Abstract
Few studies of microbial biogeography address variability across both multiple habitats and multiple seasons. Here we examine the spatial and temporal variability of bacterioplankton community composition of the Columbia River coastal margin using 16S amplicon pyrosequencing of 300 water samples collected in 2007 and 2008. Communities separated into seven groups (ANOSIM, P<0.001): river, estuary, plume, epipelagic, mesopelagic, shelf bottom (depth<350 m) and slope bottom (depth>850 m). The ordination of these samples was correlated with salinity (ρ=−0.83) and depth (ρ=−0.62). Temporal patterns were obscured by spatial variability among the coastal environments, and could only be detected within individual groups. Thus, structuring environmental factors (for example, salinity, depth) dominate over seasonal changes in determining community composition. Seasonal variability was detected across an annual cycle in the river, estuary and plume where communities separated into two groups, early year (April–July) and late year (August–Nov), demonstrating annual reassembly of communities over time. Determining both the spatial and temporal variability of bacterioplankton communities provides a framework for modeling these communities across environmental gradients from river to deep ocean.
Keywords: coastal ocean, Columbia River, gradient, spatial variability, 16S amplicon pyrosequencing, temporal variability
Introduction
Over the past few decades, aquatic microbial communities have been shown to be abundant, deeply diverse, and variable across space and time. Yet several recent studies demonstrate repeatable and predictable patterns in the composition of these communities. Spatial variability in aquatic microbial communities has been explored on scales that range from millimeters (Long and Azam, 2001) to kilometers (Hewson et al., 2006) to global (Pommier et al., 2007; Fuhrman et al., 2008). This variability is often attributed to a combination of environmental factors that influence the rate of growth of individual taxa and physical parameters that prevent different communities from interacting (Crump et al., 2004; Fuhrman et al., 2006, 2008; Lozupone and Knight, 2007; Nemergut et al., 2011). Of these factors, salinity, temperature and depth appear to be the most important in distinguishing aquatic communities over large spatial scales, in part because many environmental factors vary with salinity, temperature and depth (for example, light, nutrients, pressure), which results in separation of water masses and thereby communities (Morris et al., 2005; Fuhrman et al., 2008; Carlson et al., 2009; Treusch et al., 2009; Fortunato and Crump, 2011). On a global scale, Lozupone and Knight (2007) showed that the primary determinant of aquatic microbial community composition was salinity, whereas Fuhrman et al. (2008) found that changes in diversity of marine bacteria across a latitudinal gradient were highly correlated to temperature.
Temporal variability in marine and freshwater microbial communities is also predictable within individual environments. Seasonal shifts in microbial community composition have been demonstrated in marine environments, such as the Sargasso and Baltic Seas and the English Channel, where succession of microbial communities correlated with changes in mixed layer depth, temperature and nutrient concentrations throughout the year (Morris et al., 2005; Carlson et al., 2009; Gilbert et al., 2009; Andersson et al., 2010). Mixing, temperature and nutrient concentrations are important factors influencing communities in freshwater systems as well (Kent et al., 2007; Shade et al., 2008; Nelson, 2009; Berdjeb et al., 2011). Shade et al. (2008) found distinct communities in layers of a stratified lake, where gradients of temperature, dissolved oxygen and nutrients were present. Seasonal succession in both marine and freshwater has also been shown to be repeatable (Morris et al., 2005; Fuhrman et al., 2006; Carlson et al., 2009; Crump et al., 2009; Nelson, 2009; Andersson et al., 2010). Crump et al. (2009) showed synchronous shifts in communities of six arctic rivers strongly correlated with seasonal changes in the environment, suggesting microbial communities may shift in predictable patterns from season to season.
Microbial communities are highly diverse, but the extent and the variability of this diversity in freshwater and marine systems is uncertain. High throughput pyrosequencing of PCR-amplified 16S rRNA genes is beginning to resolve the deep diversity of these systems. Because of the large number of sequences per run (∼1 million reads), 16S amplicon pyrosequencing provides better resolution of microbial biogeographical patterns, because the depth of diversity captured with each sample is greater when compared with classical community fingerprinting techniques (for example, DGGE, T-RFLP, ARISA), which only capture the most dominant species in an environment (Sogin et al., 2006). Recent studies have used 16S amplicon pyrosequencing to determine the microbial diversity of many different environments including deep sea, arctic, soil and estuarine communities (Sogin et al., 2006; Galand et al., 2009; Gilbert et al., 2009; Lauber et al., 2009; Andersson et al., 2010).
Microbial community composition and diversity have been characterized spatially and temporally in various environments, but rarely have they been assessed over both spatial and seasonal scales. Using 16S amplicon pyrosequencing, we characterized bacterioplankton communities from 300 water samples collected across the Columbia River coastal margin over an annual cycle. The coastal waters of the Pacific Northwest are highly productive because of nutrient delivery from seasonal upwelling and from the Juan de Fuca strait and Columbia River (Hickey and Banas, 2003). The biological and physical processes of these coastal waters are complex because of variable winds, remote wind forcing, shelf width and submarine canyons (Hickey and Banas, 2003, 2008; Hickey et al., 2010), which in turn may differentially affect the composition of bacterioplankton communities along the Oregon and Washington coasts (Fortunato and Crump, 2011). The Columbia River is the second largest river in the United States with a mean annual discharge of 7300 m3 s−1 (Hickey et al., 1998). This significant release of freshwater has a strong impact on the chemical, physical and biological characteristics of the coastal ocean, including primary and secondary production within the river plume, and differentially along the Oregon and Washington coasts (Hickey et al., 2010).
In a previous study in August 2007, the community fingerprinting technique DGGE was used to broadly characterize the spatial variation of microbial communities in the Columbia River coastal margin (Fortunato and Crump, 2011). Here we used 16S amplicon pyrosequencing to expand on this earlier dataset by increasing the sample size fourfold and characterizing communities across multiple seasons using a more resolved spatial scale from the river to the deep ocean. We hypothesized that because of the large spatial scale of this study, bacterioplankton communities would separate from river to ocean, across salinity, depth and other environmental gradients that vary from fresh to marine waters. Our results indicate that spatial variability overwhelmed seasonal trends across the entire sample set, and temporal variability could only be resolved within single environment types.
Methods
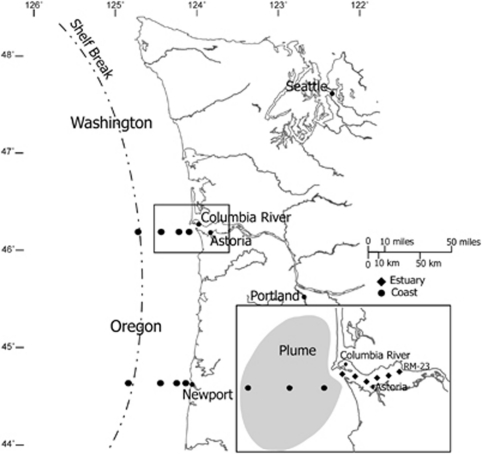
Water samples were collected from the Oregon and Washington coasts, and the Columbia River and estuary (latitude 44.652 and 47.917, longitude −123.874 and −125.929) as part of the NSF-funded Science and Technology Center for Coastal Margin Observation and Prediction. Samples were collected between 2007 and 2008 on eight cruises aboard the R/V Wecoma and R/V Barnes. Aboard the R/V Wecoma, water samples were collected from the Columbia River, estuary, plume and two coastal ocean lines (Columbia River line, Newport Hydroline) in August and November of 2007 and April, June, July and September of 2008 (Figure 1). For coastal lines, samples were taken at three depths per station (surface, within thermocline and bottom). Plume samples were taken at two depths (surface and bottom) in 2007 and four depths (surface, below plume, within thermocline and bottom) in 2008. In the estuary, samples were collected based on the location of the salt gradient in both the north and south channels of the river. Samples were collected across the salt gradient from 0 to 30. Samples were collected using a conductivity-temperature-depth (CTD) rosette water sampler with 10-liter Niskin bottles. With each CTD cast, depth profiles of salinity, temperature (°C), turbidity (NTU), oxygen (mgl−1) and chlorophyll fluorescence were recorded. Water samples aboard the R/V Barnes were collected using a high-volume low-pressure pump over salinity gradients in the estuary in August 2007 and July 2008. For all samples, surface was defined between 1 and 2 m depth, and bottom was defined between 1–5 m above sediment. Data from CTD fluorescence and temperature sensors were used to determine exact sampling depths for water collected at the chlorophyll maximum and within the thermocline.
Figure 1.
Map of the Oregon and Washington coast. Inset depicts Columbia River estuary and plume region. Dotted line denotes approximate location of shelf break.
DNA samples (1–6 l per sample) were collected, preserved and extracted as described previously (Fortunato and Crump, 2011) using methods adapted from Zhou et al. (1996) and Crump et al. (2003). Extracted DNA was PCR-amplified using primers targeting bacterial 16S ribosomal RNA genes. Each sample was assigned a uniquely barcoded reverse primer and amplified in four replicate 20-μl reactions (Hamady et al., 2008). Primers used for amplification were bacteria-specific primers focusing on the V2 region, 27F with 454B FLX linker (5′-GCCTTGCCAGCCCGCTCAGTCAGRGTTTGATYMTGGCTCAG-3′) and 338R with 454A linker and a unique 8-bp barcode, denoted by N in primer sequence (5′-GCCTCCCTCGCGCCATCAGNNNNNNNCATGCWGCCWCCCGTAGGWGT-3′) (modified from Hamady et al., 2008). Replicate amplifications were combined, purified and normalized using Invitrogen SequelPrep normalization plates (Invitrogen, Carlsbad, CA, USA). In all, 5 μl from each sample was combined into a single tube and sent for pyrosequencing on a Roche-454 FLX pyrosequencer at Engencore at the University of South Carolina (http://engencore.sc.edu/).
Sequence data were processed using two different methods: (1) Manual global alignment and removal of pyrosequencing errors using ARB (Ludwig et al., 2004) and MOTHUR software (Schloss et al., 2009), and (2) Denoising and pairwise alignment using the QIIME (v.1.2.0) software package (Caporaso et al., 2010).
For the first method, raw sequences were sorted and quality controlled (minimum length 150 bp, no ambiguous bases) using the Ribosomal Database Project Pyro tools (Cole et al., 2005). A reference sequence database was created using the community analysis program MOTHUR (Schloss et al., 2009) consisting of unique sequences from the overall dataset. These unique sequences were imported into ARB and manually aligned. Extra bases commonly added in pyrosequencing (that is, pyronoise) were placed in gaps added to the alignment. Once the manual alignment was completed, sequences were trimmed to E. coli basepair positions 136–335 and were exported using a 3% basepair frequency filter to mask insertions, but include variable bases. This reference dataset of manually aligned unique sequences was then used to align the entire dataset using MOTHUR. Our approach removed insertions from pyrosequencing, but did not repair deletions of bases, which were included in downstream analyses. Operational taxonomic units (OTUs) were determined based on 97% sequence similarity using MOTHUR.
For the second method using QIIME, sequences were quality controlled using the Split_Libraries.py script with default settings (minimum length 200, maximum length 1000, minimum mean quality score 25, maximum ambiguous bases 0, maximum homopolymer length 6, maximum primer mismatch 0). To account for pyronoise, the remaining sequences were denoised using the denoiser.py script with the ‘fast' method and default settings. Sequences were then clustered using the pick_otus.py script with the uclust method (97% sequence similarity). Potentially chimeric sequences were identified among representative sequences from each OTU with ChimeraSlayer, and a total of 3952 sequences composing 196 OTUs were eliminated from the dataset.
For both methods, relative abundance was calculated for the OTUs in each sample and used to calculate pairwise similarities among samples using the Bray–Curtis similarity coefficient (Legendre and Legendre, 1998). We also calculated pairwise similarities among samples using both weighted and unweighted UNIFRAC metrics (Lozupone et al., 2006), but the results were nearly identical to those based on Bray–Curtis, and so are not presented. Bray–Curtis similarity matrices were visualized using multiple dimensional scaling (MDS) diagrams, a form of ordination. Analysis of Similarity Statistics (ANOSIM) was calculated to test the significance of differences among a priori sampling groups based on environmental parameters. Similarity matrices, MDS diagrams and ANOSIM statistics were carried out using PRIMER v6 for Windows (PRIMER-E Ltd, Plymouth, UK).
Alpha diversity for samples was calculated using MOTHUR. The number of sequences was normalized before calculation by randomly selecting the same number of sequences per sample, based upon the sample with the least number of sequences (n=209 sequences). The taxonomy of OTUs identified was determined using the Ribosomal Database Project Classifier tool. Taxonomic assignments with less than 80% confidence were marked as unknown. A total of 306 samples were analyzed overall. This number was reduced to 300 as samples with a low number of sequences were removed.
All sequences can be downloaded from the NCBI Sequence Read Archive database under the accession number SRP006412. In addition, a Supplementary Table containing sample metadata conforming to MIMARKS standards has also been provided (Supplementary Table S1).
Results
Comparison of the two sequence analysis methods showed that the overall patterns of microbial community structure for this study are highly robust, as both spatial and temporal patterns in beta-diversity were the same for both methods. The number of OTUs identified by the QIIME analysis (8039) was slightly lower than that of the ARB/MOTHUR analysis (9389), but this was because fewer sequences passed the initial QIIME quality control step due to different quality control parameters, including maximum homopolymer length and primer mismatches. Because the patterns of community variability were comparable, the results presented are based on the QIIME sequence analysis protocol.
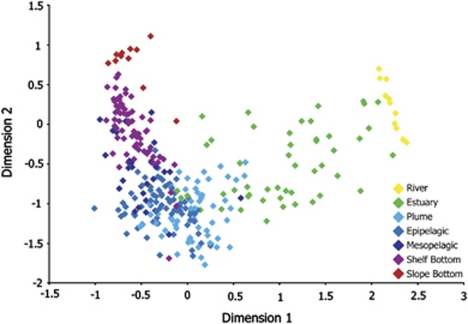
Bacterioplankton communities separated into seven distinct groups (ANOSIM, P<0.001): river, estuary, plume, epipelagic, mesopelagic, shelf bottom and slope bottom. The plume group consisted of coastal surface samples with salinity less than 31, the epipelagic group included coastal surface and chlorophyll maximum samples (average depth=8 m), the mesopelagic group consisted of coastal samples within and below the thermocline (average depth=44 m), the shelf bottom group consisted of bottom samples with depth less than 350 m and the slope bottom group consisted of bottom samples deeper than 850 m. Percent similarity for all samples was 22.9% (±15.3%) with a range from 0% to 74.8% similarity. Similarity values were higher within groups than between groups (Table 1).
Table 1. Percent similarity values within and between groups ±standard deviation (ANOSIM: P<0.001) as determined by Bray–Curtis similarity coefficient.
| River | Estuary | Plume | Epipelagic | Mesopelagic | Shelf bottom | Slope bottom | |
|---|---|---|---|---|---|---|---|
| River | 33.6±11.2 | ||||||
| Estuary | 17.8±11.7 | 24.9±11.1 | |||||
| Plume | 2.6±4.1 | 19.2±11.4 | 36.5±10.5 | ||||
| Epipelagic | 0.1±0.2 | 15.4±11.2 | 35.0±9.2 | 38.4±9.9 | |||
| Mesopelagic | 0.4±1.5 | 13.1±10.0 | 27.3±11.1 | 33.4±11.8 | 37.2±11.5 | ||
| Shelf bottom | 0.2±0.3 | 10.3±8.6 | 17.9±9.5 | 22.8±10.9 | 32.2±12.1 | 41.8±12.6 | |
| Slope bottom | 0.2±0.3 | 4.3±4.0 | 6.2±5.8 | 8.7±6.3 | 16.4±9.6 | 24.7±9.0 | 50.3±6.6 |
Abbreviation: ANOSIM, Analysis of Similarity Statistics.
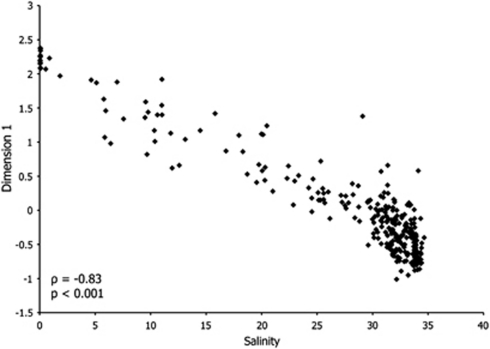
A MDS diagram of all 300 samples based on Bray–Curtis similarity values (Figure 2) depicts the seven groups based on location in the system. Groups separate along two axes that form a V-shaped arrangement of microbial communities. The first axis is clearly related to salinity and the second is related to depth. A strong correlation was shown between Dimension 1 and salinity, with a Spearman's rho value of −0.83 (P<0.001, Figure 3). A weaker relationship was observed between Dimension 2 and sample depth (ρ=−0.62, P<0.001 for Dimension 2 axis and depth), although this relationship improved when river and estuary samples were omitted (ρ=−0.76, P<0.001).
Figure 2.
Multiple dimensional scaling diagram of percent similarities for all 300 samples. Bacterioplankton communities were separated into seven groups based on location across salinity and depth gradients (ANOSIM: P<0.001, Stress: 0.12).
Figure 3.
Correlation of Dimension 1 for the 300 samples from Figure 2 and salinity. A Spearman's rho value of −0.83 (P<0.001) indicates a strong relationship between salinity and bacterial community variation.
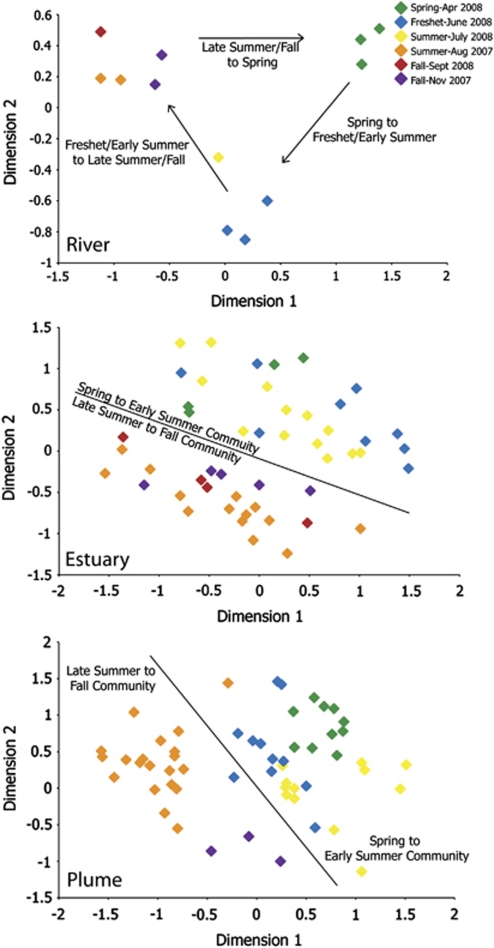
Spatial variation in communities based on sampling location is readily apparent in Figure 2. Temporal variation, however, appears to be overwhelmed by the strong spatial gradients of salinity and depth. Temporal variation was only detectable when each spatial group was analyzed separately. For river, estuary and plume samples, a seasonal trend is apparent from river to ocean (Figure 4). In the river, three communities are visible: spring, freshet-early summer and late summer-fall. In the estuary, seasonal clustering of communities was not as clear, although communities did split into two significant clusters (ANOSIM, P<0.001), an early-year community, encompassing samples from April to July, and a late-year community, encompassing samples from August to November. These same two communities, early and late, are also present in the plume (ANOSIM, P<0.001). The seasonal pattern in the other groups is less discernable. There was significant seasonal variation in the shelf bottom and epipelagic groups according to the ANOSIM statistics, but these patterns could not be discerned in the individual MDS diagrams because of the large amount of variability within each group. There was no significant temporal pattern in the slope bottom or mesopelagic groups.
Figure 4.
Seasonal multiple dimensional scaling diagram of river, estuary and plume. River displays three seasonal communites, which cluster into two communities, early (April–July) and late (August–November), in the estuary and plume. Stress=0.04, 0.15 and 0.17 for river, estuary and plume, respectively.
Most sequences in the dataset were related to the phyla Proteobacteria (44.7%) and Bacteriodetes (33.6%). Within the Proteobacteria, Alpha (21.2%), Gamma (17.0%), Beta (2.6%) and Delta (0.4%) were present. In the Bacteriodetes, Flavobacteria was the largest group, with 55 915 sequences making up 28% of the total dataset. The most abundant OTU belonged to the SAR-11 clade and consisted of 16 635 sequences. Overall, SAR-11 made up 11.3% of the dataset with a total of 22 454 sequences belonging to 208 OTUs. The second largest OTU was a Gammaproteobacteria with 13 137 sequences. Cyanobacteria was a small percentage of the total dataset, only 1.8%, but constituted as much as 19% of sequences in epipelagic samples collected off the shelf. More specific taxonomic information for each of the seven spatial groups can be found in the Supplementary Material (Table S2, Figure S1).
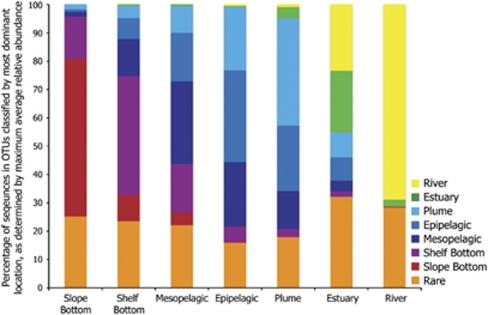
To better understand community composition, we classified each of the 8039 OTUs in this study based on the location in the system where they exhibited their maximum average relative abundance in pooled sequences (Figure 5). For example, if OTU-1 was most abundant in the plume (based on its relative abundance within each pool of sequences from the seven groups), it was classified as a plume OTU. Results suggest mixing of water masses and microbial communities from estuary to the shelf bottom. The river and slope bottom groups appear to be end members in the system, as most of the river and slope bottom sequences are found only in their respective locations. The estuary community is primarily a mix of sequences belonging to river and estuarine OTUs, with some addition from the plume and epipelagic. In the plume, however, plume sequences are mostly classified as being from plume, epipelagic and mesopelagic OTUs with few sequences coming from river or estuary OTUs.
Figure 5.
Percentage of sequences in OTUs classified by location. Slope bottom and river groups represent end members in the system. Rare category represents sequences belonging to OTU that make up to less than 0.1% of the total number of sequences from each corresponding location.
We mapped the relative abundance of the top OTU from each of the seven spatial groups (based on average relative abundance per group) using the ordination of Figure 2. These bubble plots show that the top OTUs for each group are most abundant in samples from their location and less abundant in neighboring locations (Supplementary Figure S2). The top OTUs for the estuary and the river display some seasonality, with the largest abundances occurring in only one or two seasons (for example, June and July 2008 for the estuary).
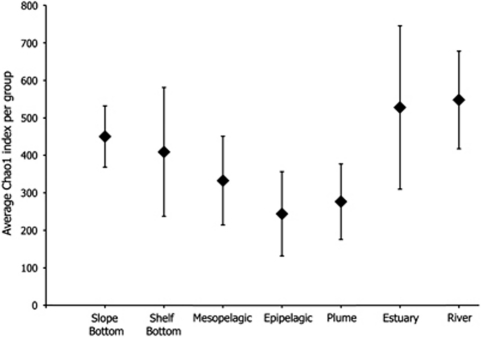
Alpha-diversity varied across the spatial groups (Figure 6). The river and slope bottom groups had the highest and third highest average diversity (Chao1=1104 and 868, respectively), indicating the presence of many more endemic taxa within these two environments, and showing further that freshwater and deep ocean represent end members in this study. As water mixes from the river to the coastal surface ocean, diversity measurements decrease to the lowest diversity in the epipelagic group (Chao1=380). Diversity then increased from surface to the deep ocean, with the mesopelagic, shelf bottom, slope bottom groups each having a higher diversity than the previous. Diversity measurements show that when water mixes from fresh to salt and from deep to surface, taxa are reduced in abundance beyond our limit of detection and thus community composition becomes more streamlined in the coastal surface.
Figure 6.
Average Chao1 index per group ± standard deviation as determined using MOTHUR (v.1.15.0). OTU number was normalized to the sample with the smallest number of sequences (n=209 sequences).
Discussion
Previous studies of variability and diversity in bacterioplankton communities are restricted to single dimensions, focusing on long-term time series, depth profiles or horizontal surveys across environmental gradients (Morris et al., 2005; Hewson et al., 2006; Lozupone and Knight, 2007; Pommier et al., 2007; Fuhrman et al., 2008; Gilbert et al., 2009; Treusch et al., 2009; Andersson et al., 2010; Nemergut et al., 2011). Here we present a dataset that compares bacterioplankton community composition in all three of these dimensions: spatially from river to surface ocean, by depth from surface to deep ocean, and through time seasonally over an annual cycle. This large-scale biogeographical analysis was enabled by the use of 16S amplicon pyrosequencing, which assesses diversity through DNA sequencing of hundreds of thousands of PCR-amplified gene copies. Previous 16S amplicon pyrosequencing studies focused on deep sampling of small numbers of samples, allowing for characterization of the ‘rare biosphere' but only at limited spatial and temporal scales (Galand et al., 2009; Gilbert et al., 2009; Andersson et al., 2010; Kirchman et al., 2010). In this study, we took a different approach to characterizing bacterioplankton communities by applying 16S amplicon pyrosequencing to ten times the number of samples seen in previous studies. Sequencing more samples produces fewer sequences per sample and limits the resolution of the rare biosphere. However, the greater number of samples in this study (n=300) led to the discovery of robust spatial patterns from river to ocean and seasonal shifts that may not have been observed if fewer samples were sequenced. Based on a previous community fingerprinting study of 71 samples from August 2007 using DGGE, we found that communities separated into just five groups defined by location across salinity and depth gradients (Fortunato and Crump, 2011). With the addition of over four times the number of samples, in this study we were able to further resolve the spatial patterns of bacterioplankton communities into seven distinct groups across steep salinity and depth gradients in addition to determining temporal variability.
Salinity and depth changed significantly from the Columbia River to the deep ocean, and these factors appear to strongly influence the composition of bacterioplankton communities. In contrast, temporal variability in bacterioplankton communities was relatively small, and was obscured by the spatial variability in communities across environments in the coastal zone. Several studies of coastal zone bacterioplankton identify time as the principle axis of community variability (Stepanauskas et al., 2003; Fuhrman et al., 2006; Kan et al., 2006; Gilbert et al., 2009), but these studies were restricted to one environmental type (for example, estuaries or a fixed coastal station) within which spatial variability of bacterioplankton communities was limited. Few studies address temporal variability across many different habitats, so it was difficult to compare our results with other studies. However, one study by Kirchman et al. (2010) identified a similar pattern among 11 surface water samples in which winter/summer differences in Arctic Ocean bacterioplankton communities was minimal compared with spatial variability across their sampling range. Thus, although temporal variability may occur within many marine habitats, it is clear that structuring environmental factors (for example, salinity, depth) dominate over seasonal changes in determining community composition.
Spatial differentiation among samples was highly correlated with salinity, confirming the observations of two global meta-analyses of microbial diversity based on 16S rRNA gene sequences (Lozupone and Knight, 2007; Tamames et al., 2010). In one of these studies, Lozupone and Knight (2007) found that salinity was the primary environmental determinant for community composition across marine, freshwater, sediment and soil environments, more so than temperature, pH or other environmental factors. In the coastal marine environment, salinity contributes to density gradients that physically separate water masses and their resident microbial communities. However, the degree to which these water masses are separated depends on the magnitude of mixing by river flow, tides, upwelling, surface winds, and so on. This mixing from fresh to marine or from surface to deep leads to the formation of communities in mixing zones that comprise bacterioplankton populations from multiple water masses. For example in the Columbia River estuary, the flushing rate exceeds the doubling time of bacterioplankton populations, thus a distinct free-living estuarine community is unable to form (Crump et al., 1999). Our study confirmed this observation, demonstrating that estuarine bacterioplankton communities are composed of populations from the river and the coastal ocean (Figure 5). We also identified significant overlap in communities across environmental gradients in the coastal ocean including the plume, epipelagic, mesopelagic and shelf bottom environments (Figure 5 and Supplementary Figure S2), although it is unclear whether this is the result of mixing or the presence of generalist organisms that thrive in different environments.
Coastal bacterioplankton communities correlated with depth from the surface to the deep ocean, despite the fact that samples were collected over multiple seasons and at sampling sites as much as 150 km apart. Salinity varies with depth, as do many other environmental parameters including temperature, light and nutrients. We therefore are treating depth here as a proxy for many factors that vary in the vertical dimension. The vertical structuring of bacterioplankton communities in the ocean has been demonstrated in many studies and has been linked to changes in hydrostatic pressure as well as water mass properties (Lee and Fuhrman, 1991; Morris et al., 2005; Blumel et al., 2007; Carlson et al., 2009; Treusch et al., 2009). For example, Treusch et al. (2009) found that Sargasso Sea bacterial communities separated into surface (upper 40 m), deep chlorophyll maximum and upper mesopelagic communities. We also observed a separation of the epipelagic and upper mesopelagic communities, but not between surface and chlorophyll maximum samples, possibly because the mixed layer depth (5–56 m) was, in general, shallower than that of the Sargasso Sea (<50–350 m)(Carlson et al., 2009; Treusch et al., 2009). Treusch et al. (2009) attributed separation of these communities to stratification and seasonal mixing in the upper water column. The coastal zone of the Pacific Northwest experiences seasonal upwelling, and thus a mixing of communities from bottom to surface. The degree of mixing is evident in Figure 5, where the mesopelagic group is actually a mix of populations from the bottom and surface. In July 2008 during strong upwelling, near-shore surface samples from the Newport Hydroline contained a higher proportion (23%) of sequences belonging to shelf bottom and slope bottom OTUs than during other times in 2008 (5%). Also during that month, the most abundant estuary-classified OTU was found in some shelf bottom samples (Supplementary Figure S2), indicating a possible exchange between these two environments.
Temporal variability could only be resolved within some environments. Seasonal changes were observed in the river, estuary and plume environments. In the river, there were three separate groups, spring, freshet-early summer and late summer-fall, corresponding to seasonal changes in Columbia River discharge, where maximum discharge occurs in late spring and is minimum in late summer to early fall (Prahl et al., 1998). In the plume, seasonal upwelling strongly influences temperature and nutrient concentrations, and thereby production in the plume (Hickey et al., 2010). Thus, plume community composition is tightly linked to the physical processes occurring along the coast. The seasonality of the estuary community then can be attributed to a combination of both river and coastal processes. The periods of maximum and minimum discharge of the river correspond to the two seasonal bacterioplankton groups seen in the estuary, early (April–July) and late (August–November). During times of high river flow, the estuarine community is shaped by the river and when river flow is at a minimum, community composition is influenced more by the plume and coastal ocean.
River and deep ocean (slope bottom group) appear to be end members in this system in that they contribute populations to nearby environments, but receive little to no contributions themselves (Figure 5). In the other five groups there was tremendous overlap in community composition from estuary to shelf bottom, suggesting dynamic exchange of communities through advection and mixing. Within each group there also appeared to be environment-specific communities, based on maximum relative abundance (Figure 5). In the plume, 37% of plume sequences were classified as belonging to plume OTUs, indicating the presence of a plume-specific community. Additionally, only 5% of plume sequences were from the river and estuary, whereas 36% came from epipelagic and mesopelagic OTUs, indicating the plume community is comprised more of coastal populations than bacteria flushed from the estuary. As mentioned previously, the plume is highly productive because of nutrient delivery from the river and coastal upwelling (Hickey et al., 2010) and as primary production increases in the plume, different epipelagic taxa could increase depending on availability and quality of organic matter. This would result in a different combination of bacterial populations and a clear distinction between the plume and epipelagic communities. We speculate then that each spatial group, from estuary to shelf bottom, contains bacterioplankton populations that are broadly distributed across environments, but each group supports a different combination of these bacteria, creating distinct communities within each environment.
16S amplicon pyrosequencing, like any other molecular technique, is prone to errors and it is important to analyze sequences in a way that accurately assesses community patterns. Analyzing 16S amplicon pyrosequencing data is difficult because of sequencing errors termed ‘pyronoise', which may artificially increase the number of OTUs observed. In Kunin et al. (2009), the authors PCR-amplified a 300 bp region of the 16S rRNA gene from a known cultured E. coli strain and then pyrosequenced it. The results returned a largely inflated number of OTUs, showing that pyrosequencing errors may lead to a gross overestimation of the number of OTUs in a sample. An increase in the number of OTUs leads to inflated alpha diversity within samples, and greater beta diversity between samples. We found that global alignment combined with manual removal of pyronoise insertions was comparable in total OTU number, alpha-diversity and beta-diversity patterns to analysis using a QIIME analysis pipeline that includes denoising (denoiser.py) and pairwise sequence alignment (uclust). We also found that removing the pyronoise is crucial for minimizing the total number of OTUs and overall sequencing errors. To demonstrate this, we globally aligned our sequences using a reference database from SILVA (Pruesse et al., 2007) and found that although our beta-diversity patterns were comparable, the OTU number and alpha diversity estimates were nearly twice than that of our previous methods (data not shown). It is important then that pyrosequencing datasets be subjected to rigorous quality checking and denoising, in order to accurately assess both the overall community patterns and the rare biosphere.
Acknowledgments
This study was carried out within the context of the Science and Technology Center for Coastal Margin Observation & Prediction supported by the National Science Foundation (grant number OCE-0424602 to Antonio Baptista). We would like to thank the captains and crews of the R/V Wecoma and R/V Barnes, chief scientists Murray Levine, Tawnya Peterson and Joe Needoba, as well as Suzanne DeLorenzo, Peter Kahn and Daniel Murphy for their help in sample collection. We also thank Joe Jones and John Busch for 454-pyrosequencing support, and Patrick Schloss and Jack Gilbert for their help in pyrosequence analysis.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Andersson AF, Riemann L, Bertilsson S. Pyrosequencing reveals contrasting seasonal dynamics of taxa within Baltic Sea bacterioplankton communities. ISME J. 2010;4:171–181. doi: 10.1038/ismej.2009.108. [DOI] [PubMed] [Google Scholar]
- Berdjeb L, Ghiglione JF, Jacquet S. Bottom-up versus top-down control of hypo- and epilimnion free-living bacterial community structures in two neighboring freshwater lakes. Appl Environ Microbiol. 2011;77:3591–3599. doi: 10.1128/AEM.02739-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumel M, Suling J, Imhoff JF. Depth-specific distribution of Bacteroidetes in the oligotrophic eastern Mediterranean Sea. Aquat Microb Ecol. 2007;46:209–224. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CA, Morris R, Parsons R, Treusch AH, Giovannoni SJ, Vergin K. Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J. 2009;3:283–295. doi: 10.1038/ismej.2008.117. [DOI] [PubMed] [Google Scholar]
- Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, et al. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump BC, Armbrust EV, Baross JA. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl Environ Microbiol. 1999;65:3192–3204. doi: 10.1128/aem.65.7.3192-3204.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump BC, Hopkinson CS, Sogin ML, Hobbie JE. Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl Environ Microbiol. 2004;70:1494–1505. doi: 10.1128/AEM.70.3.1494-1505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump BC, Kling GW, Bahr M, Hobbie JE. Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source. Appl Environ Microbiol. 2003;69:2253–2268. doi: 10.1128/AEM.69.4.2253-2268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump BC, Peterson BJ, Raymond PA, Amon RMW, Rinehart A, McClelland JW, et al. Circumpolar synchrony in big river bacterioplankton. Proc Natl Acad Sci USA. 2009;106:21208–21212. doi: 10.1073/pnas.0906149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato CS, Crump BC. Bacterioplankton community variation across river to ocean environmental gradients. Microb Ecol. 2011;62:374–382. doi: 10.1007/s00248-011-9805-z. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S. Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Natl Acad Sci USA. 2006;103:13104–13109. doi: 10.1073/pnas.0602399103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, Steele JA, Hewson I, Schwalbach MS, Brown MV, Green JL, et al. A latitudinal diversity gradient in planktonic marine bacteria. Proc Natl Acad Sci USA. 2008;105:7774–7778. doi: 10.1073/pnas.0803070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galand PE, Casamayor EO, Kirchman DL, Lovejoy C. Ecology of the rare microbial biosphere of the Arctic Ocean. Proc Natl Acad Sci USA. 2009;106:22427–22432. doi: 10.1073/pnas.0908284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Field D, Swift P, Newbold L, Oliver A, Smyth T, et al. The seasonal structure of microbial communities in the Western English Channel. Environ Microbiol. 2009;11:3132–3139. doi: 10.1111/j.1462-2920.2009.02017.x. [DOI] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewson I, Steele JA, Capone DG, Fuhrman JA. Temporal and spatial scales of variation in bacterioplankton assemblages of oligotrophic surface waters. Mar Ecol Prog Ser. 2006;311:67–77. [Google Scholar]
- Hickey BM, Banas NS. Oceanography of the US Pacific Northwest Coastal Ocean and estuaries with application to coastal ecology. Estuaries. 2003;26:1010–1031. [Google Scholar]
- Hickey BM, Banas NS. Why is the Northern end of the California current system so productive. Oceanography. 2008;21:90–107. [Google Scholar]
- Hickey BM, Kudela RM, Nash JD, Bruland KW, Peterson WT, MacCready P, et al. River influences on shelf ecosystems: introduction and synthesis. J Geophys Res-Oceans. 2010;115:1–26. [Google Scholar]
- Hickey BM, Pietrafesa LJ, Jay DA, Boicourt WC. The Columbia River plume study: subtidal variability in the velocity and salinity fields. J Geophys Res Oceans. 1998;103:10339–10368. [Google Scholar]
- Kan JJ, Crump BC, Wang K, Chen F. Bacterioplankton community in Chesapeake Bay: predictable or random assemblages. Limnol Oceanogr. 2006;51:2157–2169. [Google Scholar]
- Kent AD, Yannarell AC, Rusak JA, Triplett EW, McMahon KD. Synchrony in aquatic microbial community dynamics. ISME J. 2007;1:38–47. doi: 10.1038/ismej.2007.6. [DOI] [PubMed] [Google Scholar]
- Kirchman DL, Cottrell MT, Lovejoy C. The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ Microbiol. 2010;12:1132–1143. doi: 10.1111/j.1462-2920.2010.02154.x. [DOI] [PubMed] [Google Scholar]
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2009;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Fuhrman JA. Spatial and temporal variation of natural bacterioplankton assemblages studied by total genomic DNA cross-hybridization. Limnol Oceanogr. 1991;36:1277–1287. [Google Scholar]
- Legendre P, Legendre L. Numerical Ecology. Elsevier Science B.V.: Amsterdam; 1998. [Google Scholar]
- Long RA, Azam F. Microscale patchiness of bacterioplankton assemblage richness in seawater. Aquat Microb Ecol. 2001;26:103–113. [Google Scholar]
- Lozupone C, Hamady M, Knight R. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinform. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc Natl Acad Sci USA. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RM, Vergin KL, Cho JC, Rappe MS, Carlson CA, Giovannoni SJ. Temporal and spatial response of bacterioplankton lineages to annual convective overturn at the Bermuda Atlantic Time-series Study site. Limnol Oceanogr. 2005;50:1687–1696. [Google Scholar]
- Nelson CE. Phenology of high-elevation pelagic bacteria: the roles of meteorologic variability, catchment inputs and thermal stratification in structuring communities. ISME J. 2009;3:13–30. doi: 10.1038/ismej.2008.81. [DOI] [PubMed] [Google Scholar]
- Nemergut DR, Costello EK, Hamady M, Lozupone C, Jiang L, Schmidt SK, et al. Global patterns in the biogeography of bacterial taxa. Environ Microbiol. 2011;13:135–144. doi: 10.1111/j.1462-2920.2010.02315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier T, Canback B, Riemann L, Bostrom KH, Simu K, Lundberg P, et al. Global patterns of diversity and community structure in marine bacterioplankton. Mol Ecol. 2007;16:867–880. doi: 10.1111/j.1365-294X.2006.03189.x. [DOI] [PubMed] [Google Scholar]
- Prahl FG, Small LF, Sullivan BA, Cordell J, Simenstad CA, Crump BC, et al. Biogeochemical gradients in the lower Columbia River. Hydrobiologia. 1998;361:37–52. [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shade A, Jones SE, McMahon KD. The influence of habitat heterogeneity on freshwater bacterial community composition and dynamics. Environ Microbiol. 2008;10:1057–1067. doi: 10.1111/j.1462-2920.2007.01527.x. [DOI] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, et al. Microbial diversity in the deep sea and the underexplored ‘rare biosphere'. Proc Natl Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanauskas R, Moran MA, Bergamaschi BA, Hollibaugh JT. Covariance of bacterioplankton composition and environmental variables in a temperate delta system. Aquat Microb Ecol. 2003;31:85–98. [Google Scholar]
- Tamames J, Abellan JJ, Pignatelli M, Camacho A, Moya A. Environmental distribution of prokaryotic taxa. BMC Microbiol. 2010;10:85. doi: 10.1186/1471-2180-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treusch AH, Vergin KL, Finlay LA, Donatz MG, Burton RM, Carlson CA, et al. Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J. 2009;3:1148–1163. doi: 10.1038/ismej.2009.60. [DOI] [PubMed] [Google Scholar]
- Zhou JZ, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.