinations of these genes in different individuals. Those who have higher numbers of genes have more activating KIRs than individuals with only nine genes who would have only one activating KIR. Most individuals possessed 13 KIR genes (39.5%) followed by nine genes (26.3%), 12 (18.4%), 11 (6.6%), 14 (3.9%), 15, and 16 (both 2.6%).
Individuals have six to eight inhibitory KIR genes and from one to six activating KIR genes. KIR2DL4, which shares structural and functional features with both inhibitory and activating receptors,25–27 is categorized here as inhibitory. Equal proportion of responders and nonresponders had six inhibitory genes (25.6% vs 27%) and only one activating KIR gene (KIR2DS4); however, there was a shift in favor of more responders having eight inhibitory genes (56.4% vs 32.4%, P = 0.03) than nonresponders (Table 3). More responders had three activating genes (48.7% vs 29.75%) and more non-responders had two activating genes (27% vs 2.6%). Overall, significantly more responders had three or more activating genes than nonresponders (71.8% vs 45.9%, P = 0.035) (Table 3). This corresponded exactly with inhibitory:activating ratios (number of inhibitory genes ÷ number of activating genes) of 2.7 or less (less inhibition) and greater than 2.7 (greater inhibition).
TABLE 3.
Comparison of Percent Representation of Higher Inhibitory and Activating KIR Gene Numbers and Inhibitory:Activating Gene Ratios Between Responders and Nonresponders
| Responders (n = 39) | Nonresponders (n = 37) | Responders vs Nonresponders | |||
|---|---|---|---|---|---|
| Percent Representation | Odds Ratio | 95% Confidence Interval | P | ||
| KIR gene type and number | |||||
| Eight inhibitory genes | 56.4 | 32.4 | 2.7 | 1.06–6.87 | 0.030 |
| Three or more activating genes | 71.8 | 45.9 | 2.99 | 1.16–7.75 | 0.035 |
| Ratio inhibitory:activating genes | |||||
| 2.7 or less | 71.8 | 45.9 | 2.99 | 1.16–7.75 | 0.035 |
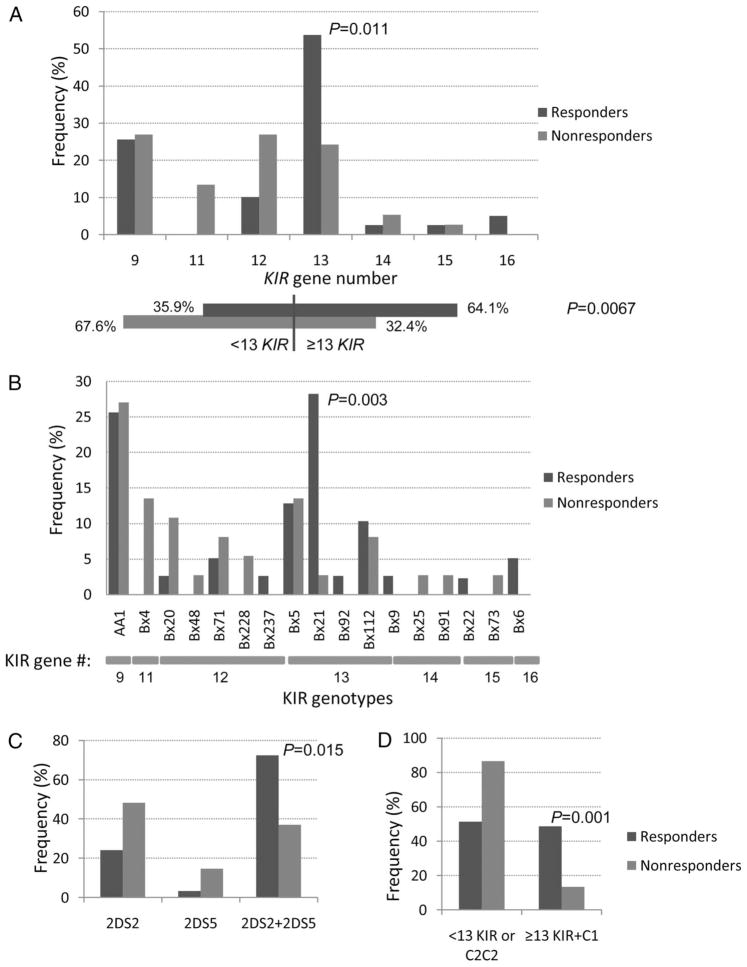
Total KIR gene number distribution among responders and nonresponders (Fig. 2A) showed that a significantly higher proportion of responders possessed 13 KIR genes than nonresponders (53.8% vs 24.3%, P = 0.011). When analyzed as individuals harboring less than 13 or 13 or more KIR genes, 64.1% of responders versus 32.4% of nonresponders had higher KIR gene numbers (P = 0.0067), overall suggesting that NK cell responses are more likely to occur among individuals with more activating KIR genes.
FIGURE 2.
Percentage representation of individual KIR gene numbers, KIR genotypes, and KIR2DS2 and/or KIR2DS5 genes among responders and nonresponders in the HIV-1-infected women. (A) Distribution of number of KIR genes (nine to 16) among responders and nonresponders. The top graph shows the representations for each KIR gene number; proportions of individuals stratified into less than 13 and 13 or greater KIR genes are shown below for both groups. (B) KIR gene haplotype (AA and Bx) representation among responders and nonresponders. KIR gene numbers corresponding to the particular KIR genotypes are indicated below the x-axis. (C) Frequencies of individuals possessing KIR genotypes that have KIR2DS2 or KIR2DS5 or both genes among responders and nonresponders. These were determined out of a total of 29 responders and 27 nonresponders (10 responders and 10 nonresponders were not included because they are AA1 genotypes, which do not possess either 2DS2 or 2DS5). (D) Frequencies of individuals who possess 13 KIR genes or more together with at least one HLA-C1 allele among responders and non-responders. The remaining individuals would have either less than 13 KIR genes or be homozygous for the HLA-C2 allele. P values for significant differences between groups are shown.
Seventeen KIR genotypes were identified in this group of 76 HIV-1-infected women (Table 1; Fig. 2B). The most prevalent genotypes (greater than 5%) were AA1 (26.3%), Bx21 (15.8%), Bx5 (13.2%), Bx112 (9.2%), and Bx4, Bx20, and Bx71 (all 6.6%). Interestingly, the 13-KIR gene-containing Bx21 genotype showed substantially higher representation among the responders (28.2% vs 2.7%, P = 0.001), virtually accounting for the entire effect seen when comparing responder and nonresponders groups on the basis of KIR gene number alone. Bx21 contains eight inhibitory genes and three activating genes (2DS2, 2DS4, 2DS5). All genotypes in this population contained KIR2DS4, highlighting the potential importance of KIR2DS2 and KIR2DS5 co-occurrence in the development of HIV-specific NK cell responses.
All KIR genotypes in our study group, with the exclusion of AA1 genotypes (26.3% of patients), possess either KIR2DS2 and/or KIR2DS5; 40.8% possessed both genes, 26.3% had KIR2DS2 alone, and 6.6% had KIR2DS5 alone. A significantly higher proportion of responders had both KIR2DS2 and KIR2DS5 (Fig 2C), as opposed to either gene alone, compared with nonresponders (72.4% vs 37%; P = 0.015), further reinforcing the need for a more activating phenotype in likelihood of detection of HIV-specific NK cell responses.
Given the importance of possessing one copy of an HLA-C1 allele, as evidenced by reduced viral load and increased NK cell response magnitude (Fig 1), we further established that a combination of at least one HLA-C1 allele together with 13 or more KIR genes was associated with NK cell responsiveness (48.7% vs 13.5%; P = 0.001) (Fig. 2D). This equated to 86.5% of nonresponders possessing either lower (less than 13) KIR gene numbers or HLA-C2C2 homozygosity.
DISCUSSION
An important role for NK cells in control of HIV-1 infection has been indicated by genetic association studies of KIR receptors that have as ligands specific HLA Class I molecules.3–5 Furthermore, our recent work showed an association of NK cell responses to HIV-1 peptides (predominantly to Env and Reg peptide pools) with lower viral loads, higher CD4 T-cell counts, and stronger T-cell responses in HIV-1 infected women13 and the association of these responses with reduced maternal–infant HIV-1 transmission.12 Collectively, all these studies prompted us to begin to question the possible role that KIR and HLA Class I B and C molecules might play in the ability of patients’ NK cells to overcome inhibitory signals sufficiently to mount responses to HIV-1 peptides. This study questions the relationships with NK cell responsiveness of the particular KIR gene repertoires and HLA of HIV-1-infected individuals that considers 1) presence or absence of a particular gene; 2) type (inhibitory or activating) and number of KIR genes; 3) KIR genotype; 4) representation of HLA Class I allotypes (C1, C2, Bw4, Bw4-80I, Bw6); and 5) specific KIR–HLA combinations. To this end, samples from 76 HIV-1-infected women were KIR and HLA-C and HLA-B genotyped and grouped as responders (a response to at least one peptide pool) and nonresponders.
Seventeen KIR genotypes were identified; to date, we have identified a total of 46 different KIR genotypes among 446 black South African mother and infant individuals.28 The genotypes in our current study group encompassed all the higher prevalence KIR genotypes found in this larger group, viz AA1, Bx21, Bx5, Bx112, Bx4, Bx20, and Bx71. Looking at the genotypes based on total KIR gene number, it was apparent that significantly more NK responders possessed 13 or more genes and that this increase was attributed to more responders having eight as opposed to seven inhibitory genes and three or more activating genes. In general, higher KIR gene number is attributed to the presence of more activating genes. Of all the genotypes, it was the Bx21 genotype, which is the most highly represented of the Bx genotypes in our South African black population, that was most strongly associated with NK cell responses to HIV-1 peptides. This particular genotype contains eight inhibitory and three activating genes. Of the three activating genes, only the effects of KIR2DS2 and KIR2DS5 could be assessed because all genotypes contained KIR2DS4. Possession of both these genes, as opposed to only one or the other, was a characteristic of NK responders. Overall, these findings suggest that a more activating phenotype is associated with the presence of HIV-specific NK cell responses, consistent with the idea that under these conditions, the balance between inhibitory and activating signals of NK cells favors activation, which in turn contributes to more effective control of HIV-1 infection.
Irrespective of what mechanism underlies NK cell responsiveness to HIV-1 peptides, an individual armed with KIR genes that allows for a greater opportunity for NK cell activation (more activating genes and in combination with at least one HLA-C1 allele) is a requirement for response ability. Because all HLA-C alleles fall into either the HLA-C1 or HLA-C2 allotype subsets, the importance of HLA-C1 in reduction of viral load and increased magnitudes of HIV-1 peptide-specific NK cell responses points to the likely importance of its KIR partners KIR2DL2, KIR2DL3, and KIR2DS2. All individuals were either homozygous for KIR2DL2 or KIR2DL3 or are KIR2DL2/KIR2DL3 heterozygotes because these are alleles of the same locus. Approximately 10% more responders than nonresponders possessed KIR2DS2 (72% vs 62%) with responders having 7% more KIR2DL3. It was the combination of KIR2DL3 plus HLA-C1 that showed a trend to an increase in the responders, the importance of homozygosity of KIR2DL3 in the absence of its ligand HLA-C1 (so C2C2 homozygosity) being more highly represented in the non-responders further emphasizing the importance of this relationship in the responders (P = 0.053). It will be important to further study the effects of allelic variation at these KIR loci to establish if particular variants are more associated with different levels of KIR expression or altered binding affinities that might affect their interactions with HLA-C1 molecules. Importantly, we found these same KIR molecules to be the most significantly involved in maternal transmission of HIV-1 and in acquisition of HIV-1 in the infant.28
Another study of South African individuals showed that among those who had both KIR2DL1 and KIR2DS1 genes, the frequency of NK cells expressing one or both of these receptors tended to decrease with increasing viral load, a trend that was not seen in individuals who had KIR2DL1 but not KIR2DS1.11 These molecules are among the KIRs that bind HLA-C molecules; the affinity of these interactions is greatest for KIR2DL1-C2>KIR2DL2-C1>KIR2DL3-C1.29 The affinity interactions of the corresponding activating receptors KIR2DS1-C2 and KIR2DS2-C1 tend to be less than their inhibitory counterparts. It has been previously suggested from genetic studies that KIR–HLA combinations associated with less inhibition might favor greater likelihood of NK cell activation as opposed to those with stronger affinity interactions, for example, the “weaker” interaction of KIR2DL3 and HLA-C1 has been associated with enhanced resolution of hepatitis C virus infection.30 In African sex workers, it has been demonstrated that possession of inhibitory genes in the absence of genes for their cognate ligands was associated with reduced HIV-1 acquisition (KIR2DL2/2DL3 heterozygotes with no HLA-C1, KIR3DL1 homozygotes with no HLA-Bw4).31 In addition, individuals with KIR genotypes having more activating KIR genes have also shown some protection.31,32 Overall, the tendency toward weaker inhibition and so greater activation seems important in control of HIV-1 infection and protection from HIV-1 acquisition.
How can peptide-specific NK cell responses measured ex vivo in the whole blood assay be explained? Peptide–HLA Class I complexes have been shown to be recognized by activating KIR (KIR2DS1) receptors in cells infected with Epstein-Barr virus.33 Furthermore, NK cells have been shown in vitro to kill their HIV-infected target cells in a receptor ligand-specific manner that involved activating KIR3DS1 and its putative ligand HLA-Bw4-80Iso.34 It can therefore be envisaged that HIV-1 peptides delivered exogenously bind specifically to HLA Class I molecules on antigen-presenting cells and that these complexes are recognized by particular KIR receptors on NK cells. Clones of NK cells that express more than one or several activating receptors, all engaged with their cognate ligands would result in the integration of several signals that ultimately culminate in NK cell activation. Any one activating signal alone may prove insufficient for activation.33,35 Peptide antagonism has recently been suggested as a possible mechanism for NK cell activation, and it was demonstrated that KIR-positive NK cells are more influenced by changes in peptide sequence than changes resulting from HLA Class I expression on target cells.36 In the context of HIV-1-specific peptides in our assay, it would seem possible that some of these interactions with HIV-1 peptides could be antagonistic in nature, resulting in abrogating inhibitory KIR interactions with HLA Class I molecules and so overcoming the threshold for activation of NK cells. Because activating and inhibitory KIR interactions show similarities in sensitivity to alterations in peptide sequences,33 it may be that a combination of binding of peptides to inhibitory KIR and to the corresponding activating KIR may together or independently result in an overall outcome of activation of NK cells, this governed by the extent to which the inhibitory-activating axis is altered. ADCC antibodies provide another means of NK cell activation by peptides in the whole blood assay,37 the triggering of NK cells occurring through engagement of the CD16 activating receptor on NK cells. It stands to reason that in some patients, this could account for the entire response or a component of the response; in other individuals, other mechanisms may dominate.
Although the exact events underlying the specific nature of activated NK cell responses to particular HIV-1 peptides in whole blood assays remain to be elucidated, it is clear that both variation at the KIR locus and dose of particular HLA-C allotypes impact on the ability of NK cells to respond to HIV-1 peptides, a feature of importance in control of HIV-1 infection.
Acknowledgments
This study was supported in part by the South African AIDS Vaccine Initiative (SAAVI) and by grants from NICHD 42402, the Wellcome Trust, and Elizabeth Glaser Pediatric AIDS Foundation. C.T.T. is a Wellcome Trust International Senior Research Fellow (076352/Z/05/Z).
Footnotes
The authors have no conflicts of interest to disclose.
References
- 1.Moretta L, Moretta A. Unravelling natural killer cell function: triggering and inhibitory human NK receptors. EMBO J. 2004;23:255–259. doi: 10.1038/sj.emboj.7600019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 3.Gaudieri S, DeSantis D, McKinnon E, et al. Killer immunoglobulin-like receptors and HLA act both independently and synergistically to modify HIV disease progression. Genes Immun. 2005;6:683–690. doi: 10.1038/sj.gene.6364256. [DOI] [PubMed] [Google Scholar]
- 4.Martin MP, Gao X, Lee JH, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 5.Martin MP, Qi Y, Gao X, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonaparte MI, Barker E. Killing of human immunodeficiency virus-infected primary T-cell blasts by autologous natural killer cells is dependent on the ability of the virus to alter the expression of major histocompatibility complex class I molecules. Blood. 2004;104:2087–2094. doi: 10.1182/blood-2004-02-0696. [DOI] [PubMed] [Google Scholar]
- 7.De Maria A, Fogli M, Costa P, et al. The impaired NK cell cytolytic function in viremic HIV-1 infection is associated with a reduced surface expression of natural cytotoxicity receptors (NKp46, NKp30 and NKp44) Eur J Immunol. 2003;33:2410–2418. doi: 10.1002/eji.200324141. [DOI] [PubMed] [Google Scholar]
- 8.Sirianni MC, Ensoli F, Alario C, et al. Distribution of the natural killer–related receptor for HLA-C during highly active antiretroviral therapy for human immunodeficiency virus infection. Hum Immunol. 2001;62:1328–1334. doi: 10.1016/s0198-8859(01)00355-x. [DOI] [PubMed] [Google Scholar]
- 9.Mavilio D, Benjamin J, Daucher M, et al. Natural killer cells in HIV-1 infection: dichotomous effects of viremia on inhibitory and activating receptors and their functional correlates. Proc Natl Acad Sci U S A. 2003;100:15011–15016. doi: 10.1073/pnas.2336091100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fogli M, Costa P, Murdaca G, et al. Significant NK cell activation associated with decreased cytolytic function in peripheral blood of HIV-1-infected patients. Eur J Immunol. 2004;34:2313–2321. doi: 10.1002/eji.200425251. [DOI] [PubMed] [Google Scholar]
- 11.Wong AH, Williams K, Reddy S, et al. Alterations in natural killer cell receptor profiles during HIV type 1 disease progression among chronically infected South African adults. AIDS Res Hum Retroviruses. 2010;26:459–469. doi: 10.1089/aid.2009.0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiemessen CT, Shalekoff S, Meddows-Taylor S, et al. Cutting Edge: Unusual NK cell responses to HIV-1 peptides are associated with protection against maternal–infant transmission of HIV-1. J Immunol. 2009;182:5914–5918. doi: 10.4049/jimmunol.0900419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tiemessen CT, Shalekoff S, Meddows-Taylor S, et al. Natural killer cells that respond to HIV-1 peptides are associated with control of HIV-1 infection. J Infect Dis. 2010;202:1444–1453. doi: 10.1086/656535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Middleton D, Menchaca L, Rood H, et al. New allele frequency database. Tissue Antigens. 2003;61:403–407. doi: 10.1034/j.1399-0039.2003.00062.x. http://www.allelefrequencies.net. [DOI] [PubMed]
- 15.Cereb N, Maye P, Lee S, et al. Locus-specific amplification of HLA class I genes from genomic DNA: locus-specific sequences in the first and third introns of HLA-A, -B, and -C alleles. Tissue Antigens. 1995;45:1–11. doi: 10.1111/j.1399-0039.1995.tb02408.x. [DOI] [PubMed] [Google Scholar]
- 16.Uitenbroek DG. SISA Binomial. Southhampton: DG Uitenbroek; 1997. [Accessed October 15, 2010]. Available at: http://www.quantitativeskills.com/sisa/distributions/binomial.htm. [Google Scholar]
- 17.Salter RD, Parham P. Mutually exclusive public epitopes of HLA-A,B,C molecules. Hum Immunol. 1989;26:85–89. doi: 10.1016/0198-8859(89)90093-1. [DOI] [PubMed] [Google Scholar]
- 18.Muller CA, Engler-Blum G, Gekeler V, et al. Genetic and serological heterogeneity of the supertypic HLA-B locus specificities Bw4 and Bw6. Immunogenetics. 1989;30:200–207. doi: 10.1007/BF02421207. [DOI] [PubMed] [Google Scholar]
- 19.Carr WH, Pando MJ, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol. 2005;175:5222–5229. doi: 10.4049/jimmunol.175.8.5222. [DOI] [PubMed] [Google Scholar]
- 20.Cella M, Longo A, Ferrara GB, et al. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med. 1994;180:1235–1242. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gumperz JE, Barber LD, Valiante NM, et al. Conserved and variable residues within the Bw4 motif of HLA-B make separable contributions to recognition by the NKB1 killer cell-inhibitory receptor. J Immunol. 1997;158:5237–5241. [PubMed] [Google Scholar]
- 22.Bjorkman PJ, Parham P. Structure, function, and diversity of class I major histocompatibility complex molecules. Annu Rev Biochem. 1990;59:253–288. doi: 10.1146/annurev.bi.59.070190.001345. [DOI] [PubMed] [Google Scholar]
- 23.Uhrberg M, Valiante NM, Shum BP, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 24.Wilson MJ, Torkar M, Trowsdale J. Genetic analysis of a highly homologous gene family. The killer cell immunoglobulin-like receptors. Methods Mol Biol. 2000;121:251–263. doi: 10.1385/1-59259-044-6:251. [DOI] [PubMed] [Google Scholar]
- 25.Selvakumar A, Steffens U, Dupont B. NK cell receptor gene of the KIR family with two IG domains but highest homology to KIR receptors with three IG domains. Tissue Antigens. 1996;48:285–294. doi: 10.1111/j.1399-0039.1996.tb02647.x. [DOI] [PubMed] [Google Scholar]
- 26.Rajagopalan S, Fu J, Long EO. Cutting edge: induction of IFN-gamma production but not cytotoxicity by the killer cell Ig-like receptor KIR2DL4 (CD158d) in resting NK cells. J Immunol. 2001;167:1877–1881. doi: 10.4049/jimmunol.167.4.1877. [DOI] [PubMed] [Google Scholar]
- 27.Faure M, Long EO. KIR2DL4 (CD158d), an NK cell-activating receptor with inhibitory potential. J Immunol. 2002;168:6208–6214. doi: 10.4049/jimmunol.168.12.6208. [DOI] [PubMed] [Google Scholar]
- 28.Paximadis M, Minevich G, Winchester R, et al. KIR–HLA and maternal–infant HIV-1 transmission in sub-Saharan Africa. Plos ONE. 2011;6:e16541. doi: 10.1371/journal.pone.0016541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajagopalan S, Long EO. Understanding how combinations of HLA and KIR genes influence disease. J Exp Med. 2005;201:1025–1029. doi: 10.1084/jem.20050499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khakoo SI, Thio CL, Martin MP, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 31.Jennes W, Verheyden S, Demanet C, et al. Cutting edge: resistance to HIV-1 infection among African female sex workers is associated with inhibitory KIR in the absence of their HLA ligands. J Immunol. 2006;177:6588–6592. doi: 10.4049/jimmunol.177.10.6588. [DOI] [PubMed] [Google Scholar]
- 32.Ravet S, Scott-Algara D, Bonnet E, et al. Distinctive NK-cell receptor repertoires sustain high-level constitutive NK-cell activation in HIV-exposed uninfected individuals. Blood. 2007;109:4296–4305. doi: 10.1182/blood-2006-08-040238. [DOI] [PubMed] [Google Scholar]
- 33.Stewart CA, Laugier-Anfossi F, Vely F, et al. Recognition of peptide–MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci U S A. 2005;102:13224–13229. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alter G, Martin MP, Teigen N, et al. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204:3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bryceson YT, Long EO. Line of attack: NK cell specificity and integration of signals. Curr Opin Immunol. 2008;20:344–352. doi: 10.1016/j.coi.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fadda L, Borhis G, Ahmed P, et al. Peptide antagonism as a mechanism for NK cell activation. Proc Natl Acad Sci U S A. 2010;107:10160–10165. doi: 10.1073/pnas.0913745107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stratov I, Chung A, Kent SJ. Robust NK cell-mediated human immunodeficiency virus (HIV)-specific antibody-dependent responses in HIV-infected subjects. J Virol. 2008;82:5450–5459. doi: 10.1128/JVI.01952-07. [DOI] [PMC free article] [PubMed] [Google Scholar]