Abstract
Chemists continually synthesize myriad new chemicals (~2000/yr), some of which make their way into the environment or otherwise pose possible threats to humans who potentially become exposed to the compounds. Regulators must determine whether these, along with the glut (~80,000) of existing, chemicals are toxic and at what exposure levels. An important component of this determination is to ascertain the mode of action (MOA) of each compound as it relates to the pathway the compound uses to induce genotoxicity. Several assays have traditionally been used to reveal these effects to the genome: the Ames test, tests with yeast and mammalian cell lines, and animal studies. Previously, we described a new multi-well plate-based method which makes use of the DT40 isogenic cell line and its dozens of available mutants knocked out in DNA repair and cell cycle pathways and we now provide a detailed protocol of the further improvement of the assay. Although the DT40 line has existed for some time and has been used in numerous studies of DNA repair pathways, little use has been made of this valuable resource for toxicological investigations. Our method introduces the XTT dye scheme determination of cell survival in a manner that greatly increases throughput and reduces cost while maintaining reasonable sensitivity. Although this new genotoxicity assay requires validation with many more mutagens before becoming an established, regulatory decision-making analysis tool, we believe that this method will be very advantageous if eventually added to the repertoire of those investigating MOAs of potentially genotoxic substances.
Keywords: DT40, DNA damage response, genotoxicity assay
Introduction
Every year, a large number of new chemicals are being added to the already vast list that is in use. Therefore, there is a growing need to be able to quickly establish the potential dangers of these compounds in terms of genotoxicity, mutagenicity and carcinogenicity. To this end a number of procedures have been developed, such as the Ames test, the yeast reverse-genetic assay, various in vitro cell-based assays and a variety of animal studies. It is clear that any new genotoxicity assay that could improve upon any weaknesses of currently available analyses should be greatly welcomed as an additional analytical method.
The reverse genetic approach provides a powerful method for the study of gene function and regulation. One such approach which has been gaining in popularity makes use of the DT40 cell line (Winding and Berchtold, 2001; Sonoda et al., 2001; Yamazoe et al., 2004; Dhar et al., 2001). DT40 cells originated from a chicken B-lymphocyte line derived from an avian leucosis virus induced bursal lymphoma originally isolated in 1985 (Baba et al., 1985). The DT40 cell line is rather unique among higher eukaryotic cells in that it exhibits a high ratio of targeted to random integration of transfected DNA (Sonoda et al., 2001; Dhar et al., 2001). Due to the facility with which the DT40 line can be manipulated genetically, and the fact that DT40 mutants are observed to show a strong phenotypic resemblance to murine mutants with respect to genes involved in DNA recombination and repair (Sonoda et al., 2001), it has seen a steady growth in its use in genetic studies including immunoglobulin diversification, DNA repair, chromosome segregation, RNA metabolism and cell signaling (Winding and Berchtold, 2001; Yamazoe et al., 2004; Dhar et al., 2001).
Another unique feature of the DT40 cells is their reported lack of functional p53 (Ulrich et al., 1992). When testing a DNA repair mutant with a functioning p53 protein, general genomic instability may cause the cell to die due to the activation of apoptosis. The absence of functional p53 in DT40 cells presents the advantage of a compromised apoptosis pathway allowing for the observance of cell death due to defect-driven repair failures rather than by the activation of apoptosis.
In the field of toxicology, the use of biomarkers has been rapidly gaining in importance. Once one knows the genotoxicity of the target compound and which DNA repair pathway is needed to alleviate the damage, the type of biomarker required tends to be more easily determined. Without such information, biomarkers are sometimes being established without knowledge of whether the target is biologically important. In an attempt to shed light on the mechanisms used to repair DNA damage induced by exogenous and endogenous agents we have made extensive use of the DT40 system and the great number of repair and cell-cycle checkpoint mutants that have been created (Table I). One such study we performed using multi-well plate-based method determined that cells deficient in the FANC/BRCA pathway and homologous recombination (HR) repair are hypersensitive to formaldehyde at concentrations found in human plasma (Ridpath et al., 2007). Until that time, formaldehyde had been determined to cause DNA-protein crosslinks but little was known about how such lesions are repaired. Our results suggest that the use of syngeneic mutant cell lines, which is a very modern technology, is capable of deciphering the physical causes of DNA damage that are missed by more classical technologies. Indeed, the use of syngeneic lines to the analytic ability of toxicology is worthy of interest to more than simply the DNA repair field.
Table I.
Example of Available DT40 Mutant Cell Lines
| BER | NER | NHEJ | HR |
| - POL β | - XPA | - KU70 | - RAD52 |
| - FEN1 | - XPG | - LIG IV | - RAD54 |
| - PARP1 | - DNAPKcs | - RAD51c | |
| DNA damage sensors | MMR | TLS | - RAD51d |
| - MSH2 | - POL K | - XRCC2 | |
| - RAD9 | - MSH3 | - POL Q | - XRCC3 |
| - RAD17 | - MSH6 | - POL Q | - BRCA1 |
| Helicase | - REV1 | - BRCA2 | |
| - BLM | - REV3 | - FANCD2 | |
| - WRN | - RAD18 |
DNA damage repair/tolerance pathways represented:
BER - base excision repair; NER – nucleotide excision repair; NHEJ – non - homologous end- joining; HR – homologous recombination; MMR – mis-match repair; TLS – trans-lesion synthesis
One downside to the method, however, is that the cells are usually grown in methylcellulose gel suspension with clonogenic assays used to rate cell survival (Simpson and Sale, 2006). Clonogenic assays are considered to be very accurate but are rather tedious. Indeed, to test just one agent at several dilutions along with controls against numerous cell lines could require hundreds of dishes with an incredible amount of manual manipulation. We felt that if the system were to be used for screening, a simpler, more rapid and less expensive method would be advantageous. Here, we describe a detailed protocol of a modified version of the DT40-based response analysis that is convenient but still a reasonably sensitive method for determining genotoxicity of chemicals by making use of the compound 2,3-bis (2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide (XTT) which is converted in respirating cells to a formazan dye with specific absorbance at 450 nm. The percent survival can then be easily and accurately determined by comparison of absorbance measurements of test wells to those of controls.
Materials and Methods
DT40 Cell Culturing and Maintenance
Fetal bovine serum (FBS) and penicillin/streptomycin were obtained from Sigma (St. Louis, MO). RPMI 1640 culture medium (+glutamine, −phenol red) and chicken serum were acquired from Invitrogen (Grand Island, NY).
DT40 cells are cultured in a manner similar to most vertebrate or mammalian cell lines that are grown in suspension. Optimal propagation is provided by growing in RPMI 1640 with 10% FBS, 1% chicken serum and 1% penicillin/streptomycin. RPMI sans phenol red should be used, as the color in the indicator may interfere during spectroscopy. FBS and chicken serum should be heat inactivated at 56 °C for 30 min. The cells are incubated at 39.5 °C and 5% CO2 with 95% humidity. Since DT40 cells have a relatively short generation time (7–10 hr), for the sake of convenience we have found that cell growth can be slowed by incubation at 37 °C or even as low as 31 °C without undue stress to adjust the growth rate during the final 16 hours of cultivation. This process allows for the avoidance of overgrowth of the cells. Also, due to the rapid growth rate, the cultures must be observed regularly to prevent overgrowth with subsequent starvation resulting in stress to or death of the cells.
Sub-culturing should be performed as needed to provide cells in the log phase of growth as an optimum test parameter and to maintain cell lines for future use. For subculture, cells are counted by hemacytometer. The target cell concentration for both subculturing and the preparation of the cell suspension for the assay that produces the most reproducible assay result lies between 0.7 million and 1.5 million cells/mL. Also, cell condition should be observed during counting. Under a light microscope, normal DT40 cells appear nearly circular. Stressed cells may appear elongated or have an uneven margin. If cells appear stressed, it is advisable to newly sub-culture before using in the assay.
For best results, a certain amount of care should be observed when handling DT40 cells. All transfers of cells should be into pre-warmed medium and mixing of the cells prior to pipetting should be very gentle but absolutely thorough – especially for cell counts or seeding for an assay. It is also advisable to use pipette tips with large bore openings such as those used in genomic studies. Our results and reproducibility improved notably with the use of these procedures. Indeed, our success rates using the assay had been very wide-ranging (from 1 to 70%) using our previous method (Ridpath et al., 2007); whereas, the current improved method provides a success rate of nearly 100%.
Although DT40 cells are considered immortal, each cell line should be restarted from frozen stock about once each month. Considering that most of our cell lines are deficient in DNA repair, this seems an appropriate choice. The cells can readily be stored under liquid nitrogen for extended periods.
Test Procedure
Cisplatin, methyl methanesulfonate (MMS), 2,3-bis[2-methoxy-4-nitro-5sulfo-phenyl]-2H-tetrazolium-5-carbox-anilide inner salt (XTT), 1-methoxy-5-methylphenazinium methyl sulfate (PMS) and DMSO were obtained from Sigma (St. Louis, MO).
The overall scheme of this assay is shown in Fig. 1. Cells are prepared in sterile centrifuge tubes by adding enough volume of cell-containing medium from the subculture dish to 6 mL warmed fresh medium to provide a cell concentration adequate to seed approximately 2500 cells in a volume of 250 μL to each of 22 wells. Before seeding, the cells should be adequately but gently mixed in the tubes. The cells are then seeded by pipetting 250 μL of the cell suspension to each of 22 wells of the 24-well plate (Table II). To the two blank wells add 250 μL plain medium. These plates should be kept in an incubator at 39.5 °C until ready for treatment with the chemical compound(s). This method provides for treatment with six dilutions of the chemical, each into three wells, with four untreated wells as controls and two as blanks Table II). Treatment is performed by adding 27.8 μL (total well volume, 277.8 μL) of the chemical dilution to each appropriate well and 27.8 μL of the chemical solvent - usually PBS - to the control and blank wells. The plates are returned to the incubator.
Figure 1. Protocol overview of XTT-based DT40 cell DNA damage response analysis using 24-well plate format.
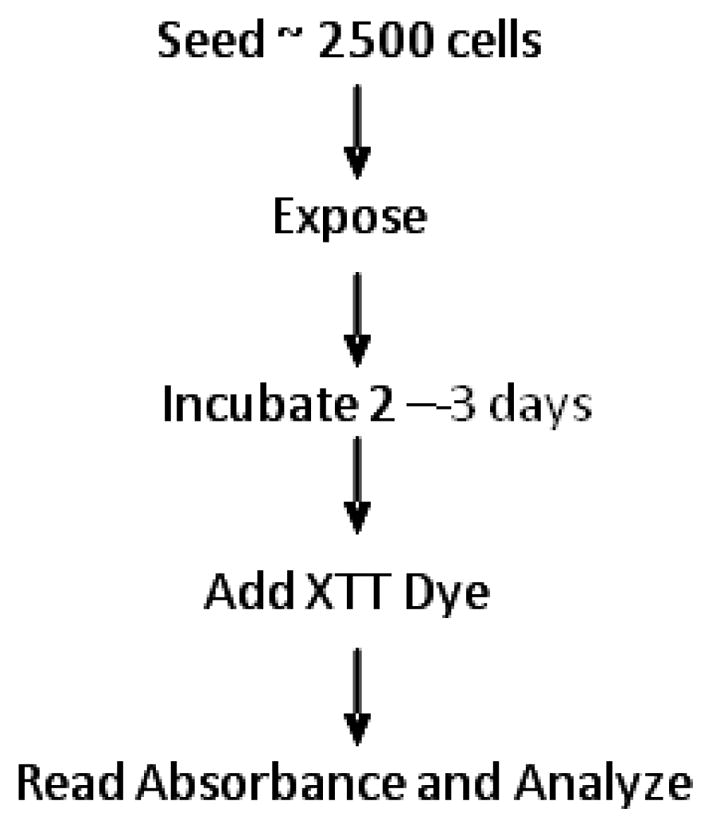
Table II.
Format for treatment of 24-well plate
| 1 | 1 | 1 | 5 | 5 | 5 |
| 2 | 2 | 2 | 6 | 6 | 6 |
| 3 | 3 | 3 | C | C | C |
| 4 | 4 | 4 | C | Bl | Bl |
Numbers represent dosing solutions: from highest concentration (1) to lowest (6); C: control (cells plus vehicle only); Bl: blank (complete medium without cells plus vehicle).
Once treated, the incubating cells should not be disturbed for 48 hr after which they should be observed microscopically to determine the growth of the cells. Once the cells in the control wells are nearly contiguous, each well (including the blanks) should be treated with XTT dye (XTT cocktail is prepared with 500 mg of the XTT salt, 10.2 mg PMS and 3.3 mL DMSO). This is a critical step as the cells should not be allowed to grow until starvation begins causing cell death. Conversely, if the number of cells is too low, an inordinate amount of time may be required for the cells to metabolize the dye and adequate contrast may be difficult to obtain. The cells are exposed to the XTT cocktail by first preparing a stock solution of XTT in complete medium (9.1 μL XTT mixture/2.5 mL medium) then treating with 100 μL of the dye preparation. The plates are returned to the incubator until the dye has developed sufficient color for absorbance to be read on the plate reader (typically 2 – 4 hr). Once the plates have developed sufficient color, they may be read immediately or stored at 4 °C in relative darkness until reading is convenient. We confirmed no statistical difference exists between readings immediately after color development and after storage of the plates at 4 °C for several days (data not shown). This represents another improvement over our previously reported version of the assay (Ridpath et al., 2007). Stored plates should be kept in sealable plastic bags or something similar to prevent drying of the medium in the wells. The ability to store the plates is yet another reason for using medium without phenol red as the indicator can change color due to temperature modification of the pH in the medium, which would interfere with accurate spectrometric readings later.
For measuring absorbance, our lab uses a Tecan Safire (Tecan Systems, San Jose, CA) plate reader with Magellen6 software (Tecan, version 6.4). The software provides a convenient methodology for reading plates with many different numbers of wells and subsequent saving of absorbance values to Microsoft Excel. Absorbance measurement is read at 450 nm with a reference of 650 nm.
All data are reported as the means ± standard deviation of at least triplicate samples. Analysis of covariance (ANCOVA) was used to test for mean intercept differences and differences in the slopes of the linear dose-response curves in cell viability analysis between wild-type and a series of mutant cells. A Student’s t-test was utilized to determine the significant differences (p<0.05) between means of two groups.
Results and Discussion
Reproducibility and sensitivity in the DT40 cell DNA damage response analysis using the XTT method
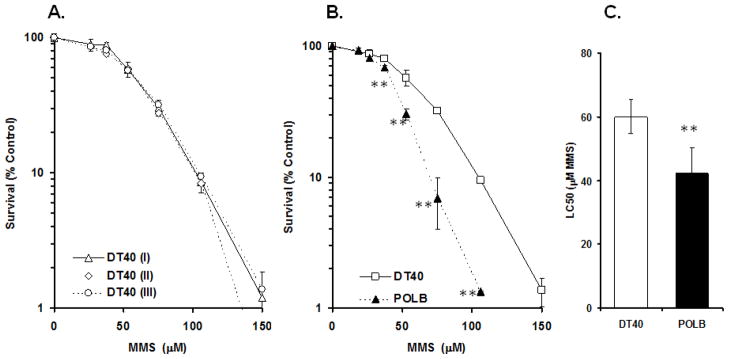
To address reproducibility of the response of DT40 parental cells to mutagens, we exposed DT40 cells to methyl methanesulfonate (MMS), a strong DNA alkylating agent whose damage is repaired by base excision repair (BER), at different concentrations in three independent experiments. Fig. 2A indicates the level of reproducibility achieved from those experiments. We tested for statistical difference between the three results for LC50 values, slope and y-axis interception. These analyses provide no significant difference, suggesting a reasonable reproducibility of our assay for measuring the response in DT40 cells to MMS in terms of cell viability. We also compared DT40 cells and POL β (a BER polymerase) mutants exposed to MMS for cell viability (Fig. 2B). The statistically significant difference in dose-response relationship between two cell lines was detectable as low as 37.5 μM of MMS. This sensitivity appears to be similar to the results obtained by clonogenic analysis (Yoshimura et al., 2006).
Figure 2. Cell survival results for DT40 cells and POLB mutants continuously exposed to MMS.
(A) Results of three independent survival experiments in DT40 cells exposed to MMS. Survival data were log-transformed giving approximate normality. Analysis of covariance (ANCOVA) was used to test for mean intercept differences and differences in the slopes of the linear dose-response curves in cell viability analysis between experiments. No significant difference (p<0.05) was detected between experiments. (B) Survival percentage for DT40 and POLB mutant cells exposed to MMS compared with concurrent control. A student’s t-test was utilized to determine the significant differences between means of two groups (DT40 (II) vs POLB exposed to MMS at same concentration, **P < 0.01). (C) Survival data from DT40 (II) and POLB cells exposed to MMS were log-transformed giving approximate normality. Each LC50 value was then calculated for each cell line. ANCOVA showed a significant difference for mean intercepts of the linear dose-response curves between wild-type and POLB cells (**P < 0.01). Bars: 95% confidence intervals for LC50 value for each cell line.
Comparison of cisplatin exposure results between the DT40 XTT dye and clonogenic methods
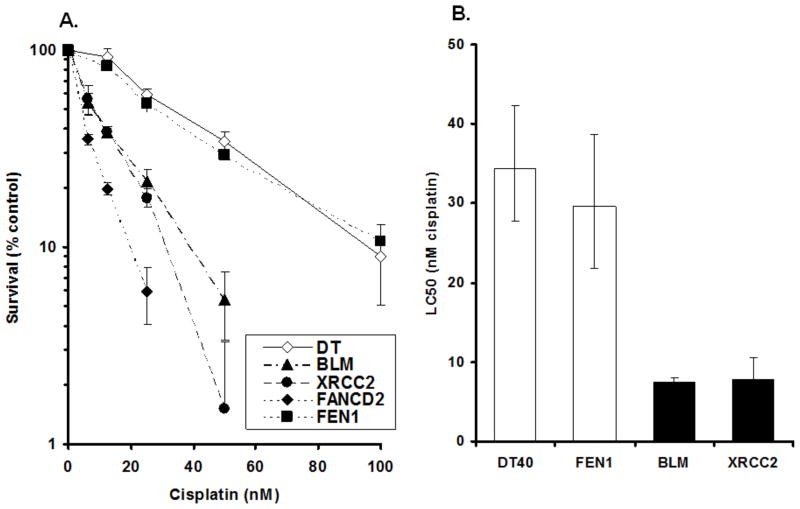
To further assess for any difference of results between the DT40 XTT dye and the clonogenic methods, cisplatin, a DNA cross-linking agent has been chosen. For the purpose of this comparison, we have chosen to report on three DT40 mutant cells lines that are known to be involved in the repair of DNA cross-links: the DNA helicase, BLM and the enzymes, XRCC2 and FANCD2. Cells lacking these proteins are known to be hypersensitive to DNA cross-linking agents. In addition, we have chosen the flap endonuclease, FEN1, which should show little, if any, additional sensitivity to cisplatin beyond the wt. As depicted in Fig. 3A the results for the BLM, XRCC2 and FANCD2 mutants indicate quite strong sensitivity for these mutants compared with the wt, while the FEN1 knockout showed little sensitivity. This result for XRCC2 shows very close agreement for relative sensitivity with the wt to that of Takata, et al., although our result indicated sensitivity for both the wt and the mutant at much lower concentrations due to continuous treatment (Takata et al., 1998). Takata, et al., using the clonogenic assay, report that XRCC2 mutants were approximately 8-fold more sensitive to cisplatin than the normal cells while we showed an approximate 5-fold increase (an LC50 of 22% compared with the wt) in sensitivity (Fig. 3B). This result for XRCC2 exposure also shows excellent agreement with Nojima, et al., who reported an approximate 25% D1 survival compared with the wt, using the colony formation assay (Nojima et al., 2005). The result for the BLM mutant using the XTT method produced an LC50 of 22% compared with the wt, whereas Nojima, et al., was somewhat higher with a D1 survival at ~ 45%. For the FANCD2 mutant exposed to cisplatin, we found a profound sensitivity where the LC50 survival was only 9% that of the normal cells. This result agrees well with that obtained by Matsushita, et al., which were ~ 5% using the clonogenic method (Matsushita et al., 2005). As expected, the XTT method with FEN1 mutants indicated an LC50 of 87% compared with the wt, while Nojima, et al., was ~ 90% in their laboratories clonogenic assay (Nojima et al., 2005).
Figure 3. Cell survival results for DT40 cells and their isogenic mutants continuously exposed to cisplatin.
(A) Survival percentage for each cell line exposed to cisplatin compared with concurrent control. (B) Survival data were log-transformed giving approximate normality. Black columns indicate significant differences analyzed by ANCOVA (p<0.01) between parental DT40 cells and mutants (BLM and XRCC2 cells). In contrast, FEN1-deficient cells showed no significant difference by ANCOVA (p>0.05). Bars: 95% confidence intervals for LC50 value for each cell line.
Taking into consideration the forgoing we conclude that, while the DT40 clonogenic assay is very accurate, it is somewhat cumbersome, costly and time consuming for use as a screen for toxicological purposes where a higher throughput may be desired. We have shown that our DT40/XTT method can provide reasonable sensitivity compared with the clonogenic method while doing so with much less tedium, cost and especially time to produce results.
Characteristics of DT40 Cells and Cells Utilized for Other Genotoxicity Analyses
The Ames test is a bacterial reverse-mutation assay designed as a screen to detect the genotoxicity of chemicals. Salmonella strains deficient in their ability to produce histidine are exposed to chemicals and mutagenicity is determined by the cells’ reversion to a wild type phenotype which is histidine-independent due to mutations caused by the chemical under test (Maron and Ames, 1983). However, although the Ames test achieves a good association with animal carcinogenicity results, it is known that a great many of the enzymes that are used by humans (nearly all of which have homologs in DT40 cells) in DNA repair pathways simply are not known to exist in bacterial cells. Prime examples would be the DT40/human BER proteins, polymerase β (Pol β) and PARP1, for which no equivalents have been found in bacteria. Added to this can be HR, for which there are numerous proteins involved in vertebrate cells which do not exist in bacterial cells and the pathways are thought to operate in a very different manner (Friedberg et al., 2005). Finally, it has been recognized that Salmonella most likely lack the non-homologous end-joining pathway used by eukaryotes (and some other prokaryotes) as an alternative to HR in the of repair DNA double-strand breaks (Bowater et al., 2006).
The use of mutant yeast strains has for decades been one of the mainstays of reverse genetic studies. However, yeasts have been found to be more tolerant to repair and cell-cycle checkpoint defects than higher eukaryotic cells (Dhar et al., 2001) leading to the possibility of underestimation of sensitivity in cell death experiments. This is perhaps due, at least in part, to the difficulty certain chemicals have in crossing the less permeable cell membrane of yeast cells. It has been recognized that there is a high level of homology between yeast genes and those of mammalian cells; whereas, function of some of the DNA damage response genes appears to be different. For example, Rainey, et al., have reported that Chk1 and Chk2 knockouts in DT40 showed that these checkpoint effector kinases control a very different range of checkpoint responses in vertebrates compared with yeast (Rainey et al., 2006). Also, the relative role of each DNA repair pathway observed in yeast and vertebrate cells appears to differ (Yamazoe et al., 2004). For example, HR repair in yeast functions at any time during the cell cycle, whereas HR in vertebrates is only active during late S to G2 (Takata et al., 1998). In fact, genes required for HR, such as Rad51 or Rad54, are not expressed in quiescent vertebrate cells after insult by genotoxic agents (Tan et al., 1999) so that non-homologous end joining would be the pathway of choice, whereas yeast could be using HR. Furthermore, vertebrate proteins directly involved in DSB repair also stimulate a cell-cycle checkpoint, and conversely, proteins involved in checkpoints promote DNA repair, whereas a checkpoint defect does not affect repair in yeast.
The BER pathways in mammalian and yeast cells exhibit many disparities (Kelley et al., 2003). Yeast cells also lack some of the enzymes for dealing with 8-oxo-dG (i.e., MYH, MTH), enzymes that exist in mammalian cells (Kolodner and Marsischky, 1999). Therefore, in yeast, adenine which has been mispaired with 8-oxo-dG is most likely removed by the mismatch repair machinery rather than BER. Also, yeast cells contain no equivalent proteins to Pol β or PARP1, both of which are used in vertebrate cells in BER (Friedberg et al., 2005). As a result, yeast perform primarily long patch BER, whereas vertebrates may be using mainly short patch BER (Frosina et al., 1996; Sobol et al., 1996; Biade et al., 1998). In summary, it seems evident that vertebrate cells such as the DT40 line provide an advantage over yeast in the examination of DNA repair pathways as well as cell cycle checkpoints.
Although many gene-targeting experiments have been performed using mammalian cells, the approach has been hampered by low efficiencies (10−2 to 10−5) in the integration of exogenous DNA through homologous recombination (HR), as most of the DNA integrates at random positions on the chromosomes (Sonoda et al., 2001). In 1991, Buerstedde and Takeda found that targeted integration frequencies in DT40 cells far exceeded those of random integration (more than 1:2) and were orders of magnitude higher than the frequencies in mammalian and other higher eukaryotic cells (Buerstedde and Takeda, 1991). For example, DT40 cells have 15 to 100 times greater targeting efficiency than murine ES cells (Dhar et al., 2001). As such, to detect one targeting event only 10–50 DT40 colonies need be screened in comparison to 100–1000 colonies for the mouse cells. The low gene-targeting efficiencies of ES cells also make it nearly impossible to produce double or triple mutants in those cells where it is relatively easy in DT40 cells. Also, many murine knockouts in recombination and DNA repair processes cause genomic instability with damage checkpoint stimulation. This genomic instability leads to a loss of viability of both the fetus and cultured cells making the cells of little use in the analysis of chromosomal processes because of their limited growth (Yamazoe et al., 2004). Furthermore, Sale reports that knockouts for HR proteins are embryonic lethal in mice (Sale, 2004), and, as this is not the case for most DT40 recombination knockouts, the DT40 cells possess a distinct advantage when investigating HR repair.
Many different knock-out mouse embryonic fibroblasts are available for genotoxicity testing. However, the different knock-outs are from various strains of mice which can make comparisons of responses to potential genotoxins somewhat more difficult. Alternatively, all DT40 mutant cell lines have originated from a single parent and are therefore isogenic. This allows for very precise comparisons between responses to DNA damaging agents.
Another advantage of DT40 cells is their inherent ability to maintain a stable karyotype which makes them particularly valuable in assessing the effect of mutagenic compounds. In contrast, murine ES cells tend to lose multipotency upon exposure to such mutagens (Dhar et al., 2001).
Finally, DT40 cells and their isogenic mutants deficient in specific DNA repair/damage checkpoint genes were used to demonstrate essential roles of DNA metabolism genes in the toleration of genotoxic stress induced by DNA damaging agents. These studies indicate that these DT40 cell lines are a useful tool in the detection of DNA damaging agents. The following mutagens/chemicals have been frequently applied to the DT40 system: MMS (Yoshimura et al., 2006); H2O2 (Yoshimura et al., 2006); IR/UV radiation (Ji et al., 2009); etoposide, camptothecin, cisplatin (Nojima et al., 2005). Extensive DNA damage response analysis has been performed for cisplatin (Nojima et al., 2005); UV/ionizing radiation (Ji et al., 2009); estrogen/tamoxifen metabolites (Mizutani et al., 2004) and NaAsO2 (Ji et al., 2009) using the clonogenic assay.
Advantages of the XTT Dye Method over the Clonogenic Method
As can be observed in Table III, there is a dramatic reduction in materials, manipulation and cost required for the XTT dye method. Also, and just as importantly, much more data can be acquired per unit time. This should be especially attractive to government agencies that wish to quickly establish correct and effective biomarkers so that more meaningful regulations may be determined on a timely basis.
Table III.
Comparison of clonogenic method to XTT dye method
| Clonogenic method (Simpson and Sale 2006) | XTT dye method | |
|---|---|---|
| Medium | Methylcellulose medium requiring overnight mixing, pH adjustment, etc. | Complete medium requiring about 5 min preparation time |
| Plates | One 6-well plate/cell line/dose totaling 20 plates for 4 cell lines and 5 doses | One 24-well plate/cell line with up to 6×3 doses on a single plate |
| Plating of samples | Separate pipetting of medium (with more stirring) and cell samples | A single addition of cell suspension to well |
| Incubation period | 10 days to 2 weeks | 3–4 days |
| Test method | Requires centrifugation, washing and resuspension of cells | No centrifugation/resuspension of cells |
| Measurement | Manual colony counting – one cell line/dose at a time. Plates should be counted promptly when ready | Absorbance reading on plate reader – one complete cell line with all doses at a time. Plates can be stored at 4 °C until convenient to read. |
| Estimated daily data accumulation | 8 cell lines with 5 doses/cell line (40 plates) | 80–100 cell lines with 6 doses/cell lines plus control |
As far as we are aware, only two studies have performed multi-well based DT40 cell DNA damage response analysis using more than 12 different cell lines. One of these reports is our previous study regarding formaldehyde (Ridpath et al., 2007). The other recently reported using DT40 mutant cells with a 12/24-well format to analyze DNA damage responses to UV/ionizing radiation and NaAsO2 (Ji et al., 2009). That particular investigation also compared their 12/24-well plate assay and the clonogenic assay with regard to sensitivity and produced very comparable results between the two methods. These data combined with results from the present study indicate that a multi-well plate-based DT40 cell assay is reasonably sensitive for the detection of genotoxicity induced by diverse agents such as formaldehyde, cisplatin, MMS, UV/ionizing radiation and NaAsO2.
Additionally, the XTT dye method may lend itself readily to automation, whereas it is doubtful the same could be said for the clonogenic method. The advantage of automation becomes obvious if one wishes to use the method as a screen to aid in the determination of the MOA of numerous environmental chemicals. In fact, the ability of the DT40 system to provide very specific information about the DNA repair pathway used to alleviate damage induced by a chemical agent could become one of the most valuable tools in the chest of those given the responsibility to ascertain the genotoxicity/mutagenicity of the agent. Also, and perhaps even more importantly, currently available DT40 mutant cell line has originated from a single parent and is therefore isogenic. This reduces greatly the uncertainty found with other systems where mutant cell lines have been created from different sources – which further increases the value of DT40 in the determination of the MOA.
It is worthwhile to note that this DT40 cell-based DNA damage response analysis has been introduced into toxicology community only recently. Therefore, this assay needs to be further characterized as to the sensitivity and reproducibility using many different types of mutagenic agents as well as non-mutagenic compounds, and compared with traditional genotoxicity assays before using for regulatory decision-making. Despite this additional needed verification, we believe this attractive, high throughput applicable assay will be one of the important genotoxicity analyses in the near future.
Acknowledgments
Grant support: Center for Environmental Health and Susceptibility grant NIEHS P30-ES10126, Superfund Basic Research Program grant NIEHS P42-ES05948, and University of North Carolina at Chapel Hill Department of Environmental Sciences and Engineering B.B. Parker Fellows Program (JRR), and Public Health Service training grant HP 01176-11-00 (JRR).
References
- Baba TW, Giroir BP, Humphries EH. Cell lines derived from avian lymphomas exhibit two distinct phenotypes. Virology. 1985;144:139–151. doi: 10.1016/0042-6822(85)90312-5. [DOI] [PubMed] [Google Scholar]
- Biade S, Sobol RW, Wilson SH, Matsumoto Y. Impairment of proliferating cell nuclear antigen-dependent apurinic/apyrimidinic site repair on linear DNA. J Biol Chem. 1998;273:898–902. doi: 10.1074/jbc.273.2.898. [DOI] [PubMed] [Google Scholar]
- Bowater R, Doherty AJ. Making ends meet: Repairing breaks in bacterial DNA by non-homologous end-joining. PLoS Genet. 2006;2:e8. doi: 10.1371/journal.pgen.0020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerstedde JM, Takeda S. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell. 1991;67:179–188. doi: 10.1016/0092-8674(91)90581-i. [DOI] [PubMed] [Google Scholar]
- Dhar PK, Sonoda E, Fujimori A, Yamashita YM, Takeda S. DNA repair studies: Experimental evidence in support of chicken DT40 cell line as a unique model. J Environ Pathol Toxicol Oncol. 2001;20:273–283. [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA repair and mutagenesis. 2. Washington, DC: ASM Press; 2005. p. 1118. [Google Scholar]
- Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DP, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. J Biol Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- Ji K, Kogame T, Choi K, Wang X, Lee J, Taniguchi Y, Takeda S. A novel approach using DNA-repair-deficient chicken DT40 cell lines for screening and characterizing the genotoxicity of environmental contaminants. Environ Health Perspect. 2009;117:1737–1744. doi: 10.1289/ehp.0900842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley MR, Kow YW, Wilson DM., 3rd Disparity between DNA base excision repair in yeast and mammals: Translational implications. Cancer Res. 2003;63:549–554. [PubMed] [Google Scholar]
- Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- Maron DM, Ames BN. Revised methods for the salmonella mutagenicity test. Mutat Res. 1983;113:173–215. doi: 10.1016/0165-1161(83)90010-9. [DOI] [PubMed] [Google Scholar]
- Matsushita N, Kitao H, Ishiai M, Nagashima N, Hirano S, Okawa K, Ohta T, Yu DS, McHugh PJ, Hickson ID, Venkitaraman AR, Kurumizaka H, Takata M. A FancD2-monoubiquitin fusion reveals hidden functions of fanconi anemia core complex in DNA repair. Mol Cell. 2005;19:841–847. doi: 10.1016/j.molcel.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Mizutani A, Okada T, Shibutani S, Sonoda E, Hochegger H, Nishigori C, Miyachi Y, Takeda S, Yamazoe M. Extensive chromosomal breaks are induced by tamoxifen and estrogen in DNA repair-deficient cells. Cancer Res. 2004;64:3144–3147. doi: 10.1158/0008-5472.can-03-3489. [DOI] [PubMed] [Google Scholar]
- Nojima K, Hochegger H, Saberi A, Fukushima T, Kikuchi K, Yoshimura M, Orelli BJ, Bishop DK, Hirano S, Ohzeki M, Ishiai M, Yamamoto K, Takata M, Arakawa H, Buerstedde JM, Yamazoe M, Kawamoto T, Araki K, Takahashi JA, Hashimoto N, Takeda S, Sonoda E. Multiple repair pathways mediate tolerance to chemotherapeutic cross-linking agents in vertebrate cells. Cancer Res. 2005;65:11704–11711. doi: 10.1158/0008-5472.CAN-05-1214. [DOI] [PubMed] [Google Scholar]
- Rainey MD, Zachos G, Gillespie DA. Analysing the DNA damage and replication checkpoints in DT40 cells. Subcell Biochem. 2006;40:107–117. doi: 10.1007/978-1-4020-4896-8_8. [DOI] [PubMed] [Google Scholar]
- Ridpath JR, Nakamura A, Tano K, Luke AM, Sonoda E, Arakawa H, Buerstedde JM, Gillespie DA, Sale JE, Yamazoe M, Bishop DK, Takata M, Takeda S, Watanabe M, Swenberg JA, Nakamura J. Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 2007;67:11117–11122. doi: 10.1158/0008-5472.CAN-07-3028. [DOI] [PubMed] [Google Scholar]
- Sale JE. Immunoglobulin diversification in DT40: A model for vertebrate DNA damage tolerance. DNA Repair (Amst) 2004;3:693–702. doi: 10.1016/j.dnarep.2004.03.042. [DOI] [PubMed] [Google Scholar]
- Simpson LJ, Sale JE. Colony survival assay. Subcell Biochem. 2006;40:387–391. doi: 10.1007/978-1-4020-4896-8_31. [DOI] [PubMed] [Google Scholar]
- Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- Sonoda E, Morrison C, Yamashita YM, Takata M, Takeda S. Reverse genetic studies of homologous DNA recombination using the chicken B-lymphocyte line, DT40. Philos Trans R Soc Lond B Biol Sci. 2001;356:111–117. doi: 10.1098/rstb.2000.0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TL, Essers J, Citterio E, Swagemakers SM, de Wit J, Benson FE, Hoeijmakers JH, Kanaar R. Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Curr Biol. 1999;9:325–328. doi: 10.1016/s0960-9822(99)80142-0. [DOI] [PubMed] [Google Scholar]
- Ulrich E, Boehmelt G, Bird A, Beug H. Immortalization of conditionally transformed chicken cells: Loss of normal p53 expression is an early step that is independent of cell transformation. Genes Dev. 1992;6:876–887. doi: 10.1101/gad.6.5.876. [DOI] [PubMed] [Google Scholar]
- Winding P, Berchtold MW. The chicken B cell line DT40: A novel tool for gene disruption experiments. J Immunol Methods. 2001;249:1–16. doi: 10.1016/s0022-1759(00)00333-1. [DOI] [PubMed] [Google Scholar]
- Yamazoe M, Sonoda E, Hochegger H, Takeda S. Reverse genetic studies of the DNA damage response in the chicken B lymphocyte line DT40. DNA Repair (Amst) 2004;3:1175–1185. doi: 10.1016/j.dnarep.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Yoshimura M, Kohzaki M, Nakamura J, Asagoshi K, Sonoda E, Hou E, Prasad R, Wilson SH, Tano K, Yasui A, Lan L, Seki M, Wood RD, Arakawa H, Buerstedde JM, Hochegger H, Okada T, Hiraoka M, Takeda S. Vertebrate POLQ and POLbeta cooperate in base excision repair of oxidative DNA damage. Mol Cell. 2006;24:115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]