Abstract
Coastal sands filter and accumulate organic and inorganic materials from the terrestrial and marine environment, and thus provide a high diversity of microbial niches. Sands of temperate climate zones represent a temporally and spatially highly dynamic marine environment characterized by strong physical mixing and seasonal variation. Yet little is known about the temporal fluctuations of resident and rare members of bacterial communities in this environment. By combining community fingerprinting via pyrosequencing of ribosomal genes with the characterization of multiple environmental parameters, we disentangled the effects of seasonality, environmental heterogeneity, sediment depth and biogeochemical gradients on the fluctuations of bacterial communities of marine sands. Surprisingly, only 3–5% of all bacterial types of a given depth zone were present at all times, but 50–80% of them belonged to the most abundant types in the data set. About 60–70% of the bacterial types consisted of tag sequences occurring only once over a period of 1 year. Most members of the rare biosphere did not become abundant at any time or at any sediment depth, but varied significantly with environmental parameters associated with nutritional stress. Despite the large proportion and turnover of rare organisms, the overall community patterns were driven by deterministic relationships associated with seasonal fluctuations in key biogeochemical parameters related to primary productivity. The maintenance of major biogeochemical functions throughout the observation period suggests that the small proportion of resident bacterial types in sands perform the key biogeochemical processes, with minimal effects from the rare fraction of the communities.
Keywords: 454 pyrosequencing, coastal seas, bacterial diversity, multivariate analysis, rare biosphere
Introduction
Marine coasts represent highly dynamic ecosystems where the atmosphere, continents and the ocean interact. Permeable sands constitute the dominant sediment type on continental shelves (Emery, 1968; Boudreau et al., 2001) and have a central role for global carbon and nutrient cycles. They act as biocatalytic filters for various types of materials advected by currents and winds, including dissolved and particulate organic matter derived from living and dead biomass of terrestrial or marine origin (de Beer et al., 2005). Sandy sediments are also constantly subjected to biotic (e.g., bioturbation) and abiotic disturbances (e.g., mixing by currents, seasonal and tidal temperature fluctuation, anoxia; Boudreau et al., 2001). They may also host human pathogens, depending on the human impact by, for example, recreational use of beaches, and temperature anomalies (Ruppert et al., 2004; Dinsdale et al., 2008).
The high diversity of niches provided by the sand habitat supports a rich community of bacteria fluctuating with environmental variations (e.g., Hunter et al., 2006; Sørensen et al., 2007; Mills et al., 2008; Rusch et al., 2009; Gaidos et al., 2011). Yet, the dynamics of environmental parameters in coastal sands could still be a challenge to many microbial populations, selecting for few, but tolerant and well-adapted resident types (Gaidos et al., 2011). Some of these produce extracellular polymeric substances to attach to sand grain surfaces. The resulting biofilm habitat can be considered as an active bioreactor accumulating diverse microbial populations, their extracellular enzymes, nutrients and organic matter from various origins (Flemming and Wingender, 2010). The pore volume between the sand grains represents a place of constant particle exchange due to turbulent flow of currents and pore-water advection, which may induce significant fluxes of particulate organic matter (Stoodley et al., 2005). It remains yet unknown whether sand grain-associated biofilms trap microbes from terrestrial sources and from the water column flushing through the sand, and if they are a source of diversity to the sea around them. As they represent a transition zone between land and ocean characterized by high mixing rates, the presence of many rare bacterial types could be expected. In coarse-grained reef sediments, it has been shown that the typical patterns of rare microbial types found in DNA-based libraries (i.e., the characteristic long tail in rank-abundance curves; Pedrós-Alió, 2007) could not be observed in RNA-based libraries, suggesting that the rare biosphere may be inactive (Gaidos et al., 2011). So far, few studies have examined the structure and ecology of microbial communities in coastal sediments, and these were mostly based on low-resolution community fingerprinting approaches (Urakawa et al., 2000; Bertics and Ziebis, 2009; Böer et al., 2009b). Although those molecular techniques have provided a first description of abundant populations, they generally did not describe rare populations that might represent considerable diversity (Acinas et al., 2004). High-throughput sequencing strategies such as shotgun sequencing (Venter et al., 2004) or pyrosequencing (Sogin et al., 2006) allow a deeper exploration of microbial diversity. Pyrosequencing of ribosomal genes has already permitted the description of the rare microbial biosphere in several types of ecosystems, such as surface and deep-sea water (Sogin et al., 2006), extreme environments (Huber et al., 2007; Brazelton et al., 2010), or the human hand surface (Fierer et al., 2008). Patterns of the rare biosphere in the Arctic Ocean have been related to differences in water masses and attributed to dispersal limitation (Galand et al., 2009). Here we focus on temporal fluctuations of resident and of rare microbial types in coastal sands, and on their link with fluctuations in environmental parameters.
Including the rare microbial biosphere in ecological analyses is challenging when considering the potential for technical noise in pyrosequencing data (Quince et al., 2009; Kunin et al., 2010). Here we apply a new analytical method to assess the effects of rarity on community structure and ecological interpretation (Gobet et al., 2010). The main objective of this study was to explore the putative factors structuring bacterial diversity in temperate coastal sands over six sampling dates within a year (2005–2006), and over different sediment depths (0–5 cm, 5–10 cm, 10–15 cm). For this purpose, we combined several data sets from the Long Term Ecological Research (LTER) Station, Sylt, Germany, including 454 massively parallel tag sequencing (MPTS) targeting the V6 region of the 16S rRNA gene (Gobet et al., 2010) and concurrent environmental surveys (e.g., nutrients, enzymatic activities, bacterial production, weather and water parameters; Böer et al., 2009a, 2009b), to obtain as complete a description as possible of the changes in the bacterial community in their environmental context. We falsified the following hypotheses: (1) bacterial communities in coastal sands have the same composition across sediment depth and in comparison with the overlying water column, (2) as a biofilm community, their temporal fluctuation is low, and (3) the rare microbial biosphere in sands does not respond to environmental changes.
Materials and methods
Study site, sampling procedures and contextual parameters
Detailed sample processing and environmental measurements have been published elsewhere (Böer et al., 2009a). Briefly, sediment samples were repeatedly collected every 2–3 months on a shallow (0.5–2.5 m), subtidal sand flat of the LTER site at the island of Sylt in the North Sea (55° 00′ 47.7″ N, 8° 25′ 59.3″ E) between February 2005 and March 2006. Sandy cores of 15 cm were directly sectioned into three 5 cm layers. Subsamples were stored at −20 °C for DNA extraction or used for the measurement of contextual parameters (extracellular enzymatic activities, nutrients, pigments, carbohydrates, bacterial cell counts; Böer et al., 2009a). Water column data consisting of chlorophyll a, pH and water temperature, were obtained from the Sylt LTER time series (van Beusekom et al., 2009), and data on wind speed was obtained from the German Weather Service (Deutsche Wetterdienst; weather station of List at Sylt).
To compare the communities of sediments, pore water and overlying water, at one time, additional samples were taken at Sylt Königshafen in April 2008 (sediments 55° 2′ 28″ N, 8° 24′ 26″ E; seawater 55° 1′ 41″ N, 8° 26′ 10″ E). Seawater of 1 l was successively filtered through 10 μm and 0.2 μm filters (type GTTP; diameter 47 mm) before DNA extraction. Pore water of the upper 5 cm of the sediment was separated from the sand grains (with bacterial biofilms) by low-speed centrifugation (10 min at 1801 g, at 4 °C) through GF-C filters (47 mm). The resulting ‘dry' sediment (about 8 g) and 2-ml pore water (filtered through 0.2-μm GTTP-type filters; diameter 47 mm) were subsequently used for DNA extraction.
DNA extraction and 454 MPTS
For samples taken in 2008, DNA was extracted using an UltraClean Soil DNA Isolation Kit (MoBio Laboratories Inc., Carlsbad, CA, USA) from (1) ca. 10 g of homogenized wet sediment separated in the dry sand and the pore water, and (2) 1 l of water filtered through 0.2-μm filters (pore water and water column) cut into pieces with a sterilized cutter. DNA was further stored in a final volume of 50–100 μl of Tris-EDTA buffer. To study temporal fluctuations and vertical community patterns in sediments, DNA from samples collected between 2005–2006 were similarly extracted from 1 g of sediment, as described previously (Böer et al., 2009b).
DNA was deposited within the International Census of Marine Microbes project (http://icomm.mbl.edu/), where it was amplified using primers targeting the V6 region of the bacterial 16S rRNA gene, and which included 454 Life Science's A or B sequencing adapter according to Sogin et al. (2006). Pyrosequencing was performed on a Genome Sequencer 20 system (Roche, Basel, Switzerland) at 454 Life Sciences (Branford, CT, USA) by primer extension (Margulies et al., 2005), yielding on average 60-bp long fragments after removing primer sequences.
Sequence analysis
Data from the 454 MPTS were retrieved from the publicly available ‘Visualization and Analysis of Microbial Populations Structure (VAMPS)' website (http://vamps.mbl.edu/; projects AB_SAND_Bv6 and ICM_FIS_Bv6) in the framework of the International Census of Marine Microbes project. Sequences were taxonomically assigned by an automatic annotation pipeline (Sogin et al., 2006), using several known databases (Entrez Genome, RDP, SILVA), allowing a taxonomic annotation of up to 94% of the data set. In our study, analyses were based on a definition of operational taxonomic units (OTU) as unique (i.e. two sequences belong to two different OTUunique, when they differ by at least one base) to keep a consistent definition throughout. Other classifications used in the study are indicated as follows: OTUall represents all, un-annotated sequences, PyroNoise0% and PyroNoise3% correspond to the PyroNoise-corrected data sets at 0% and 3% sequence identity, respectively (Quince et al., 2009), OTUall—x% rare, which represent the OTUall data set truncated of its x% rarer OTUunique according to Gobet et al. (2010).
Taxa-environment relationships
To investigate taxa-environment relationships, most of the parameters (except pH, water temperature, wind speed and salinity) were log10-transformed, whereas the community matrices (OTUall data set, and the resident single-sequence OTU relative (SSOrel) data sets or the (potential) pathogen sequence abundance matrix (including the genera Parachlamydia, Arcobacter, Francisella, Acinetobacter, Rickettsiella, Pseudomonas and Ralstonia)) were standardized by Hellinger transformation (Legendre and Gallagher, 2001). A forward selection (based on a canonical redundancy analysis algorithm and 999 Monte Carlo permutation tests) of the environmental factors was done to find the set of parameters that could best explain the variation in the community table. The best-fitting models were chosen using the Akaike Information Criterion. Canonical variation partitioning (Borcard et al., 1992; Ramette and Tiedje, 2007b) was then applied on the community data to disentangle the specific effects of environmental variables (pigments, nutrients, extracellular enzymatic activities, cell abundance) selected previously and of their covariation on microbial community structure. Statistical analyses were carried out using the R statistical environment (R version 2.10.0, R Development Core Team, 2009), using the vegan (Oksanen et al., 2009), gplots (Bolker et al., 2009) packages, and custom R scripts.
Results and discussion
The bacterial community composition of coastal sands
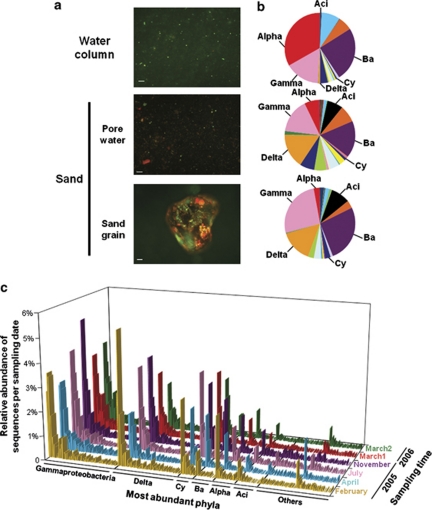
The three compartments, sand, pore water and the overlying water column, may be expected to share a large proportion of the same microbial assemblages, because the pore space of permeable sands is constantly flushed by the overlying water, trapping detritus and living cells from the water column (Boudreau et al., 2001). We compared bacterial communities from one large, well-mixed sample of each compartment (sand, pore water and water column) at one time, to investigate the similarity in community composition between those coastal compartments, which are constantly mixed by wind and waves. However, microscopic observations (Figure 1a) confirm that sand grains are covered by a biofilm consisting of cells embedded in extracellular polymeric substances, and that there is potentially little exchange with cells flowing through the sediment (Rusch et al., 2001). At any time, the pore water contains less than 0.2% of the cell abundance associated with the sand grains (this study and Rusch et al., 2003). Accordingly, at that specific sampling time, we also found a rather low proportion of shared bacterial populations between the sand and the water column, at the level of both community composition and evenness (Figures 1b and c, Supplementary Figure S1). Only 2–3% of all unique OTUunique (sequences from the original OTUall data set that have at least one nucleotide difference) were shared between the three compartments, confirming previous findings (Epstein et al., 1997; Llobet-Brossa et al., 1998).
Figure 1.
Microbial community distribution in the sand and the water column. (a) From top to bottom: acridine orange staining of bacteria in the water column, the pore water and on the surface of a sand grain (scale bar=50 μm). (b) Relative number of sequences in different compartments for the top 5-cm sand layer and in the overlying water column in April 2008. Here, the phylum level was chosen for illustrative purposes. (c) Sequence distribution in the sand over time, in which each bar represents an OTUunique (only OTUunique occurring more than 100 times in the whole data set are shown). The Proteobacteria phylum was further split into its corresponding classes, for example, Alpha, Gamma, Delta; Cy, Cyanobacteria; Ba, Bacteroidetes; Aci, Acidobacteria; Others: Actinobacteria, NA, not annotated Proteobacteria, Planctomycetes, Chloroflexi, Verrucomicrobia, WS3, Firmicutes, Lentisphaerae, Deferribacteres and Gemmatimonadetes.
The Sylt (North Sea) water column was mainly dominated by Bacteroidetes and by the Alpha- and Gamma-subdivisions of the Proteobacteria, confirming previous results on the basis of fluorescence in situ hybridization and 16S rRNA gene-based clone libraries (Glöckner et al., 1999; Eilers et al., 2000; Zubkov et al., 2002). Sequences of the phyla Verrucomicrobia and Actinobacteria were also abundant (Figure 1b). The top 5 cm of Sylt sands were dominated by Bacteroidetes, Gammaproteobacteria, Deltaproteobacteria and Planctomycetes, also in accord with previous clone libraries obtained from coastal sediments of the Wadden Sea (Llobet-Brossa et al., 1998; Musat et al., 2006), or other permeable sediments from shelf sands from South Atlantic Bight and coastal sediment in Hawaii (Hunter et al., 2006; Sørensen et al., 2007). In contrast, tropical coral reef sands were less consistent, with a dominance of Proteobacteria, Actinobacteria (Gaidos et al., 2011), or Alphaproteobacteria and Spirochaetes (Schöttner et al., 2011) in the community. Acidobacteria sequences were abundant in temperate (Figure 1b, Supplementary Figure S2) and tropical sands (Gaidos et al., 2011; Schöttner et al., 2011). At the phylum level, the pore water microbial composition resembled the sand community with a dominance of Bacteroidetes, Gammaproteobacteria, Deltaproteobacteria and Acidobacteria (Figure 1b). At the OTUunique level, substantial differences were detected between the pore water and the sand-associated bacterial community or the water column (Supplementary Figure S1C). The further analysis of temporal fluctuations was restricted to wet sands containing both the biofilm community, as well as the pore water types. Certainly, temporal variation and the role of the environment in the exchange of microbial communities between land and sea, intertidal and subtidal flats with their different physical compartments deserve further studies.
The characterization of coastal sand communities over six sampling dates (Figure 1) provided a total of 197 684 rRNA gene sequences, corresponding to 27 630 OTUunique. Per sample, sequences ranged from about 5000–19 000, corresponding to 496–2993 OTU3% (i.e., OTU defined at 3% sequence difference after denoising the V6 sequences by PyroNoise; Supplementary Table S1). Noticeably, the high bacterial richness estimates for coastal sands were comparable to 454 MPTS-based estimates reported for coarse carbonate sands from coral reefs (Gaidos et al., 2011), soils (Roesch et al., 2007), marine sediments and crusts (Sogin et al., 2006; Huber et al., 2007; Brazelton et al., 2010). A high, yet unexplored diversity in these habitats was already previously proposed based on clone libraries (Hunter et al., 2006; Mills et al., 2008; Rusch et al., 2009). In addition, here we found that total sequence and OTU numbers systematically increased with depth from 0–15 cm at most of the sampling dates (Supplementary Table S1). A positive relationship between sediment depth and bacterial diversity was previously observed using different fingerprinting techniques (Urakawa et al., 2000; Böer et al., 2009b), and explained by increasing physico-chemical stability of the habitat. The Delta- and Beta-subdivisions of the Proteobacteria, Deferribacteres, Spirochaetes and Nitrospira, all showed an increasing OTU richness with depth. In contrast, Cyanobacteria and Bacteroidetes decreased with depth (Supplementary Figure S2).
Turnover of bacterial diversity with sediment depth and time
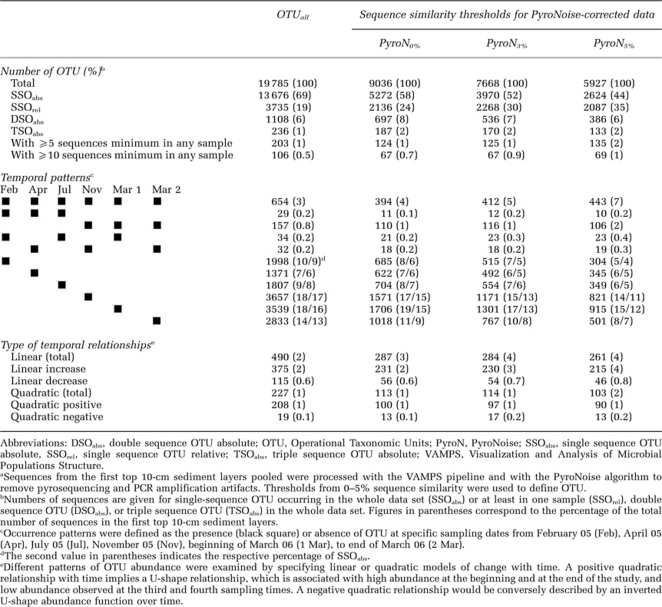
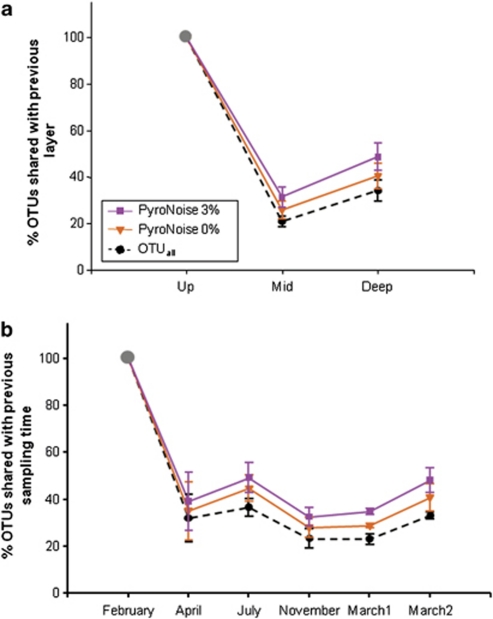
The phylum to class levels are commonly used to describe the diversity of bacterial communities on the basis of whole-cell fluorescence in situ hybridization or 16S-based clone libraries (Llobet-Brossa et al., 1998; Musat et al., 2006). At those levels, microbial community patterns in sands mostly remained unchanged over time and sediment depth (Supplementary Figure S2). However, drastic changes in bacterial community composition were evidenced when finer taxonomic resolutions were used; overall, only 152 OTUunique (0.55% of the total number of 27 630 OTUunique) were present in all samples at all times, and their sequence abundance ranged from 54–6550. Also, only 3–5% of all OTUunique within a sampling depth layer were present at all times (Table 1). The majority (77%) of these OTUunique, which we define as resident OTU, consisted of the phyla Acidobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria, Alpha-, Delta- and Gammaproteobacteria and Verrucomicrobia. For the OTUall data set (i.e., data set with all OTUunique), a low percentage of OTUunique were shared between sediment layers or any two sampling dates. Only about 20% of sequences were shared between the deeper layers and the upper layer (Supplementary Figure S3A), the communities of the two deeper layers were slightly more similar (Figure 2a). Moreover, resident OTUs seemed to be characterized by a high and relatively stable number of sequences (Figure 3b). About 70% of the resident OTUs had abundances of more than 10 sequences per sample (Table 1, Figure 3b), suggesting that they belonged to archetypal sand microbial communities.
Table 1. Temporal patterns of OTU variationa.
Figure 2.
Bacterial community turnover between two consecutive depth layers (a) or sampling times (b). The percentage of OTU shared between a sampling depth (or date) and the previous one was calculated after PyroNoise correction and OTU clustering of the 454 MPTS data set at several levels of sequence dissimilarity. Bars correspond to s.d. calculated over 4–6 sampling dates (a) or three depth layers (b), except for July and November 2005 in which two depth layers were considered. The first depth layer and sampling date (February 2005) are indicated by the grey point as 100% of OTU. OTUall represents the original data set with all OTUunique. It is used here as a reference data set to study the effects of various correction levels on the observed dynamics of the bacterial community composition.
Figure 3.
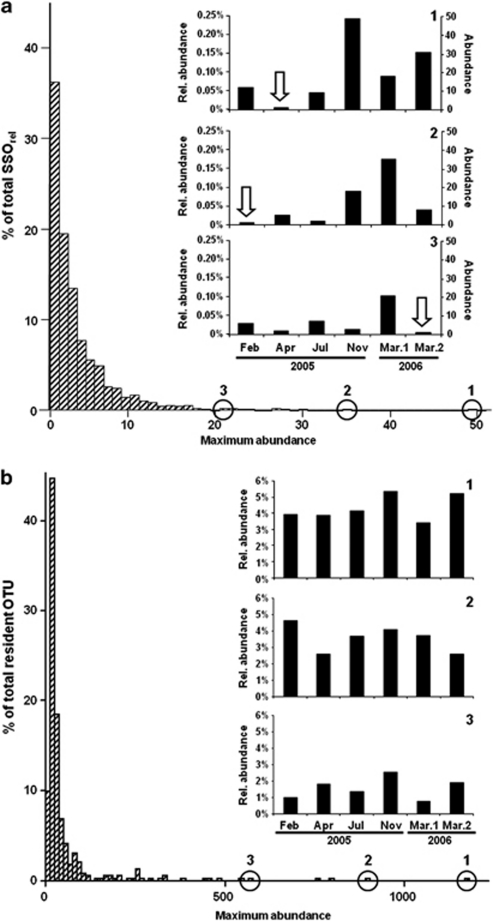
Distribution of the maximum abundance of (a) SSOrel (i.e., those OTU0% that, at least in one sample, consisted of only one sequence) and (b) resident OTU0% (i.e., OTU0% present at all times) in the top 10-cm layer. In (a), panels 1, 2 and 3 are examples of cases, in which particular high fluctuations from the single-sequence case (white arrow) to higher-sequence abundances were observed. In (b), panels 1, 2, 3 illustrate cases of particularly high fluctuations in relative abundances (no absolute abundances were calculated in this case). All data were initially processed to remove pyrosequencing noise. Rel. abundance, relative abundance to the total number of sequences per sampling time: February 2005 (13 285), April 2005 (7902), July 2005 (10 910), November 2005 (21 931), 1 March 2006 (17 302), 2 March 2006 (17 897).
When time was considered alone, only 18–37% of OTUunique were found to be shared between any two sampling times (Figure 2b, Supplementary Figure S3A). This indicates that a very large fraction of the community may be constantly replaced. Yet, the fact that the turnover rate did not increase with sampling time suggests that some populations vanished and reappeared during the investigated time period (Supplementary Figure S3). Some of the most sequence-abundant groups were found to be positively correlated with time (Figure 4); for example, Gammaproteobacteria and Planctomycetes, which followed the seasonal fluctuations in cell abundances observed in Sylt sediment (Musat et al., 2006). Remarkably, the fluctuation within a month (beginning and end of March 2006) was almost as high as within a year. This could be explained by the observed large environmental variations that occurred in March 2006, starting with cold temperatures and high nutrient levels at the end of the winter period, and ending with spring blooms and stormy conditions (Böer et al., 2009b).
Figure 4.
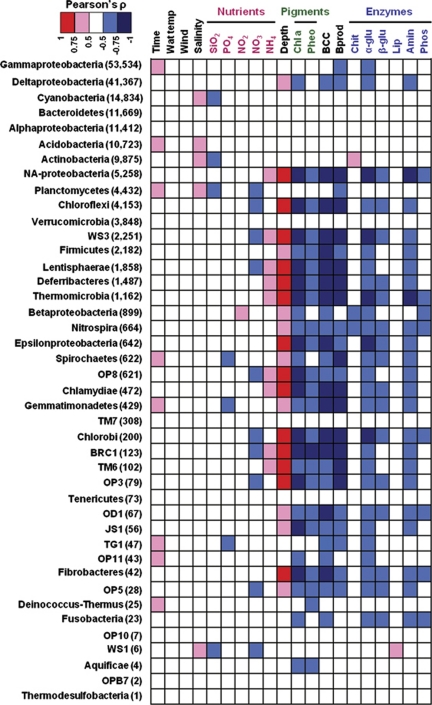
Environmental factors associated with variations of the bacterial community structure at the phylum level. Pearson's ρ indicates correlations between phylum sequence abundance and several environmental parameters. For example, a red square between sediment depth and Chloroflexi indicates a higher number of sequences with increasing depth; nonsignificant relationships between water temperature and sequence variation in any of the bacterial groups are indicated by white squares; a blue square between chlorophyll a and Chloroflexi denotes a decrease in sequences as chlorophyll a concentration gets higher (the latter being concordant with the relationship between increasing depth and Chloroflexi sequences number). The Proteobacteria phylum level was separated into its corresponding classes to obtain a higher resolution. NA-Proteobacteria are the Proteobacteria with missing class annotation. The total number of sequences in each phylum is indicated in parentheses. SiO2, silicate; PO4, phosphate; NO2, nitrite; NO3, nitrate; NH4, ammonium; Chl a, chlorophyll a; Pheo, pheophytin; BCC, bacterial abundance; Bprod, bacterial carbon production; Chit, chitinase; α-glu, α-glucosidase; β-glu, β-glucosidase; Lip, lipase; Amin, aminopeptidase; Phos, phosphatase.
An interesting aspect is the abundance and role of the resident, potentially sand-biofilm associated populations versus rare and transient populations getting advected through the sands. Indeed, in our data set, more than 50% of the OTUunique present at all times in the three depth layers were members of the Gammaproteobacteria and Deltaproteobacteria, two classes representing up to 23% and up to 10%, respectively, of the total cell counts in such sands (Musat et al., 2006). But also, 40% of all OTUunique appearing only once in a given sample or in the whole OTUall data set belonged to these classes. Noticeably, the rare members of dominant classes never became abundant at any sampling time, similarly to observations made in Arctic waters (Galand et al., 2009).
Observing a high turnover for a substantial fraction of the bacterial community in marine sands raises several questions about the ecological significance of these dynamics. Large turnover in community composition could be due to various phenomena, which are not mutually exclusive, such as: (1) migration of non-resident bacterial populations into and out of an ecosystem (Sloan et al., 2006). This could be due to the rapid physical transport processes prevailing in marine coastal sand ecosystems. (2) High adaptability of microbial communities to ever-changing and complex environments, as previously observed in the laboratory (Rosenzweig et al., 1994). (3) Emergence of latent ‘rare' prokaryotic stages that may become dominant when appropriate conditions are met (Pedrós-Alió, 2006). This fluctuation from rare to dominant types may be supported by the ‘seed bank' hypothesis (Finlay, 2002). (4) Finally, the presence of extracellular DNA could also have strongly impacted our estimates of community turnover. This could be possible because, first, sands are known to act as natural filters that concentrate particles and DNA in suspension (Naviaux et al., 2005). Second, our molecular approaches did not differentiate between DNA from living cells and extracellular DNA settling from seawater (Corinaldesi et al., 2005). However, the OTUunique dissimilarity between water column and sand described above may indicate that the described bacterial community is typical of the sand ecosystem, and that a large effect of extracellular DNA on our results is not likely. To further test hypotheses 1–3, we investigated the contribution of the rare biosphere to the overall community turnover and determined how the rare biosphere depends on environmental conditions.
Impact of the rare biosphere on community turnover
The rare biosphere has been postulated to consist of low-abundance microbial organisms that would not be subjected to predation or viral lysis, and would likely represent a huge proportion of microbial communities, as generally indicated by long distribution tails in rank-abundance curves (Pedrós-Alió, 2007). For example, among the rare bacteria in coastal sands were the potential human pathogens (including Parachlamydia, Arcobacter, Francisella, Acinetobacter, Rickettsiella, Pseudomonas and Ralstonia) with a total of 16–88 sequences corresponding to 2–54 OTUunique in the OTUall data set (0.22% of all sequences in OTUall). To understand what fraction of the community may be associated with the large diversity turnover observed in the sands, we gradually removed increasing fractions of the rare sequences in the data set starting from the rarest ones (Gobet et al., 2010); the large turnover previously described for the complete data set over both sediment depth and time was no longer observed when up to 50% of the rare sequences were removed (Supplementary Figure S3C), indicating that most of the community turnover was due to changes in the rare tail of the data set.
The rare biosphere could be identified as OTU appearing only once in a given sample (i.e., SSOrel, representing about 20% of all PyroNoise-corrected OTU0%), or as OTU appearing only once in the whole data set (i.e., SSO absolute (SSOabs), representing about 58% of the OTU0%), as compared with the 3–5% of the OTU0% that were resident (Table 1). Moreover, when only the SSOrel fraction of the data set was retained, very similar fractions of the total explained-biological variation were identified as for the total data set (Supplementary Figure S4). This study therefore reports that not only a large fraction of bacterial communities consists of rare types, as previously found in many 454 MPTS-based studies in other environmental samples (e.g., Gilbert et al., 2009; Bolhuis and Stal, 2011), which may undergo substantial replacement over few centimeters of sediment depth, as previously observed using lower resolution molecular techniques (Hewson et al., 2007), but may also dramatically change over few months of time. In addition, rare bacterial types seemed to remain rare in the bacterial community over time and sediment depth; 94% of the SSOrel had a maximum abundance of 10 sequences only (Figure 3a). In conclusion, both the presence of this large proportion of singleton OTU0% in sandy sediments and the large turnover in community composition could be explained by the dispersal of OTU from other sand locations by advective transport and physical mixing (Boudreau et al., 2001). Further research would be needed to examine more in detail the biogeography of rare and of resident bacterial types in coastal sands.
Ecological interpretation of overall microbial diversity patterns
To determine whether observed temporal patterns may be attributed to either deterministic (niche-based) or stochastic processes, or both (Ramette and Tiedje, 2007a), bacterial community structure was interpreted in concert with previously published contextual data (Böer et al., 2009a) for the investigated samples, to test the effects of time, sediment depth, cell abundance and biogeochemical gradients (i.e., pigments, nutrients and extracellular enzymes, Figure 4). A multivariate variation partitioning approach (Borcard et al., 1992; Ramette and Tiedje, 2007b) showed that biogeochemical gradients (pigments, nutrients and extracellular enzymes), cell abundance and their covariation were directly related to the major changes in community structure (Supplementary Figure S4), as previously assessed on the basis of a lower-resolution fingerprinting technique for the most abundant OTUs (Böer et al., 2009b). This study, including also the rare types, showed that significant biogeochemical variables included in the most parsimonious multivariate models were qualitatively almost the same for the resident OTU, the phylum, OTUall and SSOrel levels (Supplementary Figure S4 and Supplementary Table S2). Also, a greater amount of biological variation could be explained when a lower taxonomic resolution was used, leading to a less complex data set (Gobet et al., 2010).
The main parameters significantly influencing the variation for several OTU definition levels were chlorophyll a, activity of the extracellular enzyme phosphatase and cell abundance (Supplementary Table S2), with all three parameters being highly correlated with sediment depth. Cell abundance was also identified as an important factor influencing the variation in the different OTU data sets (Supplementary Table S2). Noticeably, community diversity and composition varied much more than microbial functions (e.g., biomass, benthic oxygen consumption and extracellular enzymatic activities). This may suggest that the few continuously abundant, resident microbes perform these main microbial functions in sandy ecosystems, and that the rare types being replaced at high rates have little effects on bulk functions. Furthermore, with regard to heterotrophic degradation of organic matter, a substantial level of functional redundancy may exist in the dominant taxa of the Sylt sands, comprising both resident and transient organisms.
Our statistical analyses showed that the temporal dynamics of bacterial communities in temperate sandy sediments were clearly distinct from the seasonally reoccurring, cyclic bacterial patterns that were observed in water column samples offshore Southern California (Fuhrman et al., 2006), in the English channel (Gilbert et al., 2009), or in the Baltic Sea (Andersson et al., 2010). Those patterns also differed from the observed stability of the Arctic bacterioplankton community through seasons (Kirchman et al., 2010). This indicates that contrasting patterns of temporal variation may be associated with different microbial habitats and ocean regions, thus inviting further studies on the main drivers of temporal fluctuation or stability in marine ecosystems.
The rare biosphere responds to environmental drivers
The biogeography (Galand et al., 2009) and the temporal dynamics over thousands of years (Brazelton et al., 2010) of the rare microbial biosphere have previously been described in different marine habitats, but not much is known about factors influencing the composition of the rare microbial biosphere. To better understand whether patterns of the rare fraction were random or environmentally driven, the proportion of SSOrel (Supplementary Table S3) in each sample was correlated with environmental parameters. Notably, fluctuations in SSOrel were not random (as determined by model selection using Monte Carlo permutation test and by selecting the lowest Akaike Information Criterion model values) and seemed to be negatively correlated with pigments (chlorophyll a, pheopigments) and bacterial carbon production in the sediments (Supplementary Figure S5). For OTU0%, the corresponding Pearson correlation coefficients between the proportions of SSOrel and chlorophyll a, pheopigments and bacterial carbon production were −0.730, −0.695 and −0.626, respectively, and were all highly significant. However, neither sediment depth nor sampling time could significantly explain the variation in the proportion of rare types. Because pigment concentration and bacterial carbon production may indicate food availability in sand ecosystems (Rusch et al., 2003), an increased proportion of rare types that is concomitant with a decrease of those parameters would indicate that rarity becomes more prevalent when the environmental conditions become harsher for microbial life. Those hypotheses are concordant with the fact that primary and secondary productions in Sylt sands reach a minimum in late winter, when temperatures are close to the freezing point and strong winds dominate (van Beusekom, personal communication). Therefore, our previous hypotheses 2 (microbial adaptability) and 3 (seed bank hypothesis) to explain the observed high turnover in sandy sediments may be both supported by this finding of significant environmental control on the fluctuations of rare microbial types. Future structural and functional community analyses are needed to examine whether the large proportion of single OTU can be linked to specific environmental conditions in other marine coastal sandy habitats and hence typical for this microbial realm dominated by physical transport.
Acknowledgments
We acknowledge Martina Alisch, Marianne Jacob, Susanne Menger and Shalin Seebah for great assistance with Sylt sampling and help with sample processing. We thank Rafael Stiens for cell microscopy. This work was supported by the Marie Curie Early Stage Training fellowship in Marine Microbiology (MarMic EST contract MEST-CT-2004-007776 to AG) and by the International Max Planck Research School of Marine Microbiology (AG), as well as by the Max Planck Society (AR, AB), Helmholtz Association (AB, JB) and Leibniz Program of the DFG (AB). CQ is supported by an Engineering and Physical Sciences Research Council Career Acceleration Fellowship (EP/H003851/1). The sequencing and bioinformatic infrastructure (VAMPS) were supported by grants from the Alfred P Sloan and the William M Keck Foundations to MLS. This is a contribution to the International Census of Marine Microbes (ICoMM).
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL, et al. Fine-scale phylogenetic architecture of a complex bacterial community. Nature. 2004;430:551–554. doi: 10.1038/nature02649. [DOI] [PubMed] [Google Scholar]
- Andersson AF, Riemann L, Bertilsson S. Pyrosequencing reveals contrasting seasonal dynamics of taxa within Baltic Sea bacterioplankton communities. ISME J. 2010;4:171–181. doi: 10.1038/ismej.2009.108. [DOI] [PubMed] [Google Scholar]
- Bertics VJ, Ziebis W. Biodiversity of benthic microbial communities in bioturbated coastal sediments is controlled by geochemical microniches. ISME J. 2009;3:1269–1285. doi: 10.1038/ismej.2009.62. [DOI] [PubMed] [Google Scholar]
- Bolhuis H, Stal LJ.2011Analysis of bacterial and archaeal diversity in coastal microbial mats using massive parallel 16S rRNA gene tag sequencing ISME Jdoi: 10.1038/ismej.2011.52 [DOI] [PMC free article] [PubMed]
- Böer S, Arnosti C, van Beusekom J, Boetius A. Temporal variations in microbial activities and carbon turnover in subtidal sandy sediments. Biogeosciences. 2009a;6 (7:1149–1165. [Google Scholar]
- Böer SI, Hedtkamp SIC, van Beusekom JEE, Fuhrman JA, Boetius A, Ramette A. Time- and sediment depth-related variations in bacterial diversity and community structure in subtidal sands. ISME J. 2009b;3:780–791. doi: 10.1038/ismej.2009.29. [DOI] [PubMed] [Google Scholar]
- Bolker B, Bonebakker L, Gentleman R, Liaw WHA, Lumley T, Maechler M, et al. 2009. gplots: Various R programming tools for plotting data. R package version 2.7.4.
- Borcard D, Legendre P, Drapeau P. Partialling out the spatial component of ecological variation. Ecology. 1992;73:1045–1055. [Google Scholar]
- Boudreau BP, Huettel M, Forster S, Jahnke RA, McLachlan A, Middelburg JJ, et al. Permeable marine sediments: overturning an old paradigm. EOS. 2001;82:133–136. [Google Scholar]
- Brazelton WJ, Ludwig KA, Sogin ML, Andreishcheva EN, Kelley DS, Shen CC, et al. Archaea and bacteria with surprising microdiversity show shifts in dominance over 1000-year time scales in hydrothermal chimneys. Proc Nat Acad Sci USA. 2010;107:1612–1617. doi: 10.1073/pnas.0905369107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer D, Wenzhofer F, Ferdelman TG, Boehme SE, Huettel M, van Beusekom JEE, et al. Transport and mineralization rates in North Sea sandy intertidal sediments, Sylt-Romo Basin, Wadden Sea. Limnol Oceanogr. 2005;50:113–127. [Google Scholar]
- Corinaldesi C, Danovaro R, Dell'Anno A. Simultaneous recovery of extracellular and intracellular DNA suitable for molecular studies from marine sediments. Appl Environ Microbiol. 2005;71:46–50. doi: 10.1128/AEM.71.1.46-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsdale EA, Pantos O, Smriga S, Edwards RA, Angly F, Wegley L, et al. Microbial ecology of four coral atolls in the Northern Line Islands. PloS ONE. 2008;3:e1584. doi: 10.1371/journal.pone.0001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers H, Pernthaler J, Glockner FO, Amann R. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl Environ Microbiol. 2000;66:3044–3051. doi: 10.1128/aem.66.7.3044-3051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery K. Relict sediments on continental shelves of the world. Am Assoc Pet Geol Bull. 1968;52:445–464. [Google Scholar]
- Epstein SS, Alexander D, Cosman K, Dompe A, Gallagher S, Jarsobski J, et al. Enumeration of sandy sediment bacteria: Are the counts quantitative or relative. Mar Ecol Prog Ser. 1997;151:11–16. [Google Scholar]
- Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Nat Acad Sci USA. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- Flemming H-C, Wingender J. The biofilm matrix. Nat Rev Micro. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S. Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Nat Acad Sci USA. 2006;103:13104–13109. doi: 10.1073/pnas.0602399103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidos E, Rusch A, Ilardo M. Ribosomal tag pyrosequencing of DNA and RNA from benthic coral reef microbiota: community spatial structure, rare members and nitrogen-cycling guilds. Environ Microbiol. 2011;13:1138–1152. doi: 10.1111/j.1462-2920.2010.02392.x. [DOI] [PubMed] [Google Scholar]
- Galand PE, Casamayor EO, Kirchman DL, Lovejoy C. Ecology of the rare microbial biosphere of the Arctic Ocean. Proc Nat Acad Sci USA. 2009;106:22427–22432. doi: 10.1073/pnas.0908284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Field D, Swift P, Newbold L, Oliver A, Smyth T, et al. The seasonal structure of microbial communities in the Western English Channel. Environ Microbiol. 2009;11:3132–3139. doi: 10.1111/j.1462-2920.2009.02017.x. [DOI] [PubMed] [Google Scholar]
- Glöckner FO, Fuchs BM, Amann R. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobet A, Quince C, Ramette A. Multivariate Cutoff Level Analysis (MultiCoLA) of large community data sets. Nucleic Acids Res. 2010;38:e155. doi: 10.1093/nar/gkq545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewson I, Jacobson/Meyers ME, Fuhrman JA. Diversity and biogeography of bacterial assemblages in surface sediments across the San Pedro Basin, Southern California Borderlands. Environ Microbiol. 2007;9:923–933. doi: 10.1111/j.1462-2920.2006.01214.x. [DOI] [PubMed] [Google Scholar]
- Huber JA, Mark Welch D, Morrison HG, Huse SM, Neal PR, Butterfield DA, et al. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- Hunter EM, Mills HJ, Kostka JE. Microbial community diversity associated with carbon and nitrogen cycling in permeable shelf sediments. Appl Environ Microbiol. 2006;72:5689–5701. doi: 10.1128/AEM.03007-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman DL, Cottrell MT, Lovejoy C. The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ Microbiol. 2010;12:1132–1143. doi: 10.1111/j.1462-2920.2010.02154.x. [DOI] [PubMed] [Google Scholar]
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2010;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- Legendre P, Gallagher ED. Ecologically meaningful transformations for ordination of species data. Oecologia. 2001;129:271–280. doi: 10.1007/s004420100716. [DOI] [PubMed] [Google Scholar]
- Llobet-Brossa E, Rossello-Mora R, Amann R. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl Environ Microbiol. 1998;64:2691–2696. doi: 10.1128/aem.64.7.2691-2696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills HJ, Hunter E, Humphrys M, Kerkhof L, McGuinness L, Huettel M, et al. Characterization of nitrifying, denitrifying, and overall bacterial communities in permeable marine sediments of the northeastern Gulf of Mexico. Appl Environ Microbiol. 2008;74:4440–4453. doi: 10.1128/AEM.02692-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musat N, Werner U, Knittel K, Kolb S, Dodenhof T, van Beusekom JEE, et al. Microbial community structure of sandy intertidal sediments in the North Sea, Sylt-Romo Basin, Wadden Sea. Syst Appl Microbiol. 2006;29:333–348. doi: 10.1016/j.syapm.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Naviaux RK, Good B, McPherson JD, Steffen DL, Markusic D, Ransom B, et al. Sand DNA—a genetic library of life at the water's edge. Mar Ecol Prog Ser. 2005;301:9–22. [Google Scholar]
- Oksanen J, Kindt R, Legendre P, O'Hara B, Simpson GL, Solymos P, et al. 2009. vegan: Community Ecology Package. R package version 1.15-2.
- Pedrós-Alió C. Marine microbial diversity: can it be determined. Trends Microbiol. 2006;14:257–263. doi: 10.1016/j.tim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Pedrós-Alió C. Dipping into the rare biosphere. Science. 2007;315:192–193. doi: 10.1126/science.1135933. [DOI] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria; 2009. [Google Scholar]
- Ramette A, Tiedje JM. Biogeography: an emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microb Ecol. 2007a;53:197–207. doi: 10.1007/s00248-005-5010-2. [DOI] [PubMed] [Google Scholar]
- Ramette A, Tiedje JM. Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem. Proc Nat Acad Sci USA. 2007b;104:2761–2766. doi: 10.1073/pnas.0610671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig RF, Sharp RR, Treves DS, Adams J. Microbial evolution in a simple unstructured environment—genetic differentiation in Escherichia coli. Genetics. 1994;137:903–917. doi: 10.1093/genetics/137.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert J, Panzig B, Guertler L, Hinz P, Schwesinger G, Felix SB, et al. Two cases of severe sepsis due to Vibrio vulnificus wound infection acquired in the Baltic Sea. Eur J Clin Microbiol Infect Dis. 2004;23:912–915. doi: 10.1007/s10096-004-1241-2. [DOI] [PubMed] [Google Scholar]
- Rusch A, Forster S, Huettel M. Bacteria, diatoms and detritus in an intertidal sandflat subject to advective transport across the water-sediment interface. Biogeochemistry. 2001;55:1–27. [Google Scholar]
- Rusch A, Hannides A, Gaidos E. Diverse communities of active Bacteria and Archaea along oxygen gradients in coral reef sediments. Coral Reefs. 2009;28:15–26. [Google Scholar]
- Rusch A, Huettel M, Reimers CE, Taghon GL, Fuller CM. Activity and distribution of bacterial populations in Middle Atlantic Bight shelf sands. FEMS Microbiol Ecol. 2003;44:89–100. doi: 10.1111/j.1574-6941.2003.tb01093.x. [DOI] [PubMed] [Google Scholar]
- Schöttner S, Pfitzner B, Grünke S, Rasheed M, Wild C, Ramette A. Drivers of bacterial diversity dynamics in permeable carbonate and silicate coral reef sands from the Red Sea. Environ Microbiol. 2011;13:1815–1826. doi: 10.1111/j.1462-2920.2011.02494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan WT, Lunn M, Woodcock S, Head IM, Nee S, Curtis TP. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ Microbiol. 2006;8:732–740. doi: 10.1111/j.1462-2920.2005.00956.x. [DOI] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, et al. Microbial diversity in the deep sea and the underexplored ‘rare biosphere'. Proc Nat Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen KB, Glazer B, Hannides A, Gaidos E. Spatial structure of the microbial community in sandy carbonate sediment. Mar Ecol Prog Ser. 2007;346:61–74. [Google Scholar]
- Stoodley P, Dodds I, De Beer D, Scott HL, Boyle JD. Flowing biofilms as a transport mechanism for biomass through porous media under laminar and turbulent conditions in a laboratory reactor system. Biofouling. 2005;21:161–168. doi: 10.1080/08927010500375524. [DOI] [PubMed] [Google Scholar]
- Urakawa H, Yoshida T, Nishimura M, Ohwada K. Characterization of depth-related population variation in microbial communities of a coastal marine sediment using 16S rDNA-based approaches and quinone profiling. Environ Microbiol. 2000;2:542–554. doi: 10.1046/j.1462-2920.2000.00137.x. [DOI] [PubMed] [Google Scholar]
- van Beusekom JEE, Loebl M, Martens P. Distant riverine nutrient supply and local temperature drive the long-term phytoplankton development in a temperate coastal basin. J Sea Res. 2009;61:26–33. [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Zubkov MV, Fuchs BM, Tarran GA, Burkill PH, Amann R. Mesoscale distribution of dominant bacterioplankton groups in the northern North Sea in early summer. Aquatic Microb Ecol. 2002;29:135–144. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.