Abstract
Heterotrophic protists are a highly diverse and biogeochemically significant component of marine ecosystems, yet little is known about their species-specific prey preferences and symbiotic interactions in situ. Here we demonstrate how these previously unresolved questions can be addressed by sequencing the eukaryote and bacterial SSU rRNA genes from individual, uncultured protist cells collected from their natural marine environment and sorted by flow cytometry. We detected Pelagibacter ubique in association with a MAST-4 protist, an actinobacterium in association with a chrysophyte and three bacteroidetes in association with diverse protist groups. The presence of identical phylotypes among the putative prey and the free bacterioplankton in the same sample provides evidence for predator–prey interactions. Our results also suggest a discovery of novel symbionts, distantly related to Rickettsiales and the candidate divisions ZB3 and TG2, associated with Cercozoa and Chrysophyta cells. This study demonstrates the power of single cell sequencing to untangle ecological interactions between uncultured protists and prokaryotes.
Keywords: protist, single cell genomics, symbiont, marine, Pelagibacter, MAST
Marine protists have a variety of fundamental ecological roles and comprise organisms with highly diverse evolutionary histories and lifestyles (Massana et al., 2006; Worden et al., 2006; Harada et al., 2007; Worden and Not, 2008; Zubkov and Tarran, 2008; Not et al., 2009; Heywood et al., 2011). Phototrophic protists contribute significantly to phytoplankton biomass and primary production (Jardillier et al., 2010), whereas heterotrophic protists have critical roles in microbial food webs by grazing on picoplankton, channeling organic carbon to higher trophic levels, controlling bacterial abundance and remineralizing nutrients (Azam et al., 1983; Fuhrman and McManus, 1984; Pernthaler, 2005; Worden and Not, 2008; Massana et al., 2009). Mixotrotrophy is also common among marine protists (Zubkov and Tarran, 2008). Furthermore, symbiotic relationships between prokaryotes and marine protists may be prevalent, but their studies so far have been limited to a few cultured model systems (Kneip et al., 2008). Because of difficulties in the cultivation of heterotrophic protists, little is known about the species-specific prey preferences of the predominant protists or the frequency and the nature of symbiotic relationships in their natural environment. Studies performed to date, using model prey and/or predator cells, suggest interactions between diverse microbial taxa (Fu et al., 2003; Jezbera et al., 2005; Massana et al., 2009). However, to the best of our knowledge, identification of both the protistan predator and its natural prey or symbiont under in situ conditions has not been accomplished to date, because of methodological limitations. This represents a major gap in our knowledge of microbial ecology.
Here we present a new approach to disentangle the complexity of protist–prokaryote interactions by sequencing SSU rRNA genes of individual protists and the prokaryotes that are physically associated with them. We collected a coastal water sample from 1 m depth in Boothbay Harbor in the Gulf of Maine, USA (43°50′39.87″N 69°38′27.49″W) on 19 July 2009 and incubated it with Lysotracker Green as described in detail (Rose et al., 2004). Lysotracker is a green fluorescing probe that stains food vacuoles in protists and has been previously used to detect heterotrophic protists from natural assemblages (Rose et al., 2004; Heywood et al., 2011). Individual cells displaying Lysotracker fluorescence were deposited into 384-well microplates by flow cytometric cell sorting, lysed using cold KOH and subjected to whole-genome multiple displacement amplification as described in detail in the Supporting Information. We refer to the multiple displacement amplification products from the sorted single cells as single amplified genomes (SAGs) that included amplified genomic DNA from prokaryotes associated with the sorted protists. The multiple displacement amplification products served as templates in real-time PCR, separately targeting the eukaryotic and the bacterial SSU rRNA genes (Supplementary Tables S1 and S2). The PCR amplicons were directly sequenced using Sanger technology. To compare the composition of bacteria associated with protists to the free bacterioplankton composition, we also generated SAGs of single bacterioplankton cells sorted from the same seawater sample (Supplementary Information).
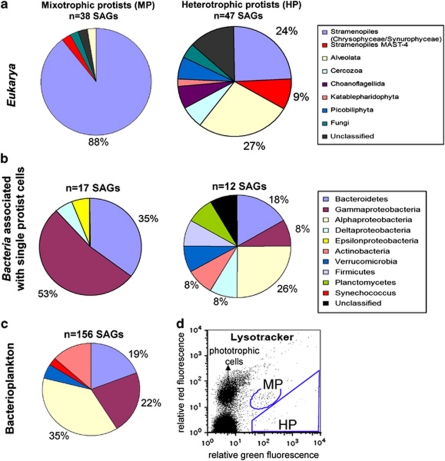
Two optically distinct groups of small protists (<20 μm) displaying Lysotracker fluorescence were detected by flow cytometry and sorted separately (Figure 1). One of the groups displayed only Lysotracker (green) fluorescence, typical of heterotrophic protists (Heywood et al., 2011), whereas another group showed both Lysotracker fluorescence and red autofluorescence from chlorophyll. The latter group may have been composed of mixotrophs and/or heterotrophs with ingested chlorophyll-containing prey. We operationally define them here as ‘mixotrophic protists'. For each of these protist populations, 315 SAGs were generated and analyzed by PCR amplification and sequencing of their 18S and 16S rRNA genes (Supplementary Tables S1 and S2). We successfully sequenced the 18S rRNA gene from 47 and 38 SAGs from the putative heterotrophic and mixotrophic protists, respectively (Supplementary Table S2). Chloroplast SSU rRNA genes were detected in 66 SAGs of the putative mixotrophs (21% of total SAGs), further supporting a mixotrophic lifestyle (Supplementary Table S2). No chloroplast sequences were recovered from the putative heterotrophs, confirming the specificity of our analytical techniques. Mitochondrial 16S rRNA gene sequences were recovered from five putative heterotrophs and 22 putative mixotrophs. The putative mixotrophs were composed mainly of chrysophytes (88% of SAGs), whereas the putative heterotrophs were highly diverse (Figure 1), consistent with our previous study on heterotrophic protists in the Gulf of Maine (Heywood et al., 2011).
Figure 1.
(a) Taxonomic composition of SAGs generated from the putative heterotrophic and mixotrophic protist populations and (b) the corresponding bacterial diversity of 16S rRNA genes found in protist cells. Key DNA tools (http://keydnatools.com/) and ARB software (Pruesse et al., 2007) were used to assign phylogenetic position of SAGs. All sequences were screened for chimeras by Mallard 1.02 (Ashelford et al., 2006) and Pintail 1.01 (Ashelford et al., 2005). (c) Taxonomic composition of marine bacterioplankton SAGs generated from the same water sample used for the protist sorting. (d) Flow cytometric sort regions for the putative mixotrophic (MP) and heterotrophic protist (HP) populations. Putative heterotrophs showed high green fluorescence from Lysotracker-stained vacuoles, whereas putative mixotrophs displayed both green and red fluorescence from Lysotracker and chlorophyll. The large population in the lower left is noise and/or heterotrophic bacteria. Side scatter was used to limit the sort to cells that were <∼20 μm in diameter (data not shown). Genbank accession numbers are as follows: 18S rRNA gene (JF488764–JF488851), 16S rRNA bacterial gene (JF488634–JF488667), 16S rRNA mitochondrial gene (JF488737–JF488763) and 16S rRNA chloroplast gene (JF488668–JF488736).
Bacterial 16S rRNA gene sequences were recovered from 12 and 17 putative heterotrophic and mixotrophic protists, respectively. In most cases they were similar to those recovered from free bacterioplankton SAGs, with the predominance of Gammaproteobacteria, Bacteroidetes and Alphaproteobacteria (Figure 1). Among the represented bacterial classes, only the frequency of Gammaproteobacteria was significantly higher in mixotrophic protist SAGs as compared with free bacterioplankton SAGs (α selectivity index for predator food preference=0.8 (Cheson, 1983; Jezbera et al., 2006); Z-test for two proportions, P=0.02). This suggests that Gammaproteobacteria were more likely than other bacterial classes to be grazed by (or attached to) protists in the studied marine sample.
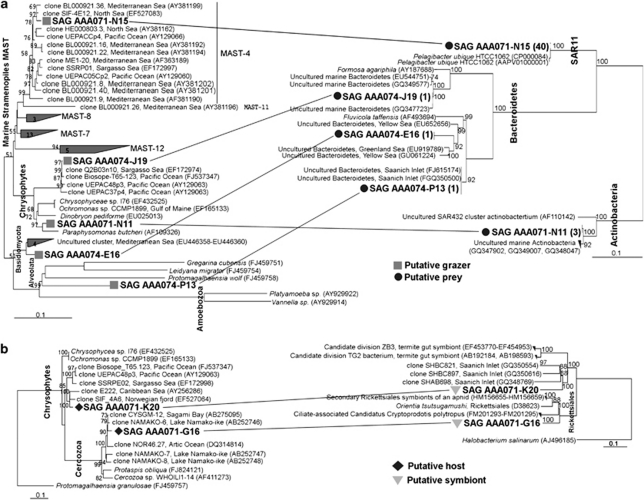
We recovered both eukaryote and bacterial SSU rRNA gene sequences from seven SAGs, in this way obtaining direct evidence for their in situ ecological interaction (Figure 2). In five of these cases, identical 16S rRNA sequences of the protist-associated bacteria were detected in the SAG library of randomly sorted bacterioplankton (Supplementary Table S2). Interestingly, we detected a Pelagibacter ubique sequence in one MAST-4 SAG (AAA071-N15), which are among the most abundant groups of marine bacterioplankton and heterotrophic protists, respectively (Massana et al., 2004; Worden and Not, 2008; Not et al., 2009). In addition, Bacteroidetes sequences were recovered from SAGs representing a widespread, uncultured marine Chrysophyte cluster (SAG AAA074-J19), an alveolate belonging to the Apicomplexa (AAA074-P13) and one protist related to an uncultured, widespread Basidiomycota cluster (AAA074-E16), previously found in oxygen-depleted regions of the Mediterranean and Arabian Seas. Finally, one protist SAG (AAA071-N11), phylogenetically related to the known heterotroph Paraphysomonas butcheri, contained a 16S rRNA gene related to Actinobacteria clones found in other coastal areas, and the Actinobacteria SAR432 cluster. Although Bacteroidetes are often found associated with (Jasti et al., 2005) or attached to (Sapp et al., 2007) phytoplankton, we are not aware of such associations being reported between bacteria and heterotrophic protists. Furthermore, Pelagibacter and SAR432 are known free-living taxonomic groups and their finding in/on protists most likely is a result of protist grazing activity. Thus, we interpret these results as a likely indication of bacteria being recently ingested by the sorted protists. Further studies will be required to unequivocally determine whether these associations represent predator–prey interactions or bacterial attachment on the protist cell surfaces.
Figure 2.
Best maximum likelihood trees (1000 bootstrap) showing the taxonomic identities for both the protist (left) and the associated bacteria (right) obtained by single cell sequencing. Bootstrap values ⩾50 are displayed. Phylogenetic analysis was performed with RAxML version 7.0.3 (Stamatakis, 2006) implemented in ARB package using the reference ARB database 102 (http://www.arb-silva.de). The core tree was calculated with the closest reference sequences and then partial eukaryotic and bacterial sequences from SAGs (736–949 nucleotide positions) were added using the ARB parsimony tool. SAG names ‘AAA071' indicate SAGs generated from the putative heterotrophic protist population, whereas ‘AAA074' were from the putative mixotrophic protist population. (a) Phylogenetic tree showing the identity of both the protist grazer and its putative bacterial prey. In parentheses is the number of SAGs with the identical 16S rRNA gene detected in the bacterioplankton SAG library generated from the same water sample used for protist sorting. (b) Phylogenetic tree showing the taxonomic identities of two single cell protists harboring putative bacterial symbionts.
Protist SAGs AAA071-K20 (Chrysophyta) and AAA071-G16 (Cercozoa) contained bacterial 16S rRNA genes that were not related to known marine bacterioplankton. In fact, the closest entries in Genbank displayed <89% of the SSU rRNA gene similarity and belonged to Rickettsiales and the candidate divisions ZB3 and TG2. The latter group is formed by endosymbionts of aphids and termites (Figure 2b), whereas rickettsia are often intracellular parasites or pathogens of different hosts, including protists (Ferrantini et al., 2009). This suggests the discovery of novel symbionts/parasites in the uncultured, widely distributed marine picoeukaryote groups, although further verification is needed to confirm these findings.
The use of single cell sequencing to assess the composition of natural heterotrophic protist assemblages was recently proven a reliable approach (Heywood et al., 2011). Because of the stringent cell sorting conditions applied here, random co-sorting of multiple cells is unlikely, as demonstrated in prior studies using our methodology (Stepanauskas and Sieracki, 2007; Woyke et al., 2009; Heywood et al., 2011; Fleming et al., 2011; Hess et al., 2011). The limitations of the method applied here include primer-target mismatches in the PCR screens of SAGs, the presence of PCR template secondary structures (Potvin and Lovejoy, 2009), incomplete cell lysis and uneven amplification of genomic DNA regions during multiple displacement amplification (Zhang et al., 2006; Woyke et al., 2009). All these factors may contribute to the relatively low observed frequency of 18S and 16S rRNA gene co-recovery from the same SAG. Other plausible causes include rapid degradation of prey DNA in food vacuoles, and the competition of bacterial and organelle 16S rRNA genes in PCR reactions. It is also important to recognize that some protists may have had multiple prokaryote taxa associated with them, which would have been detected only by a deep sequencing of the 16S rRNA genes or genomic DNA of individual SAGs.
In this pilot study, we demonstrate how single cell sequencing enables the analysis of ecological interactions between uncultured protists and prokaryotes, opening novel opportunities to examine microbial grazing and symbiotic relationships in situ. The PCR-based SAG sequencing used here should be viewed as the first step, enabling the formulation of specific hypotheses, which can be tested by complementary studies, such as protist SAG whole genome sequencing, fluorescence in situ hybridization of detected groups of interest and manipulative experiments.
Acknowledgments
We thank Wendy Korjeff-Bellows for fieldwork assistance. This research was supported by the NSF Grants OCE-0623288, EF-826924 and OCE-821374 to RS and MES, and by a Maine Technology Institute research infrastructure grants to the Bigelow Laboratory.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol. 2006;72:5734–5741. doi: 10.1128/AEM.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol. 2005;71:7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam F, Fenchel T, Field JG, Meyer-Reil LA, Thingstad F. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser. 1983;10:257–263. [Google Scholar]
- Cheson J. The estimation and analysis of preference and its relationship to foraging models. Ecology. 1983;64:1297–1304. [Google Scholar]
- Ferrantini F, Fokin SI, Modeo L, Andreoli I, Dini F, Görtz HD, et al. “Candidatus Cryptoprodotis polytropus,” a novel Rickettsia-like organism in the ciliated protist Pseudomicrothorax dubius (Ciliophora, Nassophorea) J Eukaryot Microbiol. 2009;56:119–129. doi: 10.1111/j.1550-7408.2008.00377.x. [DOI] [PubMed] [Google Scholar]
- Fleming EJ, Langdon AE, Martinez-Garcia M, Stepanauskas R, Poulton N, Masland D, et al. What's new is old: resolving the identity of Leptothrix ochracea using single cell genomics, pyrosequencing and FISH. PLoS ONE. 2011;6:e17769. doi: 10.1371/journal.pone.0017769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, O'Kelly C, Sieracki M, Distal DL. Protistan grazing analysis by flow cytometry using prey labeled by in vivo expression of fluorescent proteins. Appl Environ Microbiol. 2003;69:6848–6855. doi: 10.1128/AEM.69.11.6848-6855.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, McManus GB. Do bacteria-sized marine eukaryotes consume significant bacterial production. Science. 1984;224:1257–1260. doi: 10.1126/science.224.4654.1257. [DOI] [PubMed] [Google Scholar]
- Harada A, Ohtsuka S, Horiguchi T. Species of the parasitic genus Duboscquella are members of the enigmatic marine Alveolate group I. Protist. 2007;158:337–347. doi: 10.1016/j.protis.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Hess M, Sczyrba A, Egan R, Kim T-W, Chokhawala H, Schroth G, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331:463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- Heywood JL, Sieracki ME, Bellows W, Poulton NJ, Stepanauskas R. Capturing diversity of marine heterotrophic protists: one cell at a time. ISME J. 2011;5:674–684. doi: 10.1038/ismej.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardillier L, Zubkov MV, Pearman J, Scanlan DJ. Significant CO2 fixation by small prymnesiophytes in the subtropical and tropical northeast Atlantic Ocean. ISME J. 2010;4:1180–1192. doi: 10.1038/ismej.2010.36. [DOI] [PubMed] [Google Scholar]
- Jasti S, Sieracki ME, Poulton NJ, Giewat MW, Rooney-Varga JN. Phylogenetic diversity and specificity of bacteria closely associated with Alexandrium spp. and other phytoplankton. Appl Environ Microbiol. 2005;71:3483–3494. doi: 10.1128/AEM.71.7.3483-3494.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezbera J, Horòák K, Šimek K. Food selection by bacterivorous protists: insight from the analysis of the food vacuole content by means of fluorescence in situ hybridization. FEMS Microbiol Ecol. 2005;52:351–363. doi: 10.1016/j.femsec.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Jezbera J, Hornák K, Simek K. Prey selectivity of bacterivorous protists in different size fractions of reservoir water amended with nutrients. Environ Microbiol. 2006;8:1330–1339. doi: 10.1111/j.1462-2920.2006.01026.x. [DOI] [PubMed] [Google Scholar]
- Kneip C, Vobeta C, Lockhart P, Maier U. The cyanobacterial endosymbiont of the unicellular algae Rhopalodia gibba shows reductive genome evolution. BMC Evol Biol. 2008;8:30. doi: 10.1186/1471-2148-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana R, Castresana J, Balague V, Guillou L, Romari K, Groisillier A, et al. Phylogenetic and ecological analysis of novel marine stramenopiles. Appl Environ Microbiol. 2004;70:3528–3534. doi: 10.1128/AEM.70.6.3528-3534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana R, Terrado R, Forn I, Lovejoy C, Pedrós-Alió C. Distribution and abundance of uncultured heterotrophic flagellates in the world oceans. Environ Microbiol. 2006;8:1515–1522. doi: 10.1111/j.1462-2920.2006.01042.x. [DOI] [PubMed] [Google Scholar]
- Massana R, Unrein F, Rodriguez-Martinez R, Forn I, Lefort T, Pinhassi J, et al. Grazing rates and functional diversity of uncultured heterotrophic flagellates. ISME J. 2009;3:588–596. doi: 10.1038/ismej.2008.130. [DOI] [PubMed] [Google Scholar]
- Not F, del Campo J, Balagué V, de Vargas C, Massana R. New insights into the diversity of marine picoeukaryotes. PLoS ONE. 2009;4:e7143. doi: 10.1371/journal.pone.0007143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernthaler J. Predation on prokaryotes in the water column and its ecological implications. Nat Rev Micro. 2005;3:537–546. doi: 10.1038/nrmicro1180. [DOI] [PubMed] [Google Scholar]
- Potvin M, Lovejoy C. PCR-based diversity estimates of artificial and environmental 18S rRNA gene libraries. J Eukaryot Microbiol. 2009;56:174–181. doi: 10.1111/j.1550-7408.2008.00386.x. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JM, Caron DA, Sieracki ME, Poulton N. Counting heterotrophic nanoplanktonic protists in cultures and aquatic communities by flow cytometry. Aquat Microb Ecol. 2004;34:263–277. [Google Scholar]
- Sapp M, Schwaderer A, Wiltshire K, Hoppe H-G, Gerdts G, Wichels A. Species-specific bacterial communities in the phycosphere of microalgae. Microb Ecol. 2007;53:683–699. doi: 10.1007/s00248-006-9162-5. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stepanauskas R, Sieracki ME. Matching phylogeny and metabolism in the uncultured marine bacteria, one cell at a time. Proc Natl Acad Sci USA. 2007;104:9052–9057. doi: 10.1073/pnas.0700496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden AZ, Cuvelier ML, Bartlett DH. In-depth analyses of marine microbial community genomics. Trends Microbiol. 2006;14:331–336. doi: 10.1016/j.tim.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Worden ZA, Not F.2008Ecology and diversity of picoeukaryotesIn Kirchman DL (ed),Microbial Ecology of the Oceans John Wiley & Sons, Inc: Hoboken, New Jersey [Google Scholar]
- Woyke T, Xie G, Copeland A, Gonzalez JM, Han C, Kiss H, et al. Assembling the marine metagenome, one cell at a time. PLoS One. 2009;4:e5299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Martiny A, Reppas NB, Barry KW, Malek J, Chrisholm SW, et al. Sequencing genomes from single cells by polymerase cloning. Nat Biotech. 2006;24:680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]
- Zubkov MV, Tarran GA. High bacterivory by the smallest phytoplankton in the North Atlantic Ocean. Nature. 2008;455:224–226. doi: 10.1038/nature07236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.