Abstract
A fundamental issue in microbial and general ecology is the question to what extent environmental conditions dictate the structure of communities and the linkages with functional properties of ecosystems (that is, ecosystem function). We approached this question by taking advantage of environmental gradients established in soil and sediments of small stream corridors in a recently created, early successional catchment. Specifically, we determined spatial and temporal patterns of bacterial community structure and their linkages with potential microbial enzyme activities along the hydrological flow paths of the catchment. Soil and sediments were sampled in a total of 15 sites on four occasions spread throughout a year. Denaturing gradient gel electrophoresis (DGGE) was used to characterize bacterial communities, and substrate analogs linked to fluorescent molecules served to track 10 different enzymes as specific measures of ecosystem function. Potential enzyme activities varied little among sites, despite contrasting environmental conditions, especially in terms of water availability. Temporal changes, in contrast, were pronounced and remarkably variable among the enzymes tested. This suggests much greater importance of temporal dynamics than spatial heterogeneity in affecting specific ecosystem functions. Most strikingly, bacterial community structure revealed neither temporal nor spatial patterns. The resulting disconnect between bacterial community structure and potential enzyme activities indicates high functional redundancy within microbial communities even in the physically and biologically simplified stream corridors of early successional landscapes.
Keywords: bacterial community structure, biodiversity and ecosystem function, Chicken Creek, enzyme activity, soil, stream sediment
Introduction
A widespread tenet in microbial and general ecology is that structure determines function. Here, structure refers to the composition and diversity of biological communities, and function relates to the processes that the communities drive. Both are directly determined by the environmental setting; in addition, functions can be indirectly affected through altered community structure. Experimental tests with assembled bacterial communities at strictly controlled diversity levels have demonstrated that changes in community attributes such as diversity can affect ecosystem processes in various cases (Bell et al., 2009; Langenheder et al., 2010; Peter et al., 2011), one of them quite spectacular (Bell et al., 2005). These results are in line with evidence from a body of similar experiments conducted with higher plants, invertebrates, algae and other microorganisms (Gessner et al., 2010; Cardinale et al., 2011). They contrast with outcomes of indirect microbial community manipulations, for instance, by means of dilution series (Langenheder et al., 2005, 2006; Wertz et al., 2006; Östman et al., 2010) or fumigation (Griffiths et al., 2000), which have failed to detect similar relationships. The discrepancy can be reconciled by acknowledging that key differences exist between the two approaches (Bell et al., 2009). Among the most notable ones are the vastly divergent diversity levels considered, and the fact that manipulations such as fumigation or dilution series generate microbial communities that are subsets of the taxa originally present, whereas experiments with assembled communities can independently vary in diversity and taxonomic composition. A drawback of the latter approach, however, is the unknown degree to which the simplified assembled communities represent natural situations.
A third approach that can yield insights into relationships between community structure and ecosystem function consists in simultaneously analyzing communities and processes across a gradient of environmental conditions in space or time and examining the data for common patterns (Findlay et al., 2003; Docherty et al., 2006; Findlay and Sinsabaugh, 2006; Comte and del Giorgio, 2009). This strategy is akin to experimentally modifying an environmental factor (for example, the type, source or concentration of organic carbon or nutrients) before assessing differences in both functional and structural community parameters (Langenheder et al., 2005, 2006). Four different outcomes are possible in such studies: (1) neither community structure nor function responds to environmental changes, which is suggestive of resistance of both structure and function; (2) ecosystem function but not community structure responds, suggesting greater sensitivity of ecosystem function than community structure; (3) community structure is affected whereas ecosystem function is not, indicating functional redundancy or compensatory effects (Kritzberg et al., 2006; Allison and Martiny, 2008); and (4) both community structure and ecosystem function are altered, in which case it remains unclear whether the latter changes are because of direct environmental influences or altered communities and their functional capacities. Although the uncertainty associated with the last point is unsatisfactory, the approach can yield valuable insights, especially if one of the first three outcomes is observed.
Stream corridors are heterogeneous environments composed of a mosaic of patches of different metabolic activities (Larned et al., 2010; Winemiller et al., 2010). Microbial activities in these patches also vary over time in response to temporal variation of environmental conditions (Jones and Lock, 1993; Wilczek et al., 2005; Sinsabaugh and Follstad Shah, 2010), changes in water availability (Romaní et al., 2006a, 2006b; Artigas et al., 2009) and varying interactions among diverse microbial groups (for example, bacteria and algae; Romaní et al., 2006a, 2006b; Ylla et al., 2009; Pohlon et al., 2010). This creates a spatiotemporal template that provides scope for gradients in environmental conditions along hydrological flow paths from upland sites to stream channels and longitudinally in stream corridors (Larned et al., 2010).
One reason evoked to explain the frequent lack of functional consequences to community changes in field studies is that microbial diversity in natural environments is extremely high. As a result, effects of community change on ecosystem functions can be easily masked. However, this masking effect could be alleviated by taking advantage of the physically and biologically simple structure of nascent ecosystems formed by volcanism (Vitousek, 2004), glacier retreat (Milner et al., 2007; Brankatschk et al., 2011) or humans (Gerwin et al., 2009; Gerull et al., 2011), where species numbers are reduced but artifacts caused by greatly simplified laboratory systems are avoided.
Another potential reason for the frequent inability to detect functional consequences arising from community changes is that the broad ecosystem processes commonly assessed (for example, microbial respiration or nitrogen mineralization) are too coarse-grained because they integrate across many individual processes. Specific metabolic functions, such as particular biogeochemical reactions catalyzed by microbial enzymes, are likely to be more tightly linked to the particular populations constituting a given microbial community (Langenheder et al., 2006). Therefore, one way to increase sensitivity of tests for effects of altered community structure on ecosystem functions is to assess activities of a suite of enzymes involved in the biogeochemical cycling of important elements such as carbon, nitrogen and phosphorus.
The objectives of the present study were to determine patterns of, and linkages between, bacterial community structure and microbial metabolic activities in soil and sediment along hydrologic flow paths in small nascent stream corridors of a recently created catchment. We hypothesized that long-term water availability and vertical exchange of water between the hyporheic zone and the streambed would be major drivers of change of both bacterial community structure and microbial metabolic activity reflected in potential enzyme activities. Accordingly, we expected to find: (1) spatial variation of bacterial community structure and potential enzyme activities in soil and sediments along the environmental gradient defined by the hydrologic flow path in the catchment, (2) similar changes of community structure and potential enzyme activities over time and (3) systematic relationships between community structure and patterns of the potential enzyme activities.
Materials and methods
Study site
The study was conducted in an experimental catchment near Cottbus in eastern Germany (51°36′N, 14°16′E), which is referred to as Chicken Creek catchment (Gerwin et al., 2009). It was deliberately created to study ecosystem succession. Average annual precipitation and air temperature are 559 mm and 9.3 °C, respectively (record 1971–2000, Meteorological Station Cottbus, German Weather Service). The catchment covering 6 ha was created between 2004 and 2005 (Gerwin et al., 2009). A 1–2 m base layer of Tertiary clay was overlain by a 2–4 m top layer of Pleistocene sand. Conditions typically associated with mining (for example, low pH or high metal concentrations) were prevented during construction. The upper portions of the stream network that developed were characterized by ephemeral flow, whereas short sections downstream had permanent flow (Figure 1). Sediments in both the stream channels and soil were dominated by fine to medium sand (grain size of 200–300 μm). Further physico-chemical characteristics of the catchment and its streams are summarized in Table 1.
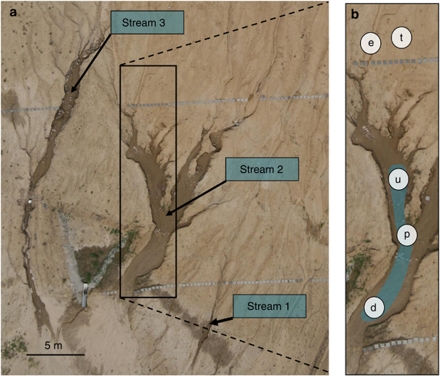
Figure 1.
Aerial view of the three main stream networks in the Chicken Creek catchment (a) and position of sampling sites along the hydrological flow path, as illustrated for stream 2 (b): terrestrial soil (t) and ephemeral (e), upwelling (u), perched-flow (p) and downwelling (d) sites in the stream channels.
Table 1. Characteristics of the three main stream corridors in the Chicken Creek catchment.
| Parameter | Stream 1 | Stream 2 | Stream 3 |
|---|---|---|---|
| Catchment area (m2) | 950 | 7473 | 20 496 |
| Mean discharge (l s−1) | 0.02 | 0.13 | 0.37 |
| Length of perennial stream section (m) | 20 | 38 | 43 |
| Length of ephemeral stream channel (m) | 103 | 171 | 249 |
| Permanently wetted streambed area (m2) | 4 | 99 | 68 |
| Ephemeral streambed area (m2) | 30 | 390 | 432 |
| Channel slope (%) | 6.5 | 5.1 | 3.9 |
| Channel width at ground level (m) | 1.2 (0.6–1.9) | 2.6 (1.0–5.8) | 2.2 (0.2–5.4) |
| Organic matter in sediment or soil (mg g−1) | 2.5±0.1 | 1.7±0.1 | 2.3±0.2 |
| Chlorophyll a in sediment or soil (mg m−2) | 35.1±0.3 | 23.8±0.2 | 11.9±0.1 |
| Dissolved organic carbon in surface water (mg l−1) | 16.3±0.8 | 11.2±0.7 | 15.9±1.1 |
| Dissolved organic carbon in pore water (mg l−1) | 18.3±0.9 | 10.8±0.2 | 14.0±0.5 |
| Dissolved organic nitrogen in surface water (μg l−1) | 18±6 | 19±5 | 42±12 |
| Dissolved organic nitrogen in pore water (μg l−1) | 17±5 | 24±7 | 34±13 |
| NH4+ in surface water (μg N l−1) | 116±26 | 74±10 | 146±33 |
| NH4+ in pore water (μg N l−1) | 98±10 | 62±3 | 122±39 |
| PO43− in surface water (μg P l−1) | 4.3±0.6 | 4.1±0.3 | 6.2±0.5 |
| PO43− in pore water (μg P l−1) | 5.9±1.3 | 4.9±0.6 | 7.5±1.6 |
Data on the stream channels refer to the main stem of the drainage networks.
Most data from Gerull et al. (2011).
Values are means±1 s.e. (n=9–12) with ranges in parentheses.
Sampling
Soil and sediment samples were collected once in each season (late November 2007, April 2008, August 2008 and October 2008) in the three major sub-catchments of the Chicken Creek catchment (Supplementary Figure S1 shows views of streams 1 and 3 in November 2007 and October 2008; Gerull et al., 2011). Five sites differing in their hydrological characteristics were selected along each of the three stream corridors (Figure 1): (1) an upland terrestrial site adjacent to the stream channel, (2) an upstream ephemeral channel site, (3) a permanently wet upwelling site, (4) a site with perched surface flow and (5) a downwelling site. Three replicate samples from the aerobic surface soil or sediment (top 2 cm) were taken at each site (Gerull et al., 2011) and immediately preserved. Approximately 3 g of soil or sediment was preserved in 10 ml of 2% formalin containing 0.1% pyrophosphate and stored at 4 °C for determination of bacterial abundance. Subsamples for measuring potential enzyme activities were frozen at −20 °C as recommended by Sinsabaugh et al. (1991) in case immediate analyses are not possible. Subsamples for molecular analyses were frozen in liquid nitrogen and stored at −80 °C until DNA was extracted. However, during shipping on dry ice, the winter samples were delivered with delay, thawed, and were therefore discarded.
Enzyme assays
A total of 10 potential enzyme activities were measured with substrate analogs linked to fluorescent molecules, 4-methylumbelliferone (MUB), 7-amino-4-methylcoumarin (AMC), or to 3,4-dihydroxyphenylalanine (L-DOPA) for colorimetric assays of phenol oxidase and peroxidase. Enzymes were chosen based on their metabolic function and use in previous studies (Table 2; Sinsabaugh et al., 1991, 2008). Enzyme assays were performed as follows: 10 g of sediment sample was placed in 300 ml of acetate buffer (32.1 m sodium acetate, 0.1% glacial acetic acid, pH 5, autoclaved). The slurry was stirred and volumes of 200 μl were pipetted into 96-well microplates. The 10 different substrate analogs were added to the wells (50 μl of 200 μ stock solutions) and the microplates incubated for 90 min at 12 °C for winter samples, 15 °C for spring and autumn samples and 18 °C for summer samples. There were eight analytical replicates. A volume of 10 μl of 0.5 N NaOH was added to increase pH in the wells before shaking the microplates and measuring fluorescence or absorbance on a microplate reader (Tecan Infinite 200, Männedorf, Switzerland). The fluorescence emission wavelengths were 445 and 450 nm for the cleaved MUB and AMC substrates, respectively. The excitation wavelength was 365 nm for both types of substrate. Absorbance in the phenol oxidase and peroxidase assays was measured at 460 nm using the same microplate reader. Potential enzyme activities were expressed in μmol or mmol substrate per gram sediment or soil dry mass per hour. Potential enzyme activities determined in winter (12 °C) and summer (18 °C) were normalized to the spring and autumn temperature of 15 °C based on a Q10 of 2 (Davidson and Janssens, 2006).
Table 2. Extracellular enzymes analyzed, substrate analogs used to determine their potential activities and biogeochemical function of the enzymes according to Sinsabaugh et al. (2008).
| Enzyme | Substrate analog | Function |
|---|---|---|
| Carbon-acquiring enzymes | ||
| β-Glucosidase | 4-MUB-β--glucopyranoside | Cellulose degradation |
| Cellobiosidase | 4-MUB-β-cellobioside | Cellulose degradation |
| β-Xylosidase | 4-MUB-β--xylopyranoide | Hemicellulose degradation |
| Nitrogen-acquiring enzymes | ||
| Chitinase | 4-MUB-N-acetyl-β--glucosaminide | Degradation of β-1,4 glucosamines |
| Leucine-AP | -Leucine-AMC | Peptide degradation |
| Glutamate-AP | -Glutamic acid γ-AMC | Peptide degradation |
| Aspartate-AP | Aspartic acid-AMC | Peptide degradation |
| Phosphorus-acquiring enzyme | ||
| Phosphatase | 4-MUB-phosphate | Phosphomonoester degradation |
| Lignin-degrading enzymes | ||
| Phenol oxidase | 3,4-dihydroxyphenylalanine (-DOPA) | Polyphenol oxidation |
| Peroxidase | 3,4-dihydroxyphenylalanine (-DOPA) with H2O2 | Polyphenol oxidation |
Abbreviations: AMC, 7-amino-4-methylcoumarin; AP, aminopeptidase; MUB, 4-methylumbelliferone.
Bacterial abundance
Bacterial abundance was determined by flow cytometry. Cells were detached with a sonifier probe (Branson Digital Sonifier 250, Danbury, CT, USA; flat tip, actual output of 38 W for 3 × 20 s) as desribed by Buesing and Gessner (2002). After homogenization, a 1-ml subsample of the cell suspension was placed on top of 0.5 ml of Histodenz solution (1.3 g ml−1; Sigma-Aldrich, Buchs, Switzerland). Samples were then centrifuged (90 min at 4 °C, 17 135 g) and the entire upper layer on top of the Histodenz layer was quantitatively removed. The bacterial cells were stained with SYBRGreen I (Promega, Dübendorf, Switzerland) in anhydrous dimethylsulfoxide and incubated in the dark for 15 min. The samples were diluted 1:10 or 1:100 with filtered (0.22 m Millex-GP, Millipore, Wohlen, Switzerland) mineral water (Evian, France) such that the cell concentration did not exceed 106 per ml. Samples were analyzed with a CyFlow space Flow Cytometer System (Partec, Görlitz, Germany) equipped with a 200 mW solid-state laser emitting light at 488 nm. Green and red fluorescence were measured at 520 nm (FL1 channel) and 630 nm (FL3 channel). The flow cytometer was set as follows: gain FL1=495, gain FL3=50, speed=4 (implying an event rate never exceeding 1000 events per second). Counts were recorded as logarithmic signals and were triggered on the green fluorescence channel (FL1). Data were processed with Flowmax software (Partec), using electronic gating to separate the desired events. Presentation of the data as FL1/FL3 dot plots allows for optimal distinction between stained intact microbial cells and instrument noise or sample background (Hammes and Egli, 2005).
Bacterial community fingerprinting
DNA was extracted from frozen soil and sediment (0.5 g fresh mass) by mechanically disrupting cells, enzymatic digestion and final DNA purification (Supplementary Methods). The purified DNA was stored at −20 °C. The V3 region of the 16S rDNA gene was amplified by PCR using primers Eub338f and Eub518r. There were 31 amplification cycles. Denaturing gradient gel electrophoresis (DGGE) of 16S rDNA was performed using a D-code electrophoresis system (Bio-Rad Laboratories, Reinach, Switzerland) (Flury and Gessner, 2011). An 8% (w/v) polyacrylamide gel (acrylamide/bisacrylamide ratio of 37.5:1) was loaded with 600 ng of PCR product. The denaturing gradient increased linearly in the gel from 45% to 60% (top to bottom) of denaturant with 100% of denaturant corresponding to 40% formamide and 7 urea. The gels were run at 60 °C and 38 V for 16 h. The bands stained with SYBRGreen II (Promega) and images were taken in a gel documentation system (Gel Doc XR, Bio-Rad). GelCompar II software (Applied Maths NV, Sint-Martens-Latem, Belgium) was used to analyze the fingerprinting profiles.
Data analysis
Analysis of variance was used with the program PASW 18.0 (SPSS Inc., IBM, Armonk, NY, USA) to test for differences in potential enzyme activities and bacterial abundance among sampling dates, streams and sites. Sampling date and site were treated as fixed factors, and stream as random factor. Samples taken at different times were considered independent because the study site is highly dynamic and samples were taken several months apart. QQ plots and frequency histograms indicated that residuals did not meet assumptions required for parametric tests. Therefore, variables (x) were transformed according to ln(x+c), where c=q0.25x2/q0.75x and q=quantile (Statistical Seminar, ETH Zurich, personal communication). Analysis of variance indicated that the three-way interactions (sampling date × site × stream) were not significant for most of the enzyme activities, except for aspartate aminopeptidase, phenol oxidase and phenol peroxidase. However, with this design, the assumptions of the model were not met, except for peroxidase. When excluding the three-way interaction from the design, the resulting model better fit the data and the underlying assumptions were met. Graphical displays showed no indication that the three-way interaction was significant, and including or excluding this interaction term produced similar results except for peroxidase and phenol oxidase. Therefore, a simplified model was used for the final analyses, in which the three-way interaction was removed for all enzymes.
Nonmetric multidimensional scaling analyses were performed on a matrix of DGGE band information and a matrix including information on all potential enzyme activities. These analyses were performed with the function meta.mds of the vegan package in the software R (R Development Core Team, 2011) based on Bray–Curtis distances and 1000 permutations. Multivariate analysis of variance was performed with the function adonis to test for significant differences among clusters, again based on Bray–Curtis distances.
Results
Bacterial abundance
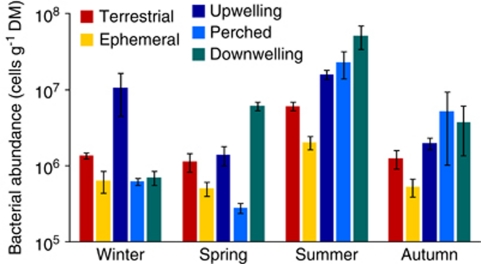
Bacterial cell abundance varied among sites and sampling dates without showing any striking pattern (Figure 2). These variations were reflected in a significant interaction between site and sampling date (analysis of variance, F12,140=6.52, P<0.001). Nevertheless, bacterial cell numbers at all sites were highest in summer (F3,6=47.48, P<0.01) and they were also higher at all sampling dates in the terrestrial and upwelling sites than at the ephemeral sites of the stream channels (F4,8=7.05, P=0.01).
Figure 2.
Bacterial abundances (mean±1 s.e., n=9) in soil and sediments collected at four sampling dates and five sites along the hydrological flow path of each of three small stream corridors. Note the logarithmic scale.
Enzyme activities
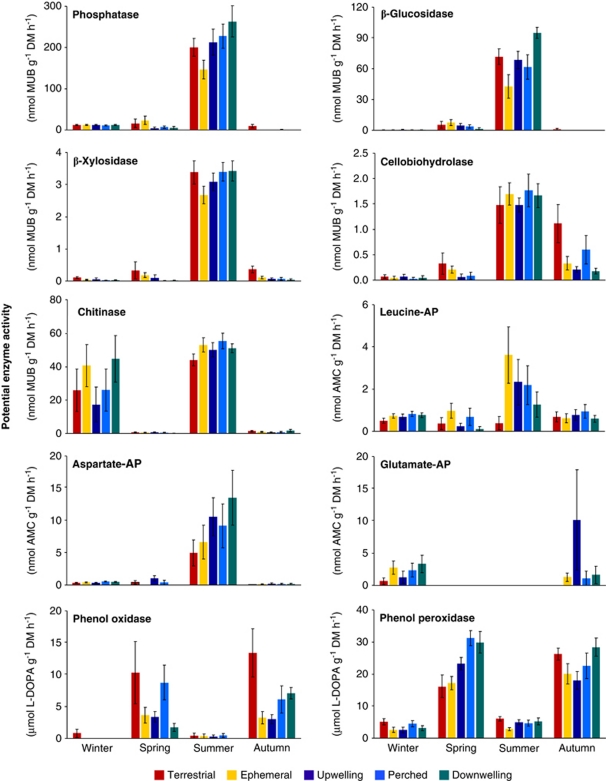
Potential activities of the 10 enzymes analyzed varied little among sites and streams in the Chicken Creek catchment (Figure 3). Only the potential activity of glutamate aminopeptidase differed among sites (F4,8=6.55, P=0.012), that of cellobiohydrolase among streams (F2,4=7.45, P=0.049) and that of β-xylosidase among both sites (F4,8=5.60, P=0.02) and streams (F2,7=6.68, P=0.03), and these spatial differences did not reveal any clear systematic pattern. Potential peroxidase activity showed significant interactions between site and stream (F8,144=2.62, P=0.01) and, in the full model, also between site, stream and sampling date (F24,120=6.02, P<0.001).
Figure 3.
Patterns of potential enzyme activities (mean±1 s.e., n=9) in soil and sediments collected at four sampling dates and five sites along the hydrological flow path of each of three small stream corridors.
In contrast to rather small spatial differences, temporal differences were large for all enzymes tested except leucine aminopeptidase (Figure 3). However, different enzymes showed markedly different temporal patterns. The potential activity of phosphatase was the highest in summer, with average values up to 250 nmol MUB g−1 h−1, differing greatly from potential activities at other sampling dates (F3,6=477, P<0.001). The polysaccharide-degrading enzymes (β-glucosidase, cellobiohydrolase and β-xylosidase) also showed greater potential activities in summer (F3,6=311, F3,6=76.2 and F3,6=79.4, respectively, P<0.001), although absolute levels differed among the three enzymes. The potential activity of β-glucosidase in summer (43–95 nmol MUB g−1 h−1) far exceeded those of cellobiohydrolase and β-xylosidase (1.5–3.4 nmol MUB g−1 h−1). Chitinase potential activity was also increased in summer (F3,6=24.5, P<0.01), ranging from 44 to 55 nmol MUB g−1 h−1, although at some sites, levels of this enzyme were also elevated in winter, when they ranged from 17 to 45 nmol MUB g−1 h−1. Potential peptidase activities (that is, leucine, glutamate and aspartate aminopeptidase) were rather variable overall and the temporal differences were less pronounced than for the other types of enzymes (Figure 3). Aspartate aminopeptidase potential activity ranged from undetectable to 13 nmol AMC g−1 h−1 and was highest in summer (F3,6=48.4, P<0.01). Glutamate aminopeptidase activity was very low, ranging from undetectable to 0.33 nmol AMC g−1 h−1, with the highest activities occurring in winter and autumn (F3,6=40.76, P<0.01). Leucine aminopeptidase, in contrast, showed slightly higher potential activities in summer, although temporal differences were not significant (F3,6=3.6, P=0.085). Phenol oxidase and phenol peroxidase, unlike all other enzymes, showed higher potential activities in spring and autumn compared with winter and summer (F3,6=41.0 and F3,6=36.0, respectively, P<0.01). The potential activities of both enzymes ranged from undetectable to 31 μmol -DOPA g−1 h−1. Significant interactions between sampling date and site or stream were observed in 4 of 20 cases, but overall these effects were weak.
Temperature-corrected enzyme activities based on a Q10 of 2 (see, for example, Davidson and Janssens, 2006) showed similar temporal patterns as uncorrected activities, although temporal differences tended to be slightly reduced (data not shown). Similarly, the same temporal patterns emerged when enzyme activities were expressed per gram of soil or sediment organic matter, rather than per gram of soil or sediment dry mass, because the organic matter content in soil and sediment fluctuated little over time. Temporal differences in cell-specific enzyme activities were less pronounced than those shown in Figure 3, although still significant for each of the 10 enzymes tested (F4,8−9=7.3 to 82, P<0.01). Thus, the overall picture that emerges is that temporal differences in potential enzyme activities were pronounced and variable among the 10 enzymes tested, whereas spatial differences were nearly absent.
Community structure and functional diversity
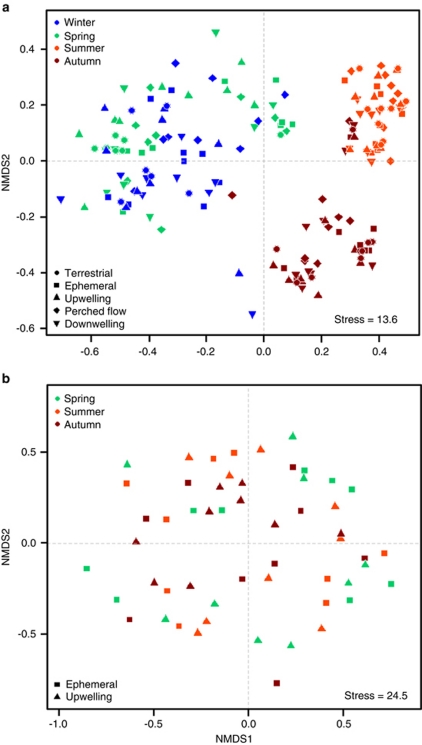
Nonmetric multidimensional scaling analysis of potential enzyme activities revealed three clusters regrouping most samples taken in winter and spring, in summer and in autumn, respectively (Figure 4a; F3,141=135.4, P=0.001), although six autumn samples grouped close to the summer cluster. A similar pattern was not found in the biplot calculated from DGGE banding patterns, including samples from all three streams, two sites in each (ephemeral and upwelling sites) and three sampling dates (once in spring, summer and autumn), which were arranged completely randomly in the nonmetric multidimensional scaling ordination (Figure 4b). As a result, a link between microbial community structure and enzyme activities was not apparent.
Figure 4.
Nonmetric multidimensional scaling (NMDS) ordination of (a) potential activities of 10 different enzymes, and (b) bacterial community structure inferred from DGGE bands of samples taken at three sampling dates (winter samples were lost during transport) and two types of sites (ephemeral and upwelling sites).
Discussion
Spatial variability of potential enzyme activities
An important result of our analysis of enzymes in the early successional Chicken Creek catchment is that potential activities varied little across sites. This resemblance for all of the 10 enzymes studied is unexpected because sites, ranging from dry soil crusts to permanently submerged stream sediments, differed widely in terms of water availability, a key factor determining microbial metabolism. The unresponsiveness of potential enzyme activities to differences in site characteristics is in accordance with observations on general microbial metabolism measured as respiratory activity at various sites in a barren desert of Antarctica (McMurdo Valley) (Zeglin et al., 2009) and in the early successional Chicken Creek catchment (Gerull et al., 2011). This suggests, contrary to our hypotheses, that neither permanence of water availability (perennial versus ephemeral sites) nor direction of vertical water exchange (groundwater upwelling versus channel water downwelling) control potential enzyme activities in early successional catchments with scarce vegetation cover.
A feature of sediments and soils that often correlates with potential enzyme activities is organic matter content (Romaní and Marxsen, 2002; Sinsabaugh et al., 2008; Artigas et al., 2009). This is corroborated by higher activities of cellulose- and hemicellulose-degrading enzymes in hyporheic sediments of an upwelling zone of a lowland stream with more organic matter than in a downwelling zone (Rulik and Spácil, 2004). However, in line with the lack of spatial variation in potential enzyme activities, organic matter content varied little in the Chicken Creek catchment, and although it was slightly higher at the terrestrial sites (Gerull et al., 2011), this difference was small and apparently insufficient to alter potential enzyme activities.
Temporal variability of potential enzyme activities
More striking than the spatial similarities in potential enzyme activities are the highly congruent changes we observed over time. Temporal temperature fluctuations accounted but for a small fraction of the activity increases of many enzymes in the summer, as revealed by persistent temporal patterns when the data were normalized to a standard temperature. Furthermore, temperature can evidently not account for the often-elevated potential activities in spring, autumn and, in one case (chitinase), in winter. This limited temperature response is consistent with data from surface water (Sinsabaugh and Follstad Shah, 2010), fine sediments (Wilczek et al., 2005) and biofilms (Jones and Lock, 1993) of lowland streams and rivers and suggests important, seasonally varying determinants of enzyme activities in addition to temperature.
One such factor is light. Although neither particularly strong (r=0.24–0.33) nor necessarily indicative of cause-and-effect relationships, highly significant correlations between chlorophyll-a content in soil and sediments (data from Gerull et al., 2011) and potential activities of phosphatase and all carbon-acquiring hydrolytic enzymes might suggest that algae were instrumental in increasing potential enzyme activities in summer. This could be due directly to algal enzyme production, especially in the case of phosphatase. Two alternative mechanisms involving heterotrophs are more likely, however, at least for the carbon-acquiring enzymes: heterotrophs may excrete enzymes using carbon compounds of algal origin as the main substrate, or the organic carbon supplied by algae serves as priming agent facilitating the enzymatic cleavage of recalcitrant carbon compounds from other sources (Rier et al., 2007; Guenet et al., 2010). Such priming might indeed be an important mechanism in the Chicken Creek catchment, because dissolved organic carbon is abundant (∼14 mg l–1) but highly recalcitrant (Gerull et al., 2011). Particulate organic carbon also appears to be recalcitrant in the catchment, as suggested by significant correlations between sediment organic matter content and potential activities of ligninolytic (phenol oxidase and peroxidase; r=0.19 and 0.26, P<0.02) but not polysaccharide-degrading enzymes (r⩽0.3, P>0.7, except for β-xylosidase, P=0.08), suggesting that priming could influence the utilization of particulate organic carbon as well. Irrespective of the predominant mechanism (that is, direct use of algal carbon or priming), the increased bacterial abundances we observed in summer indicate that heterotrophs contributed to the elevated potential activities of many of the enzymes determined.
The tight synchrony of potential enzyme activities observed across sites is further amplified by stark contrasts in the temporal patterns of the 10 individual enzymes tested. This demonstrates that measures of general metabolic activity of microbes, such as respiration, merely reflect average responses of microbial activities to environmental conditions or microbial community structure. Mechanistic understanding of the link between individual metabolic activity and ecosystem process rates might require analyses with finer resolution. The potential enzyme activities measured here are more informative in this regard than bulk respiration measurements. Nevertheless, they might still be too coarse, given that proteomic analysis has revealed that even individual microbial strains on decomposing leaf litter can simultaneously express nearly two dozen enzymes of a given class (for example, cellulases; Schneider and Riedel, 2010), let alone the number of enzymes expressed by diverse microbial communities.
Temporal variation of two nitrogen-acquiring enzymes used in our study (aspartate- and glutamate-aminopeptidase) showed strikingly opposite temporal patterns, although the basic function and molecular structure of the two enzymes is very similar. This discrepancy points to the complexity of relationships between microbial activities and environmental conditions and further underlines the difficulty of interpreting spatiotemporal patterns of potential activities of individual enzymes in terms of broad ecosystem processes.
Temporal patterns of oxidases capable of attacking aromatic rings were markedly different from those of all other enzymes. With algal photosynthesis declining in the autumn, the supply of easily degradable organic matter should also decrease, and eventually be completely consumed by heterotrophic microbes. This would leave behind a recalcitrant organic matter pool that includes a large fraction of aromatic compounds. High potential activities of phenol oxidase and peroxidase at this time could reflect the degradation of these compounds. Subsequent declines in winter (along with those of all other enzymes except chitinase) but reestablishment of high activities in spring appear to arise from temperature limitation during the cold season when ice sheets regularly formed in the streams at night.
Bacterial community structure and ecosystem function
A particularly intriguing finding of our analysis is that potential activities of the broad suite of carbon-, nitrogen- and phosphorus-acquiring enzymes that we measured were unrelated to the structure of bacterial communities in the early successional Chicken Creek catchment. Phrased differently, community structure was unrelated to function (see also Burke et al., 2011). This disconnect arose because of a lack of clear spatial or temporal patterns in bacterial communities, whereas the activities of all 10 enzymes measured showed distinct temporal changes (see above). This finding is surprising in view of the general paradigm in ecology and biology that structure determines function.
The complete absence of temporal or spatial community pattern suggests stochastic distributions of populations following the neutral community assembly model (Sloan et al., 2006). Accordingly, community structure in environmental patches is shaped only by population growth or declines, and by random immigration of individuals of new populations. High dispersal and invasion ability of bacteria in a uniform environment can then lead to ubiquity of communities across space and time (Van Der Gucht et al., 2007; Urban and De Meester, 2009). Our results indicate that high dispersal can override the influence of even substantial environmental differences. This includes strong spatial and temporal variation in water availability, which arguably is one of the most critical constraints of microbial activities.
The striking disconnect between bacterial community structure and potential enzyme activities that we observed also appears to be counter to results from controlled experiments with assembled communities, where bacterial diversity (that is, a component of community structure) has been found to matter for ecosystem functioning (Bell et al., 2005; Langenheder et al., 2010; Peter et al., 2011). However, those experiments primarily focused on assessing effects of varying numbers of species or strains rather than the composition of communities and abundance of dominant strains or species as in the present study. Moreover, in line with our results, field studies that have indirectly manipulated microbial communities have not typically found evidence for strong relationships between community structure and rates of ecosystem processes (Langenheder et al., 2005, 2006; Wertz et al., 2006; Östman et al., 2010).
One reason for these differences might be methodological, specifically the inability of DGGE to sufficiently resolve the structure of diverse communities (Woodcock et al., 2006) that could have rapidly established even in the early successional Chicken Creek catchment. DGGE tends to detect dominant taxa only, resulting in a greatly simplified blueprint of the communities characterized with this kind of fingerprinting methods (Casamayor et al., 2000; Lindström, 2000; Woodcock et al., 2006). This explanation would imply, however, that patterns of ecosystem functioning (that is, potential enzyme activities) are primarily driven by subordinate as opposed to the dominant species or strains that are best detected by DGGE. Conversely, one cannot rule out the possibility that DGGE detected a significant fraction of dormant bacteria, which have been estimated to account for an average of ∼45% of all bacterial cells in fresh waters (Lennon and Jones, 2011). However, both of these scenarios are rather unlikely.
Alternatively, the lack of congruent patterns between bacterial community structure (DGGE fingerprint pattern) and function (potential enzyme activities) in space and time can arise from functional redundancy of the bacterial communities. If the few taxa dominating in communities are generalists in terms of the functions they perform in a given ecosystem, then shifts in community structure would induce only weak effects on functional characteristics (Östman et al., 2010). This might be particularly relevant in nascent ecosystems, such as the Chicken Creek catchment, where low resource supply could favor bacterial communities composed of a large fraction of generalists that profit from a broad range of scarce resources (Egli, 1995) and, when switching among metabolic pathways, excrete different kinds of enzymes. It is noteworthy in this context that although bacterial diversity was found to affect rates of ecosystem processes (determined as rates of substrate utilization), for example, in the experiment by Langenheder et al. (2010), there was pronounced functional redundancy even at the low bacterial richness levels examined in that study. The functional redundancy hypothesis thus appears to be the most likely explanation for the disconnect between bacterial community structure and function (that is, potential enzyme activities) in early successional environments akin to the Chicken Creek catchment, a conclusion that has recently been drawn also from another microbial system (Burke et al., 2011).
Acknowledgments
We thank Vattenfall Europe Mining AG for providing the research site; A Ling, R Ender, T Wolburg, M Weber, C Kessler, M Seidel, J Westphal, K Westphal and M Stange for field or laboratory assistance; G Lippert, U Abel, R Müller and G Franke for water chemical analyses; F Hammes and colleagues for support for the use of flow cytometry; L Rosinus at the Seminar for Statistics of ETH Zurich for statistical advice; and R Freimann for critically important comments on the manuscript. This study is a contribution to the Transregional Collaborative Research Centre 38 (SFB/TRR 38) funded by the German Research Council (DFG, Bonn) and the Brandenburg Ministry of Science, Research and Culture (MWFK, Potsdam).
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allison SD, Martiny JBH. Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci USA. 2008;105:11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artigas J, Romaní AM, Gaudes A, Muñoz I, Sabater S. Organic matter availability structures microbial biomass and activity in a Mediterranean stream. Freshwat Biol. 2009;54:2025–2036. [Google Scholar]
- Bell T, Newman JA, Silverman BW, Turner SL, Lilley AK. The contribution of species richness and composition to bacterial services. Nature. 2005;436:1157–1160. doi: 10.1038/nature03891. [DOI] [PubMed] [Google Scholar]
- Bell T, Gessner MO, Griffiths RI, McLaren JR, Morin PJ, van der Heijden M, et al. 2009Microbial biodiversity and ecosystem functioning under controlled conditions and in the wildIn: Naeem S, Bunker DE, Hector A, Loreau M, Perrings C (eds).Biodiversity, Ecosystem Functioning, and Human Wellbeing: An Ecological and Economic Perspective Oxford University Press: Oxford; 121–133. [Google Scholar]
- Brankatschk R, Töwe S, Kleineidam K, Schloter M, Zeyer J. Abundances and potential activities of nitrogen cycling microbial communities along a chronosequence of a glacier forefield. ISME J. 2011;5:1025–1037. doi: 10.1038/ismej.2010.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buesing N, Gessner MO. Comparison of detachment procedures for direct counts of bacteria associated with sediment particles plant litter and epiphytic biofilms. Aquat Microb Ecol. 2002;27:29–36. [Google Scholar]
- Burke C, Steinberg P, Rusch D, Kjelleberg S, Thomas T. Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci USA. 2011;108:14288–14293. doi: 10.1073/pnas.1101591108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale BJ, Matulich KL, Hooper DU, Byrnes JE, Duffy E, Gamfeldt L, et al. The functional role of producer diversity in ecosystems. Am J Bot. 2011;98:572–592. doi: 10.3732/ajb.1000364. [DOI] [PubMed] [Google Scholar]
- Casamayor EO, Schäfer H, Bañeras L, Pedrós-Alió C, Muyzer G. Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulfurous lakes: comparison by microscopy and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2000;66:499–508. doi: 10.1128/aem.66.2.499-508.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comte J, del Giorgio PA. Links between resources, C metabolism and the major components of bacterioplankton community structure across a range of freshwater ecosystems. Environ Microbiol. 2009;11:1704–1716. doi: 10.1111/j.1462-2920.2009.01897.x. [DOI] [PubMed] [Google Scholar]
- Davidson EA, Janssens IA. Temperature sensitivity of soil carbon decomposition and feedbacks to climate change. Nature. 2006;440:165–173. doi: 10.1038/nature04514. [DOI] [PubMed] [Google Scholar]
- Docherty KM, Young KC, Maurice PA, Bridgham SD. Dissolved organic matter concentration and quality influences upon structure and function of freshwater microbial communities. Microb Ecol. 2006;52:378–388. doi: 10.1007/s00248-006-9089-x. [DOI] [PubMed] [Google Scholar]
- Egli T. The ecological and physiological significance of the growth of heterotrophic microorganisms with mixtures of substrates. Adv Microb Ecol. 1995;14:305–386. [Google Scholar]
- Findlay S, Sinsabaugh RL. Large-scale variation in subsurface stream biofilms: a cross-regional comparison of metabolic function and community similarity. Microb Ecol. 2006;52:491–500. doi: 10.1007/s00248-006-9095-z. [DOI] [PubMed] [Google Scholar]
- Findlay SEG, Sinsabaugh RL, Sobczak WV, Hoostal M. Metabolic and structural response of hyporheic microbial communities to variations in supply of dissolved organic matter. Limnol Oceanogr. 2003;48:1608–1617. [Google Scholar]
- Flury S, Gessner MO. Experimentally simulated global warming and nitrogen enrichment effects on microbial litter decomposers in a marsh. Appl Environ Microbiol. 2011;77:803–809. doi: 10.1128/AEM.01527-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerull L, Frossard A, Gessner MO, Mutz M. Variability of heterotrophic metabolism in small stream corridors of an early successional watershed. J Geophys Res Biogeosci. 2011;116:G02012. [Google Scholar]
- Gerwin W, Schaaf W, Biemelt D, Fischer A, Winter S, Hüttl RF. The artificial catchment “Chicken Creek” (Lusatia, Germany) - a landscape laboratory for interdisciplinary studies of initial ecosystem development. Ecol Eng. 2009;35:1786–1796. [Google Scholar]
- Gessner MO, Swan CM, Dang CK, McKie BG, Bardgett RD, Wall DH, et al. Diversity meets decomposition. Trends Ecol Evol. 2010;25:372–380. doi: 10.1016/j.tree.2010.01.010. [DOI] [PubMed] [Google Scholar]
- Griffiths BS, Ritz K, Bardgett RD, Cook R, Christensen S, Ekelund F, et al. Ecosystem response of pasture soil communities to fumigation-induced microbial diversity reductions: an examination of the biodiversity-ecosystem function relationship. Oikos. 2000;90:279–294. [Google Scholar]
- Guenet B, Danger M, Abbadie L, Lacroix G. Priming effect: bridging the gap between terrestrial and aquatic ecology. Ecology. 2010;91:2850–2861. doi: 10.1890/09-1968.1. [DOI] [PubMed] [Google Scholar]
- Hammes FA, Egli T. New method for assimilable organic carbon determination using flow-cytometric enumeration and a natural microbial consortium as inoculum. Environ Sci Technol. 2005;39:3289–3294. doi: 10.1021/es048277c. [DOI] [PubMed] [Google Scholar]
- Jones SE, Lock MA. Seasonal determination of extracellular hyrolytic activities in heterotrophic and mixed heterotrophic/autotrophic biofilms from two contrasting rivers. Hydrobiologia. 1993;257:1–16. [Google Scholar]
- Kritzberg ES, Langenheder S, Lindström ES. Influence of dissolved organic matter source on lake bacterioplankton structure and function - implications for seasonal dynamics of community composition. FEMS Microbiol Ecol. 2006;56:406–417. doi: 10.1111/j.1574-6941.2006.00084.x. [DOI] [PubMed] [Google Scholar]
- Langenheder S, Lindström ES, Tranvik LJ. Weak coupling between community composition and functioning of aquatic bacteria. Limnol Oceanogr. 2005;50:957–967. [Google Scholar]
- Langenheder S, Lindström ES, Tranvik LJ. Structure and function of bacterial communities emerging from different sources under identical conditions. Appl Environ Microbiol. 2006;72:212–220. doi: 10.1128/AEM.72.1.212-220.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenheder S, Bulling MT, Solan M, Prosser JI. Bacterial biodiversity-ecosystem functioning relations are modified by environmental complexity. PLoS ONE. 2010;5:1–9. doi: 10.1371/journal.pone.0010834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larned ST, Datry T, Arscott DB, Tockner K. Emerging concepts in temporary-river ecology. Freshwat Biol. 2010;55:717–738. [Google Scholar]
- Lennon JT, Jones SE. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol. 2011;9:119–130. doi: 10.1038/nrmicro2504. [DOI] [PubMed] [Google Scholar]
- Lindström ES. Bacterioplankton community composition in five lakes differing in trophic status and humic content. Microb Ecol. 2000;40:104–113. doi: 10.1007/s002480000036. [DOI] [PubMed] [Google Scholar]
- Milner AM, Fastie CL, Chapin FS, III, Engstrom DR, Sharman LC. Interactions and linkages among ecosystems during landscape evolution. BioScience. 2007;57:237–247. [Google Scholar]
- Östman Ö, Drakare S, Kritzberg ES, Langenheder S, Logue JB, Lindström ES. Regional invariance among microbial communities. Ecol Lett. 2010;13:118–127. doi: 10.1111/j.1461-0248.2009.01413.x. [DOI] [PubMed] [Google Scholar]
- Peter H, Beier S, Bertilsson S, Lindström ES, Langenheder S, Tranvik LJ. Function-specific response to depletion of microbial diversity. ISME J. 2011;5:351–361. doi: 10.1038/ismej.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlon E, Marxsen J, Küsel K. Pioneering bacterial and algal communities and potential extracellular enzyme activities of stream biofilms. FEMS Microbiol Ecol. 2010;71:364–373. doi: 10.1111/j.1574-6941.2009.00817.x. [DOI] [PubMed] [Google Scholar]
- Rier ST, Kuehn KA, Francoeur SN. Algal regulation of extracellular enzyme activity in stream microbial communities associated with inert substrata and detritus. J N Am Benthol Soc. 2007;26:439–449. [Google Scholar]
- Romaní AM, Marxsen J. Extracellular enzymatic activities in epilithic biofilms of the Breitenbach: microhabitat differences. Arch Hydrobiol. 2002;155:541–555. [Google Scholar]
- Romaní AM, Vazquez E, Butturini A. Microbial availability and size fractionation of dissolved organic carbon after drought in an intermittent stream: biogeochemical link across the stream-riparian interface. Microb Ecol. 2006a;52:501–512. doi: 10.1007/s00248-006-9112-2. [DOI] [PubMed] [Google Scholar]
- Romaní AM, Fischer H, Mille-Lindblom C, Tranvik LJ. Interactions of bacteria and fungi on decomposing litter: differential extracellular enzyme activities. Ecology. 2006b;87:2559–2569. doi: 10.1890/0012-9658(2006)87[2559:iobafo]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Rulik M, Spácil R. Extracellular enzyme activity within hyporheic sediments of a small lowland stream. Soil Biol Biochem. 2004;36:1653–1662. [Google Scholar]
- Schneider T, Riedel K. Environmental proteomics: analysis of structure and function of microbial communities. Proteomics. 2010;10:785–798. doi: 10.1002/pmic.200900450. [DOI] [PubMed] [Google Scholar]
- Sinsabaugh RL, Follstad Shah JJ. Integrating resource utilization and temperature in metabolic scaling of riverine bacterial production. Ecology. 2010;91:1455–1465. doi: 10.1890/08-2192.1. [DOI] [PubMed] [Google Scholar]
- Sinsabaugh RL, Antibus RK, Linkins AE. An enzymic approach to the analysis of microbial activity during plant litter decomposition. Agric Ecosys Environ. 1991;34:43–54. [Google Scholar]
- Sinsabaugh RL, Lauber CL, Weintraub MN, Ahmed B, Allison SD, Crenshaw C, et al. Stoichiometry of soil enzyme activity at global scale. Ecol Lett. 2008;11:1252–1264. doi: 10.1111/j.1461-0248.2008.01245.x. [DOI] [PubMed] [Google Scholar]
- Sloan WT, Lunn M, Woodcock S, Head IM, Nee S, Curtis TP. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ Microbiol. 2006;8:732–740. doi: 10.1111/j.1462-2920.2005.00956.x. [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2011R: a language and environment for statistical computing Vienna, Austria; (http://www.R-project.org). [Google Scholar]
- Urban MC, De Meester L. Community monopolization: local adaptation enhances priority effects in an evolving metacommunity. Proc Roy Soc B. 2009;276:4129–4138. doi: 10.1098/rspb.2009.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Gucht K, Cottenie K, Muylaert K, Vloemans N, Cousin S, Declerck S, et al. The power of species sorting: local factors drive bacterial community composition over a wide range of spatial scales. Proc Natl Acad Sci USA. 2007;104:20404–20409. doi: 10.1073/pnas.0707200104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitousek PM. Nutrient Cycling and Limitation: Hawai'i as a Model System. Princeton University Press, Princeton, NJ; 2004. [Google Scholar]
- Wertz S, Degrange V, Prosser JI, Poly F, Commeaux C, Freitag T, et al. Maintenance of soil functioning following erosion of microbial diversity. Environ Microbiol. 2006;8:2162–2169. doi: 10.1111/j.1462-2920.2006.01098.x. [DOI] [PubMed] [Google Scholar]
- Wilczek S, Fischer H, Pusch MT. Regulation and seasonal dynamics of extracellular enzyme activities in the sediments of a large lowland river. Microb Ecol. 2005;50:253–267. doi: 10.1007/s00248-004-0119-2. [DOI] [PubMed] [Google Scholar]
- Winemiller KO, Flecker AS, Hoeinghaus DJ. Patch dynamics and environmental heterogeneity in lotic ecosystems. J N Am Benthol Soc. 2010;29:84–99. [Google Scholar]
- Woodcock S, Curtis TP, Head IM, Lunn M, Sloan WT. Taxa-area relationships for microbes: the unsampled and the unseen. Ecol Lett. 2006;9:805–812. doi: 10.1111/j.1461-0248.2006.00929.x. [DOI] [PubMed] [Google Scholar]
- Ylla I, Borrego C, Romaní AM, Sabater S. Availability of glucose and light modulates the structure and function of a microbial biofilm: research article. FEMS Microbiol Ecol. 2009;69:27–42. doi: 10.1111/j.1574-6941.2009.00689.x. [DOI] [PubMed] [Google Scholar]
- Zeglin LH, Sinsabaugh RL, Barrett JE, Gooseff MN, Takacs-Vesbach CD. Landscape distribution of microbial activity in the McMurdo dry valleys: linked biotic processes, hydrology, and geochemistry in a cold desert ecosystem. Ecosystems. 2009;12:562–573. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.