Abstract
Background
The completion of the grape genome sequencing project has paved the way for novel gene discovery and functional analysis. Aldehyde dehydrogenases (ALDHs) comprise a gene superfamily encoding NAD(P)+-dependent enzymes that catalyze the irreversible oxidation of a wide range of endogenous and exogenous aromatic and aliphatic aldehydes. Although ALDHs have been systematically investigated in several plant species including Arabidopsis and rice, our knowledge concerning the ALDH genes, their evolutionary relationship and expression patterns in grape has been limited.
Methodology/Principal Findings
A total of 23 ALDH genes were identified in the grape genome and grouped into ten families according to the unified nomenclature system developed by the ALDH Gene Nomenclature Committee (AGNC). Members within the same grape ALDH families possess nearly identical exon-intron structures. Evolutionary analysis indicates that both segmental and tandem duplication events have contributed significantly to the expansion of grape ALDH genes. Phylogenetic analysis of ALDH protein sequences from seven plant species indicates that grape ALDHs are more closely related to those of Arabidopsis. In addition, synteny analysis between grape and Arabidopsis shows that homologs of a number of grape ALDHs are found in the corresponding syntenic blocks of Arabidopsis, suggesting that these genes arose before the speciation of the grape and Arabidopsis. Microarray gene expression analysis revealed large number of grape ALDH genes responsive to drought or salt stress. Furthermore, we found a number of ALDH genes showed significantly changed expressions in responses to infection with different pathogens and during grape berry development, suggesting novel roles of ALDH genes in plant-pathogen interactions and berry development.
Conclusion
The genome-wide identification, evolutionary and expression analysis of grape ALDH genes should facilitate research in this gene family and provide new insights regarding their evolution history and functional roles in plant stress tolerance.
Introduction
Plants are exposed to many types of abiotic stresses during their life-cycle, such as drought, salinity, and low temperature [1]. Plants adapt to abiotic stresses by the expression of a wide range of stress-responsive genes, which are thought to play key roles in stress tolerance and survival [2]. Endogenous aldehyde molecules are intermediates or by-products of several fundamental metabolic pathways, and they are also excessively generated in response to environmental stresses such as salinity, dehydration, desiccation, cold and heat shock [3], [4]. Although aldehydes are associated with common biochemical pathways, the compounds can be extremely toxic when produced in excess because of their inherent chemical reactivity [5]. Aldehyde dehydrogenases (ALDHs) comprise a gene superfamily encoding NAD(P)+-dependent enzymes that catalyze the irreversible oxidation of a wide range of endogenous and exogenous aromatic and aliphatic aldehydes [6]. ALDHs are responsible for efficient detoxification of aldehydes by converting them to carboxylic acids [6]. Additionally, they also carry out a broad range of other metabolic functions including (i) participating in intermediary metabolism, such as amino acid and retinoic acid metabolism; (ii) providing protection from osmotic stress by generating osmoprotectants, such as glycine betaine [7], [8]; and (iii) generating NAD(P)H [9]. In plants, the ALDH genes are expressed throughout various different tissues and in response to a wide variety of stressors [10], [11].
Throughout all taxa, ALDHs have been classified into 24 distinct families to date. These families are numbered according to the criteria established by the ALDH Gene Nomenclature Committee (AGNC) [12]. ALDHs with amino acid sequences that are more than 40% identical to previously identified ALDH sequences comprise a family, while those with sequences more than 60% identity comprise a subfamily. ALDHs with sequences less than 40% identity represent a new family. Among the 24 ALDH families, 14 (ALDH2, ALDH3, ALDH5, ALDH6, ALDH7, ALDH10, ALDH11, ALDH12, ALDH18, ALDH19, ALDH21, ALDH22, ALDH23 and ALDH24) contain members from plant species and seven (ALDH11, ALDH12, ALDH19, ALDH21, ALDH22, ALDH23 and ALDH24) are unique to plants. Much work has been carried out on the ALDH gene family in prokaryotes and mammals [6], [13], whereas the research on plant ALDHs is relatively limited. Furthermore, most of the analyses on plant ALDHs have been performed in model species such as Arabidopsis [6] and rice [11], with little attention paid to woody species like grape.
Grapevine (Vitis vinifera) is economically the most important perennial fruit crop worldwide, and the fourth angiosperm species, the second woody species, and the first fruit crop to have a fully sequenced genome [14], [15]. Compared to other perennials, the genome size of V. vinifera is relatively small, 475 Mb, which is similar to rice (Oryza sativa, 430 Mb) [16], barrel medic (Medicago truncatula, 500 Mb, http://medicago.org/) and black cottonwood poplar (Populus trichocarpa, 465 Mb) [17]. In addition, the grapevine genome has not undergone a recent whole genome duplication (WGD), thus enabling the discovery of ancestral traits and genetic divergence occurring during the course of flowering plant evolution [14]. The release of grape genome data allows us for the first time to carry out the genome-wide identification and analysis of ALDH gene families in a woody species. Here we systematically identified 23 ALDH genes belonging to ten different families in the grape genome. Phylogenetic and synteny analyses revealed segmental and tandem duplication events have contributed to grape ALDH evolution. We further analyzed expression profiles of grape ALDH genes under various abiotic and biotic stresses, in response to different phytohormone treatments, and during berry development and ripening, through mining publicly available microarray datasets. The results obtained from our study provided a foundation for evolutionary and functional characterization of ALDH gene families in grape and other plant species.
Results and Discussion
Genome-wide identification of ALDH gene families in grape
From the grape genome, we identified a total of 23 putative ALDH genes and grouped them into ten families based on their protein sequence identities (Table 1). Eight of the ten grape ALDH families are represented by more than one gene (ALDH2, three members; ALDH3, four members; ALDH5 and ALDH6, three members; and ALDH7, ALDH10, ALDH11, and ALDH18, two members), whereas the remaining two (ALDH12 and ALDH22) are single gene families. Among the 23 grape ALDH genes, one (VvALDH18B1) corresponds to a previously published gene (GenBank Acc#: AJ005686) [18], ten are supported by cDNA sequences that contain the full coding regions (their corresponding GenBank Acc# are VvALDH2B8: FQ382277; VvALDH5F1: FQ382545; VvALDH10B1: FQ384094; VvALDH3H1: FQ391752; VvALDH2B4: FQ391821; VvALDH11B1: FQ392316; VvALDH2B9: FQ392766; VvALDH10A9: FQ393912; VvALDH3H5: FQ394868; VvALDH6B3: FQ394961; VvALDH7B5: FQ395151), and another ten are supported by at least one EST sequence available in GenBank dbEST database; while only one (VvALDH7D1) is lack of support by EST or mRNA sequences.
Table 1. Grape ALDH genes and superfamilies.
| Family | Gene ID | Gene Locus ID | Accession No. | Putative function | CDS (bp) | ORF (aa) |
| Family 2 | VvALDH2B4_v1 | GSVIVG01007784001 | XM_002283096 | Mitochondrial ALDH | 1617 | 538 |
| VvALDH2B4_v2 | GSVIVG01007784001 | JN381165 | Cytosolic ALDH | 1434 | 477 | |
| VvALDH2B4_v3 | GSVIVG01007784001 | JN381166 | Mitochondrial ALDH | 1578 | 525 | |
| VvALDH2B8 | GSVIVG01020224001 | XM_002263443 | Mitochondrial ALDH | 1617 | 538 | |
| VvALDH2B9 | GSVIVG01032500001 | XM_002274827 | Mitochondrial ALDH | 1608 | 535 | |
| Family 3 | VvALDH3F1 | GSVIVG01018842001 | XM_002273322 | Variable substrate ALDH | 1458 | 485 |
| VvALDH3H1 | GSVIVG01008845001 | XM_002285830 | Variable substrate ALDH | 1467 | 488 | |
| VvALDH3H5 | GSVIVG01022356001 | XM_002273694 | Variable substrate ALDH | 1467 | 488 | |
| VvALDH3J1 | GSVIVG01025276001 | XM_002285430 | Variable substrate ALDH | 1458 | 485 | |
| Family 5 | VvALDH5F1 | GSVIVG01036719001 | XM_002265478 | Succinic semialdehyde dehydrogenase | 1593 | 530 |
| VvALDH5F2 | GSVIVG01036720001 | XM_002265366 | Succinic semialdehyde dehydrogenase | 1482 | 493 | |
| VvALDH5F3 | GSVIVG01036721001 | XM_002265318 | Succinic semialdehyde dehydrogenase | 1476 | 491 | |
| Family 6 | VvALDH6B3 | GSVIVG01000336001 | XM_002266354 | Methylmalonate semi-aldehyde dehydrogenase | 1620 | 539 |
| VvALDH6B5 | GSVIVG01000338001 | XM_002266580 | Methylmalonate semi-aldehyde dehydrogenase | 1716 | 571 | |
| VvALDH6B7 | GSVIVG01003951001 | XM_002266343 | Methylmalonate semi-aldehyde dehydrogenase | 3096 | 1031 | |
| Family 7 | VvALDH7B5 | GSVIVG01015062001 | XM_002278057 | Antiquitin | 1527 | 508 |
| VvALDH7D1 | GSVIVG01016734001 | XM_002272508 | Antiquitin | 1593 | 530 | |
| Family 10 | VvALDH10A9 | GSVIVG01007829001 | XM_002283654 | Betaine-aldehyde dehydrogenase | 1512 | 503 |
| VvALDH10B1 | GSVIVG01032588001 | XM_002281948 | Betaine-aldehyde dehydrogenase | 1500 | 499 | |
| Family 11 | VvALDH11A3 | GSVIVG01035891001 | XM_002285250 | NADH-dependent glyceraldehyde-3-Phosphate dehydrogenase | 1491 | 496 |
| VvALDH11B1 | GSVIVG01023590001 | XM_002279338 | 1491 | 496 | ||
| Family 12 | VvALDH12A1 | GSVIVG01008047001 | XM_002273533 | Δ1-pyrroline-5-carboxylate dehydrogenase | 1668 | 555 |
| Family 18 | VvALDH18B1 | GSVIVG01016467001 | XM_002282319 | Δ1-pyrroline-5-carboxylate synthetase | 2289 | 762 |
| VvALDH18B3 | GSVIVG01034097001 | XM_002273220 | Δ1-pyrroline-5-carboxylate synthetase | 2145 | 714 | |
| Family 22 | VvALDH22A1 | GSVIVG01035003001 | XM_002277707 | Novel ALDH | 1782 | 593 |
We have previously identified three alternatively spliced variants of VvALDH2B4 in wild Chinese grape, V. pseudoreticulata [19]. We subsequently confirmed these three splice variants using RT-PCR in other grape cultivars including Pinot Noir, Chardonnay, V. quinquangularis clone ‘shang-24’, V. pseudoreticulata clone ‘Hunan-1’, and V. piazezkii Maxim. clone ‘Meixian-6’. We named the three alternatively spliced transcripts of VvALDH2B4 as VvALDH2B4_v1, VvALDH2B4_v2 and VvALDH2B4_v3 according to nomenclature guidelines for alternative transcriptional variants of ALDH genes [20]. VvALDH2B4_v1 and VvALDH2B4_v3 have different 3′ splice acceptor sites in the third exon, while VvALDH2B_v2 has an intron retention which leads to a different translation initiation site (Fig. S1). As a result, VvALDH2B4_v2 encodes a 477-residue protein with NH2-terminal truncated, compared to 538- and 525-residue of VvALDH2B4_v1 and VvALDH2B4_v3, respectively. To date all characterized plant species possess two types of ALDH2 proteins: mitochondrial and cytosolic [6], [21], [22] and they all contain two mitochondrial ALDH2 proteins [6], [23]. Analysis with the PSORT program [24] showed that VvALDH2B4_v1 and VvALDH2B4_v3 protein sequences each contained a predicated N-terminal mitochondrial targeting signal, whereas VvALDH2B4_v2 was predicted to function in cytoplasm. In mammals, these enzymes play a role in detoxifying lipid peroxidation-derived aldehydes produced during oxidative stress as well as acetaldehyde produced during ethanol metabolism [25]. However, the specific functions of both mitochondrial and cytosolic ALDH2 proteins in plants remain to be determined.
Comparative analysis of ALDH gene families from various organisms
In the present study, we summarized numbers of gene family members for each individual ALDH family in V. vinifera and seven other plant species (A. thaliana [6], Zea mays [21], O. sativa [22], Physcomitrella patens, Chlamydomonas reinhardtii and Ostreococcus tauri [26]), three mammals (Homo sapiens, Mus musculus and Rattus norvegicus; http://www.aldh.org/), and fungi [27] (Table 2). Plant ALDHs are present in 13 families: ALDH2, ALDH3, ALDH5, ALDH6, ALDH7, ALDH10, ALDH11, ALDH12, ALDH18, ALDH21, ALDH22, ALDH23 and ALDH24. ALDH19 is also unique to plants and to date, has only been identified within the tomato genome and is thought to encode a γ-glutamyl phosphate reductase which may play a role in the biosynthesis of proline from glutamate [28]. ALDH21 and ALDH23 are unique to mosses and ALDH24 is unique to C. reinhardtii. Grape and other studied vascular plants share ten common core ALDH families (ALDH2, ALDH3, ALDH5, ALDH6, ALDH7, ALDH10, ALDH11, ALDH12, ALDH18, and ALDH22), suggesting that these ten families evolved prior to the monocot/eudicot divergence. Eight of the ten core families (ALDH2, ALDH3, ALDH5, ALDH6, ALDH10, ALDH11, ALDH12 and ALDH22) are also shared by terrestrial plants and algae, suggesting that these families have ancient origins predating the transition of aquatic plants onto land.
Table 2. Number of ALDH family members identified in various organisms.
| Organism | ALDH family | |||||||||||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | |
| V. vinifera | − | 3 | 4 | − | 3 | 3 | 2 | − | − | 2 | 2 | 1 | − | − | − | − | − | 2 | − | − | − | 1 | − | − |
| A. thaliana | − | 3 | 3 | − | 1 | 1 | 1 | − | − | 2 | 1 | 1 | − | − | − | − | − | 2 | − | − | − | 1 | − | − |
| Z.mays | − | 6 | 5 | − | 2 | 1 | 1 | − | − | 3 | 1 | 1 | − | − | − | − | − | 3 | − | − | − | 1 | − | − |
| O .sativa | − | 5 | 5 | − | 1 | 1 | 1 | − | − | 2 | 1 | 2 | − | − | − | − | − | 2 | − | − | − | 1 | − | − |
| P. patens | − | 2 | 5 | − | 2 | 1 | 1 | − | − | 1 | 5 | 1 | − | − | − | − | − | − | − | − | 1 | − | 1 | − |
| C. reinhardtii | − | 1 | − | − | 2 | 1 | − | − | − | 1 | 1 | 1 | − | − | − | − | − | − | − | − | − | − | − | 1 |
| O. tauri | − | − | 1 | − | 1 | − | − | − | − | 1 | 1 | 1 | − | − | − | − | − | − | − | − | − | 1 | − | − |
| H. sapiens | 6 | 1 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | − | − | − | − | − | − | 1 | − | 1 | − | − | − | − | − | − |
| M. musculus | 7 | 1 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | − | − | − | − | − | − | 1 | − | 1 | − | − | − | − | − | − |
| R. norvegicus | 7 | 1 | 4 | − | 1 | 1 | 1 | 1 | 1 | − | − | − | − | − | − | 1 | − | 1 | − | − | − | − | − | − |
| Fungi | + | − | − | + | + | − | − | − | − | + | − | − | − | + | + | + | − | + | − | − | − | − | − | − |
+ and − represent presence and absence, respectively, of the ALDH gene family in corresponding organisms.
It is worth noting that several previous studies indicated that A. thaliana genome lacks the ALDH18 family [6], [21], [22], while other reports indicated the existence of two unique ALDH18 genes in Arabidopsis genome [29], [30]. In this study, we performed a search for ALDH18 genes in ‘The Arabidopsis Information Resource’ (TAIR, http://www.arabidopsis.org/) and confirmed that A. thaliana genome does contain two ALDH18 genes located on chromosome 2 and 3, respectively.
It is worth noting that the apparent lack of ALDH1 and ALDH4 gene family members in plants is the result of nomenclature errors made when the genes were originally identified. The plant ALDH2 genes should be included in the ALDH1 family according to AGNC nomenclature guidelines. Both ALDH4 and ALDH12 encode delta-1-pyrroline-5-carboxylate dehydrogenases which play an important role in the degradation of proline to glutamate [31]. They also should be grouped into one single family.
ALDHs have been reported to play important roles in plant responses to various environmental stresses [6]. Plants, especially the higher plants like V. vinifera and Z. mays, seem to have more ALDH genes than animals and fungi. Unlike mammals, plants can not move and are therefore more susceptible to environmental insults, as a result they may require additional stress-response proteins such as ALDHs, to protect them when exposed to stress conditions. Compared to other well characterized plant ALDHs, grape ALDH families are the second most expanded with 23 genes, compared to 24 in Z. mays, 21 in O. sativa, 16 in A. thaliana, 20 in P. patens, eight in C. reinhardtii, and six in O. tauri.
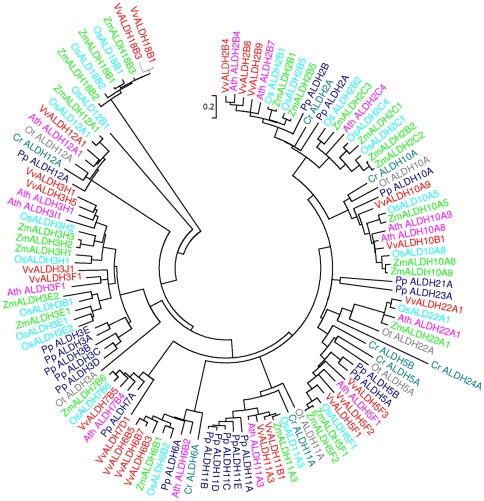
We then extracted protein sequences of ALDH genes identified in V. vinifera and six other plant species, including A. thaliana [6], P. patens, C. reinhardtii, O. tauri [26], Z. mays [21] and O. sativa [22] and constructed a phylogenetic tree (Fig. 1). The tree indicated that the majority of V. vinifera ALDH families are more closely related to those in A. thaliana. ALDH genes from lower plants P. patens, C. reinhardtii and O.tauri diverged early on from their homologues in higher plants. This was followed by a relatively recent monocot/eudicot split. The result is consistent with the current understanding of plant evolutionary history.
Figure 1. Phylogenetic analysis of grape and other plant ALDHs.
Phylogenetic tree was constructed with ALDH protein sequences from V. vinifera (Vv), Z. mays (Zm), O. sativa (Os), A. thaliana (Ath), P. patens (Pp), C. reinhardtii (Cr), and O. Tauri (Ot).
As expected ALDH proteins from same families tend to cluster together. Though evolutionary relationships could not be clarified for all families, the phylogenetic analysis revealed some interesting observations. ALDH18, for example, is the phylogenetically most distantly related family. This is consistent with previous research in rice which indicated that two OsALDH18 proteins had the greatest degree of sequence divergence from other ALDH families and did not contain the conserved ALDH active sites [11].
Phylogenetic and structural analysis of grape ALDH genes
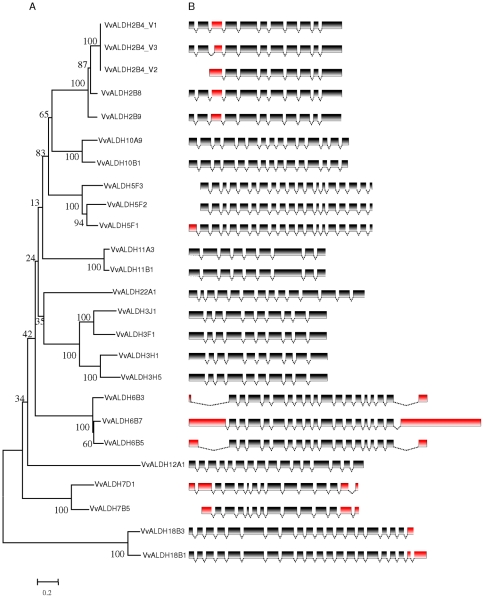
We constructed the phylogenetic tree of the 23 grape ALDH genes based on their amino acid sequences (Fig. 2a). The topology was similar to that constructed with ALDH genes from the seven plant species (Fig. 1) and, similarly, ALDH proteins from the same families are clustered together. Furthermore, the exon-intron structures of the ALDH genes were examined to gain more insights into their possible structural evolution. Exon-intron structural divergence within families plays a pivotal role in the evolution of multiple gene families. Our result showed that genes in the same family generally had similar exon-intron structures. ALDH genes in each of the families 2, 3, 6, 10 and 11 have the same number of exons and also exhibited nearly identical exon lengths, except for the first and last exons of the ALDH6 genes (Fig. 2b). The high degree of sequence identity and similar exon-intron structures of ALDH genes within each family suggests that grape ALDH families have undergone gene duplications throughout evolution, resulting in ALDH gene families containing multiple copies that are partially or completely overlapping in function. A previous study demonstrated that ALDH genes from rice and Arabidopsis had highly conserved exon-intron structures [11]. In this study, we also compared the exon-intron structures of ALDH genes identified in the grape genome with those found in Arabidopsis and rice. The results indicated that the exon-intron structures were not only conserved within a species but also conserved across these three species (data not shown). Nonetheless, we did identify losses or gains of exons during the evolution of several ALDH genes. One such example is the ALDH5 gene family. ALDH5 genes in rice and Arabidopsis, as well as VvALDH5F1 in grape, all have 20 exons; whereas grape VvALDH5F2 and VvALDH5F3 contain 19 exons, losing the first exon during evolution. Other examples include grape VvALDH7D1 and VvALDH18B1, which have acquired one additional exon in 5′- and 3′-end, respectively, during evolution.
Figure 2. Phylogenetic analysis (A) and exon-intron structures (B) of grape ALDH genes.
Numbers above or below branches of the tree indicate bootstrap values. Only coding exons, represented by black or red boxes, were drawn to scale. Dashed lines connecting two exons represent introns. Exons with different structures among the ALDH genes in same families were marked in red.
Expansion patterns of ALDH gene families in grape
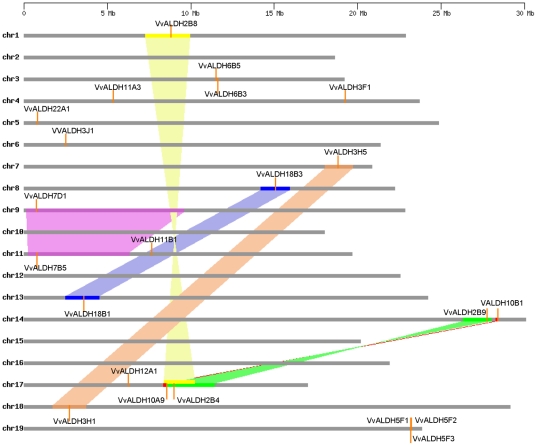
Segmental and tandem duplications are two of the main reasons for gene family expansions [32]. Two tandem ALDH gene duplications have been reported in rice (OsALDH2-1/OsALDH2-2 and OsALDH3-1/OsALDH3-2) [11]. In the present study, we also identified tandem duplications in two grape ALDH gene families (VvALDH5F1/VvALDH5F2/VvALDH5F3 and VvALDH6B3/VvALDH6B5) (Fig. 3). We then examined the duplicated blocks within the grape genome and found that 11 grape ALDH genes from five families (VvALDH2B4/VvALDH2B9/VvALDH2B8, VvALDH3H1/VvALDH3H5, VvALDH7B5/VvALDH7D1, VvALDH10A9/VvALDH10B1, and VvALDH18B1/VvALDH18B3) were located in six pairs of duplicated genome regions (Fig. 3). In summary, seven out of eight grape multi-member ALDH gene families (Table 2) are associated with either segmental or tandem duplication events, indicating that segmental and tandem duplications have played important roles in the expansion of grape ALDH genes.
Figure 3. Distribution and synteny of ALDH genes on grape chromosomes.
Chromosomes 1–19 (chr1–19) are depicted as horizontal gray bars. ALDH genes are indicated by vertical orange lines. Colored bars denote syntenic regions of the grape genome; the twisted colored bar indicates that the syntenic regions are in reverse orientation. VvALDH6B7, which is not assigned to any known chromosomes, is not shown.
Evolutionary relationship of ALDH gene families between grape and Arabidopsis
By comparing the sequences of all genes between genomes from different taxa and within each genome, it is, in principle, possible to reconstruct the evolutionary history of each gene in its entirety (within the set of sequenced genomes) [33]. To further explore the origin and evolutionary process of grape ALDH genes, we analyzed the comparative synteny map between grape and Arabidopsis genomes. Genomic comparison is a quick way to transfer genomic knowledge acquired in one taxon to a less-studied taxon [34]. Arabidopsis is the most important model plant species and the functions of most Arabidopsis ALDH genes have been well characterized. Thus, through comparative genomics analysis we could confidently infer the functions of grape ALDHs based on their Arabidopsis homologues.
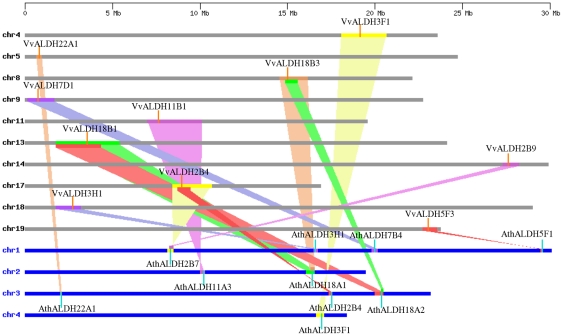
Large-scale syntenies containing orthologs from seven ALDH families (ALDH2, ALDH3, ALDH5, ALDH7, ALDH11, ALDH18 and ALDH22) in both grape and Arabidopsis genomes were identified (Fig. 4). Regarding the single grape-to-Arabidopsis ALDH gene correspondences, the syntenies were unambiguous and included the following ortholog pairs: VvALDH3H1-AthALDH3H1, VvALDH7D1-AthALDH7B4, VvALDH3F1-AthALDH3F1, VvALDH11B1-AthALDH11A3, VvALDH5F3-AthALDH5F1 and VvALDH22A1-AthALDH22A1 (Fig. 4), indicating these genes/families should have been in the genome of last common ancestor of grape and Arabidopsis. More challenging for syntenic interpretation are cases where duplicated grape genes corresponded to two Arabidopsis genes. These included VvALDH2B4/VvALDH2B9-AthALDH2B4/AthALDH2B7 and VvALDH18B3/VvALDH18B1-AthALDH18A1/AthALDH18A2. VvALDH6B3 is also located in the syntenic regions, whereas its syntenic Arabidopsis counterpart has been lost. The remaining two families (10 and 12) could not be mapped into any synteny blocks. However, we could not conclude that these two families from grape and Arabidopsis did not share a common ancestor. This may be explained by the fact that after speciation of grape and Arabidopsis, their genomes have undergone multiple rounds of significant chromosomal rearrangement and fusions, followed by selective gene loss, which can severely obscure the identification of chromosomal syntenies.
Figure 4. Synteny analysis of ALDH genes between grape and Arabidopsis.
Grape and Arabidopsis chromosomes are depicted as horizontal gray and blue bars, respectively. Grape and Arabidopsis ALDH genes are indicated by vertical orange and blue lines, respectively. Colored bars denote syntenic regions between grape and Arabidopsis chromosomes; the twisted colored bar indicates that the syntenic regions are in reverse orientation.
Expression profiles of ALDH genes under various stress conditions and during berry development and ripening
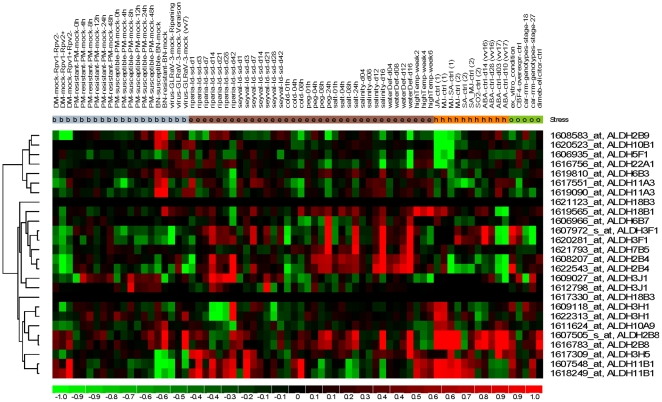
Different approaches have been undertaken to increase plant stress tolerance including manipulating and reprogramming the expression of endogenous stress-related genes. Therefore, identification and functional characterization of potential stressed-related genes provide fundamental information for future improvement of plant stress tolerance. The expression of most plant ALDH genes seems to have a common ‘stress response’ pattern within several divergent plant species from mosses to angiosperms [6]. In the present study, we investigated the responses of grape ALDH genes to various abiotic and biotic stress conditions as well as their expression patterns during grape berry development and ripening, through mining publicly available grape microarray datasets. A total of 19 experiments containing 430 hybridizations from the Affymetrix grape genome array were obtained. After manual curation, 76 comparisons between different experimental conditions and during berry development were constructed (Table S1). From the Affymetrix grape genome array, we identified 18 ALDH genes corresponding to 26 probe sets. Detailed expression of these ALDH genes is provided in Table S1. Heatmap representation of expression profiles of these ALDH genes is shown in Fig. 5, revealing that a large number of grape ALDH genes are highly responsive to certain types of abiotic or biotic stresses.
Figure 5. Hierarchical clustering of ALDH genes.
Details of the experimental conditions are provided in Table S1. Log2 based fold changes was used to create the heatmap. Differences in gene expression changes are shown in color as per the lower scale.
Abiotic stress
Abiotic stresses, such as drought, salinity and extreme temperatures are serious threats to plant growth and crop production. ALDH genes play critical roles in the adaptation of plants to various abiotic stresses [4], [35]. The microarray data analyzed included hybridizations generated from plants exposed to polyethylene glycol (PEG), and under cold, high temperature, high salinity and water-deficit stresses.
Drought and salinity are two major environmental factors determining plant productivity and distribution. It has been demonstrated that exposure to drought or salinity leads to the rapid and excessive accumulation of reactive oxygen species (ROS) in plant cells which in turn affects cellular structure and metabolism and homeostasis [36], [37]. ROS induce lipid peroxidation within lipid membranes which generates chemically reactive cleavage products, largely represented by aldehydes [38], [39]. Enhancement of ALDH activity is considered as an efficient strategy to eliminate the toxic aldehydes caused by ROS and oxidative stress [40], [41]. ALDH genes that are induced under high salinity and drought conditions have been identified in many plant species, indicating that they may play critical roles in plant adaptation to these stresses [10]. In the present study, our analysis of publicly available microarray datasets indicated that expressions of 13 grape ALDH genes were differentially expressed in at least one of the four osmotic treatments (short-term PEG, short-term salinity, long-term salinity and long-term water-deficit) (Table S1). Among them, nine (VvALDH2B4, VvALDH2B8, VvALDH3F1, VvALDH3H5, VvALDH6B3, VvALDH6B7, VvALDH7B5, VvALDH11A3 and VvALDH18B1) were up-regulated by long-term salinity and water-deficit treatments; whereas four genes (VvALDH2B9, VvALDH5F1, VvALDH11B1 and VvALDH22A1) were down-regulated (Table S1). It has been reported that plant ALDH3 genes may be an important component of ABA-dependent stress response pathways [42]. In addition, both ALDH3 and ALDH7 genes are involved in stress-regulated detoxification pathways, and ALDH7 genes are also turgor-responsive [6], [43]. In Arabidopsis, the ALDH11A3 gene encodes a non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPDH) which generates NADPH required for biosynthetic processes [6]. However, the exact function of ALDH11A3 during dehydration and salt stress remains unclear. ALDH18 genes encode P5CS (Δ1-pyrroline-5-carboxylate synthetase), a key regulatory enzyme that plays a crucial role in proline biosynthesis. Recent studies indicated that ALDH18 genes were also abiotic stress-responsive [30], [44]. Our findings are largely consistent with studies in Arabidopsis and rice that indicated ALDH genes from families 2, 3, 7, and 18 showed significant inductions in osmotically stressed plants [10], [11], [30], [35]. However, our analysis identified osmotic stress-induced genes from one additional ALDH family (ALDH6) in grape. ALDH6 genes encode methylmalonate semialdehyde dehydrogenases (MM-ALDH, EC 1.2.1.27). Mammalian ALDH6 isozymes play a role in the catabolism of valine and pyrimidines [25]. This enzyme has not been extensively studied in plants but studies have revealed that ALDH6 is an auxin-responsive gene in rice, implying its possible role in cell differentiation and organ development [45]. Further functional studies are required to reveal the exact role of these genes in grape adaptation to osmotic stress.
Cold stress, which includes chilling (<20°C) and freezing (<0°C) temperatures, adversely affects plant growth and development [46]. Under cold conditions (5°C), two ALDH genes (VvALDH11B1 and VvALDH18B1) showed increased expression while five (VvALDH2B4, VvALDH3F1, VvALDH10A9, VvALDH11A3 and VvALDH22A1) showed decreased expression. These seven cold-stress-responsive genes, except VvALDH10A9, were also regulated by drought stress, thus there may be a crosstalk between the osmotic- and cold-stress signaling pathways that regulate the expression of grape ALDH genes (Table S1). The relationship between ALDH gene expression and cold stress has not been previously documented in plants. Our analysis of ALDH genes in grape provides initial insights pertaining to cold stress and important candidates for future functional analysis. Under the heat stress, we found that none of the ALDH genes displayed significantly changed expression levels (Table S1).
Biotic stress
Little attention has been paid to the investigation on expression patterns of ALDH genes under biotic stress conditions. It has been shown that ALDH2 gene expression is regulated by powdery mildew infection in Chinese wild Vitis pseudoreticulata, suggesting potential roles of ALDH genes during plant pathogen responses [19].
Plasmopara viticola is the causal agent of downy mildew, one of the world's most catastrophic and baffling diseases of grapevine. Our microarray data analysis revealed that in a grape line (Rpv1−/Rpv2+) that is highly resistant to P. viticola, the expression of eight ALDH genes was significantly changed upon the inoculation of P. viticola, among which seven (VvALDH2B4, VvALDH2B9, VvALDH3F1, VvALDH3H1, VvALDH6B7, VvALDH7B5 and VvALDH10A9) were down-regulated and one (VvALDH11B1) was up-regulated. In the partially resistant line (Rpv1+/Rpv2−), four ALDHs (VvALDH2B4, VvALDH2B9, VvALDH7B5, VvALDH10A9) and two (VvALDH2B8 and VvALDH11B1) showed decreased and increased expressions, respectively, upon P. viticola infection; while in the susceptible line (Rpv1−/Rpv2−), none of the ALDH genes showed significant changes in their expression (Table S1). These results suggested that ALDHs could play important roles in the interaction between grapevine and P. viticola.
Powdery mildew, caused by an obligate biotrophic fungus, Uncinula necator [Schw.] Burr., is another economically important disease of grapevines. Array data analysis results indicated that the expression levels of most ALDH genes were not significantly altered upon the U. necator infection for both disease-resistant V. aestivalis genotype ‘Norton’ and disease-susceptible genotype V. vinifera ‘Cabernet sauvignon’. However, one gene, VvALDH11A3, was found to be significantly down-regulated at 4 hours post the infection in the disease-susceptible genotype (Table S1), indicating its potential role in powdery mildew development in grapevines.
Bois Noir phytoplasma is an emerging disease of V. vinifera in several regions of the world. In grape cultivar Manzoni, which is moderately resistant to Bois Noir phytoplasma, the expression of two genes (VvALDH10A9 and VvALDH11A3) was significantly increased after infection; while in Chardonnay, a highly susceptible cultivar, three genes (VvALDH2B4, VvALDH3H5 and VvALDH11B1) were significantly down-regulated and one gene (VvALDH6B3) was up-regulated (Table S1).
Viral diseases also have a serious impact on grapevine productivity and fruit quality. Among the more than 40 different viruses known to infect grapevines, the leaf roll-associated closeter-ovirus-3 (GLRaV-3) is one of the most widespread viruses [47]. Berry transcriptomes in two stages of development (veraison and ripening) in cultivar Cabernet Sauvignon infected with GLRaV-3 were analyzed. The expression of seven ALDH genes (VvALDH2B4, VvALDH2B9, VvALDH3F1, VvALDH3H1, VvALDH3H5, VvALDH11A3 and VvALDH11B1) was significantly decreased in ripening berries when infected with GLRaV-3. However, none of the ALDH genes showed significantly changed expression in the veraison stage (Table S1).
In summary, our analysis of publicly available array datasets indicated potential roles of ALDH genes in plant responses to pathogen infection. Although elucidating exact roles of these ALDH genes in plant-pathogen interactions requires further functional analysis, our findings provide a valuable increase in our knowledge base.
Hormone treatment
Plant hormones salicylic acid (SA), jasmonic acid (JA) and ethylene (ET) play central roles in biotic stress signaling upon pathogen infection [48]. Methyl Jasmonate (MJ) also affects stress responses and has a well documented role in biotic stress and wounding responses [49]. By contrast, abscisic acid (ABA) is extensively involved in responses to abiotic stresses such as drought, low temperature, and osmotic stress [48]. Analysis of expression data of grape cell-suspension cultures and berries exposed to JA, SA, ABA, MJ, or a combination of SA and MJ indicated that all 18 ALDH genes present on the array except VvALDH6B7 and VvALDH18B3 showed significantly changed expression in at least one treatment of these signaling molecules (Table S1). The expression of VvALDH2B8 was induced by all these treatments, suggesting its important role in plant stress tolerances.
ABA plays a key role in plant adaptation to adverse environmental conditions [49]. However, several studies have suggested that both ABA-dependent and ABA-independent regulatory systems are involved in stress-responsive gene expression [50]. The majority of the 13 grape ALDH genes showing significantly changed expressions in response to drought or salinity stress were also ABA-responsive. However, two genes (VvALDH6B3 and VvALDH7B5) were apparently not regulated by ABA (Table S1), confirming ABA-independent stress signaling pathways during osmotic responses.
Developmental and environmental cues
Grape berry development and ripening is a coordinated regulatory process involving genetically, hormonally, and environmentally controlled interactions of complex gene expression patterns, which ultimately leads to changes in color, texture, flavor, and aroma of the berry. The development and maturation of grape berries has been studied intensely and significant progress has been made during recent years toward elucidating the regulatory networks that determine fruit and wine quality [51]. However, the relationship between grape ALDH genes and berry development and maturation has not been reported. Our analysis of microarray data identified a number of grape ALDHs whose expression was significantly changed during berry development and ripening, e.g., the expression of VvALDH2B8, VvALDH3H5 and VvALDH18B1 was significantly increased, while the expression of VvALDH2B4 and VvALDH5F1 was significantly decreased during grape berry development and ripening (Table S1), indicating that ALDH genes could play important roles in grape berry development.
Day length is an important environmental cue for synchronizing plant growth, flowering, and dormancy with seasonality [52]. We found 15 of 18 grape ALDH genes on the array were differentially expressed during long and short photoperiods in either V. riparia or V. spp. ‘Seyval’, indicating that the expression of ALDH genes could be regulated by the photoperiod.
Conclusion
The aldehyde dehydrogenases (ALDHs) comprise a gene superfamily encoding NAD(P)+-dependent enzymes that catalyze the irreversible oxidation of a wide range of endogenous and exogenous aromatic and aliphatic aldehydes. Significant progress has been made toward the identification and characterization of ALDH gene families in model plants, with little attention paid to ALDH gene families in woody species. In the present study we identified 23 ALDHs in the V. vinifera genome, which were further grouped into ten families, and provided a unified nomenclature for the deduced ALDH polypeptides using the criteria established by the ALDH Gene Nomenclature Committee (AGNC). Our gene structure analysis showed that ALDHs from the same families contained highly similar exon-intron structures. Three alternatively spliced transcripts of ALDH2B4 were also identified. We further showed that segmental and tandem duplications have contributed substantially to the expansion of grape ALDH genes. Comparative synteny analysis between V. vinifera and Arabidopsis genomes showed that the majority of grape and Arabidopsis ALDH genes were located in syntenic regions, indicating that these ALDH genes had common ancestors. Finally, we analyzed expression profiles of grape ALDH genes in responses to various abiotic and biotic stress conditions and during grape berry development, and identified novel candidate ALDH genes that are potentially involved in grape tolerances to environmental and biotic stresses and berry development and ripening.
Methods
Identification and annotation of grape ALDH genes
Previously identified Arabidopsis ALDH sequences [6], Pfam domain PF00171 (ALDH family), PS00070 (ALDH cysteine active site), PS00687 (ALDH glutamic acid active site), KOG2450 (aldehyde dehydrogenase), KOG2451 (aldehyde dehydrogenase), KOG2453 (aldehyde dehydrogenase) and KOG2456 (aldehyde dehydrogenase) were used as queries to search in the GenBank non-redundant protein database and the Grape Genome Database (12X) (http://www.genoscope.cns.fr). Protein motifs were additionally queried against the Pfam, PROSITE, and CDD (Conserved Domain Database) [53] databases. The identified grape ALDH proteins were annotated using the criteria established by the ALDH Gene Nomenclature Committee (AGNC) [12]. Briefly, ALDH proteins with amino acid sequences more than 40% identical to previously identified ALDH sequences comprise a family, those with sequences more than 60% identity comprise a subfamily, and those with sequences less than 40% identity represent a new family.
Sequence alignments and phylogenetic analyses
Multiple alignments of ALDH protein sequences from grape, Arabidopsis [6], rice [22], maize [21], P. patens, C. reinhardtii and O. tauri [26], were performed using the ClustalW program [54]. Phylogenetic trees were constructed with the MEGA 4.0 software [55] using the neighbor-joining (NJ) method and the bootstrap test was replicated 1000 times.
Exon-intron structure analysis of grape ALDH genes
The exon-intron structures of grape ALDH genes were determined from the alignments of their coding sequences to the corresponding genomic sequences using the est2genome program [56]. The diagrams of exon-intron structures were obtained using the online program FancyGene [57].
Tandem duplication and synteny analysis
Tandem duplications of ALDH genes in the grape genome were identified by checking their physical locations in individual chromosomes. Tandem duplicated genes were defined as adjacent homologous ALDH genes on the grape chromosomes, with no more than one intervening gene. For synteny analysis, synteny blocks within the grape genome and between grape and Arabidopsis genomes were downloaded from the Plant Genome Duplication Database [58] and those containing grape ALDH genes were identified.
Expression analysis of grape ALDH genes
Affymetrix grape microarray data were downloaded from ArrayExpress [59] and PLEXdb [60] databases. A total of 19 experiments were used for our gene expression analyses (Table S1). For each microarray experiment, GCRMA method [61] was applied to perform background adjustment and normalization. The detection calls (present, marginal, or absent) for each probe set were obtained using the mas5calls function in the Affy package [62]. Genes that have absent or marginal calls across the entire arrays of an experiment were not included in the downstream statistical analysis. P-values between treatment and control conditions or during berry development for each experiment were calculated using the Limma package [63] and raw p-values of multiple tests were corrected using the False Discovery Rate (FDR) [64]. Genes with adjusted p-values (FDR) less than 0.05 were identified as differentially expressed genes. Hierarchical clustering of expression profiles of grape ALDH genes was performed using dChip [65].
Supporting Information
Alternatively spliced transcripts of VvALDH2B4 . (A) Exon-intron structure of alternatively spliced transcripts of VvALDH2B4; (B) Alignment of the 5′-open reading frame (ORF) sequences of the three alternative splice variants of VvALDH2B4. Translational initiation sites are marked with blue boxes. The 113 bp retained intron of VvALDH2B4_v2 causes a frame shift in translation and a different translational initiation site.
(PDF)
Details of publicly available grape array datasets and grape ALDH expression profiles.
(XLS)
Acknowledgments
The authors thank Dr. William Black (University of Colorado) for helpful suggestions and Prof. Simeon Kotchoni (Rutgers University) for providing the maize ALDH gene sequences.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the National Natural Science Foundation of China (30871701, 31071782, and 30671446), Program for New Century Excellent Talents in University (NCET). Additional support was provided, in part, by the National Institutes of Health (NIH) grants R01EY17963 (VV) and F31AA018248 (CB). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wang W, Vinocur B, Altman A. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta. 2003;218:1–14. doi: 10.1007/s00425-003-1105-5. [DOI] [PubMed] [Google Scholar]
- 2.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartels D. Targeting detoxification pathways: an efficient approach to obtain plants with multiple stress tolerance? Trends Plant Sci. 2001;6:284–286. doi: 10.1016/s1360-1385(01)01983-5. [DOI] [PubMed] [Google Scholar]
- 4.Kotchoni S, Bartels D. Water stress induces the up-regulation of a specific set of genes in plants: aldehyde dehydrogenase as an example. Bulg J Plant Physiol Special Issue. 2003:37–51. [Google Scholar]
- 5.Jakobi W, Ziegler D. The enzymes of detoxification. J Biol Chem. 1990;265:20715–20719. [PubMed] [Google Scholar]
- 6.Kirch HH, Bartels D, Wei YL, Schnable PS, Wood AJ. The ALDH gene superfamily of Arabidopsis. Trends Plant Sci. 2004;9:371–377. doi: 10.1016/j.tplants.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Ishitani M, Nakamura T, Han SY, Takabe T. Expression of the betaine aldehyde dehydrogenase gene in barley in response to osmotic-stress and abscisic-acid. Plant Mol Biol. 1995;27:307–315. doi: 10.1007/BF00020185. [DOI] [PubMed] [Google Scholar]
- 8.Brocker C, Lassen N, Estey T, Pappa A, Cantore M, et al. Aldehyde dehydrogenase 7A1 (ALDH7A1) is a novel enzyme involved in cellular defense against hyperosmotic stress. J Biol Chem. 2010;285:18452–18463. doi: 10.1074/jbc.M109.077925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelly GJ, Gibbs M. A mechanism for the indirect transfer of photosynthetically reduced nicotinamide adenine dinucleotide phosphate from chloroplasts to the cytoplasm. Plant Physiol. 1973;52:674–676. doi: 10.1104/pp.52.6.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirch HH, Schlingensiepen S, Kotchoni S, Sunkar R, Bartels D. Detailed expression analysis of selected genes of the aldehyde dehydrogenase (ALDH) gene superfamily in Arabidopsis thaliana. Plant Mol Biol. 2005;57:315–332. doi: 10.1007/s11103-004-7796-6. [DOI] [PubMed] [Google Scholar]
- 11.Gao CX, Han B. Evolutionary and expression study of the aldehyde dehydrogenase (ALDH) gene superfamily in rice (Oryza sativa). Gene. 2009;431:86–94. doi: 10.1016/j.gene.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Vasiliou V, Bairoch A, Tipton KF, Nebert DW. Eukaryotic aldehyde dehydrogenase (ALDH) genes: human polymorphisms, and recommended nomenclature based on divergent evolution and chromosomal mapping. Pharmacogenet Genom. 1999;9:421–434. [PubMed] [Google Scholar]
- 13.Jackson B, Brocker C, Thompson DC, Black W, Vasiliou K, et al. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum Genomics. 2011;5:283–303. doi: 10.1186/1479-7364-5-4-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 15.Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, et al. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE. 2007;2:e1326. doi: 10.1371/journal.pone.0001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goff SA, Ricke D, Lan TH, Presting G, Wang R, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 17.Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 18.Stines AP, Naylor DJ, Hoj PB, van Heeswijck R. Proline accumulation in developing grapevine fruit occurs independently of changes in the levels of delta1-pyrroline-5-carboxylate synthetase mRNA or protein. Plant Physiol. 1999;120:923–931. doi: 10.1104/pp.120.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Wang Y, Hao W. cDNA cloning and characterization of the novel genes related to aldehyde dehydrogenase from wild Chinese grape (Vitis pseudoreticulata WT Wang). DNA Sequence. 2007;18:9–18. doi: 10.1080/10425170600724618. [DOI] [PubMed] [Google Scholar]
- 20.Black WJ, Stagos D, Marchitti SA, Nebert DW, Tipton KF, et al. Human aldehyde dehydrogenase genes: alternatively spliced transcriptional variants and their suggested nomenclature. Pharmacogenet Genom. 2009;19:893–902. doi: 10.1097/FPC.0b013e3283329023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jimenez-Lopez JC, Gachomo EW, Seufferheld MJ, Kotchoni SO. The maize ALDH protein superfamily: linking structural features to functional specificities. BMC Struct Biol. 2010;10:43. doi: 10.1186/1472-6807-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotchoni SO, Jimenez-Lopez JC, Gao DY, Edwards V, Gachomo EW, et al. Modeling-dependent protein characterization of the rice aldehyde dehydrogenase (ALDH) superfamily reveals distinct functional and structural features. PLoS ONE. 2010;5:e11516. doi: 10.1371/journal.pone.0011516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu F, Schnable PS. Functional specialization of maize mitochondrial aldehyde dehydrogenases. Plant physiology. 2002;130:1657–1674. doi: 10.1104/pp.012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horton P, Park KJ, Obayashi T, Fujita N, Harada H, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4:697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood AJ, Duff RJ. The aldehyde dehydrogenase (ALDH) gene superfamily of the moss Physcomitrella patens and the algae Chlamydomonas reinhardtii and Ostreococcus tauri. Bryologist. 2009;112:1–11. [Google Scholar]
- 27.Sophos NA, Vasiliou V. Aldehyde dehydrogenase gene superfamily: the 2002 update. Chem-Biol Interact. 2003;143:5–22. doi: 10.1016/s0009-2797(02)00163-1. [DOI] [PubMed] [Google Scholar]
- 28.García-Ríos M, Fujita T, LaRosa PC, Locy RD, Clithero JM, et al. Cloning of a polycistronic cDNA from tomato encoding γ-glutamyl kinase and γ-glutamyl phosphate reductase. Proc Natl Acad Sci U S A. 1997;94:8249–8254. doi: 10.1073/pnas.94.15.8249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strizhov N, Abraham E, Okresz L, Blickling S, Zilberstein A, et al. Differential expression of two P5CS genes controlling proline accumulation during salt stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J. 1997;12:557–569. doi: 10.1046/j.1365-313x.1997.00557.x. [DOI] [PubMed] [Google Scholar]
- 30.Szekely G, Abraham E, Csepl A, Rigo G, Zsigmond L, et al. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 2008;53:11–28. doi: 10.1111/j.1365-313X.2007.03318.x. [DOI] [PubMed] [Google Scholar]
- 31.Lehmann S, Funck D, Szabados L, Rentsch D. Proline metabolism and transport in plant development. Amino Acids. 2010;39:949–962. doi: 10.1007/s00726-010-0525-3. [DOI] [PubMed] [Google Scholar]
- 32.Cannon S, Mitra A, Baumgarten A, Young N, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 34.Lyons E, Pedersen B, Kane J, Alam M, Ming R, et al. Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol. 2008;148:1772–1781. doi: 10.1104/pp.108.124867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kotchoni SO, Kuhns C, Ditzer A, Kirch HH, Bartels D. Overexpression of different aldehyde dehydrogenase genes in Arabidopsis thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant Cell Environ. 2006;29:1033–1048. doi: 10.1111/j.1365-3040.2005.01458.x. [DOI] [PubMed] [Google Scholar]
- 36.Bartels D, Sunkar R. Drought and salt tolerance in plants. Crit Rev Plant Sci. 2005;24:23–58. [Google Scholar]
- 37.Zhu JK. Plant salt tolerance. Trends Plant Sci. 2001;6:66–71. doi: 10.1016/s1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
- 38.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 39.Witz G. Biological interactions of alpha,beta-unsaturated aldehydes. Free Radic Biol Med. 1989;7:333–349. doi: 10.1016/0891-5849(89)90137-8. [DOI] [PubMed] [Google Scholar]
- 40.Sunkar R, Bartels D, Kirch HH. Overexpression of a stress inducible aldehyde dehydrogenase gene from Arabidopsis thaliana in transgenic plants improves stress tolerance. Plant J. 2003;35:452–464. doi: 10.1046/j.1365-313x.2003.01819.x. [DOI] [PubMed] [Google Scholar]
- 41.Brocker C, Cantore M, Failli P, Vasiliou V. Aldehyde dehydrogenase 7A1 (ALDH7A1) attenuates reactive aldehyde and oxidative stress induced cytotoxicity. Chem-Biol Interact. 2011;191:269–277. doi: 10.1016/j.cbi.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kirch HH, Nair A, Bartels D. Novel ABA- and dehydration-inducible aldehyde dehydrogenase genes isolated from the resurrection plant Craterostigma plantagineum and Arabidopsis thaliana. Plant J. 2001;28:555–567. doi: 10.1046/j.1365-313x.2001.01176.x. [DOI] [PubMed] [Google Scholar]
- 43.Stiti N, Missihoun TD, Kotchoni S, Kirch HH, Bartels D. Frontiers: aldehyde dehydrogenases in Arabidopsis thaliana: biochemical requirements, metabolic pathways, and functional analysis. Front Plant Physiol. 2011;2:65. doi: 10.3389/fpls.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su M, Li XF, Ma XY, Peng XJ, Zhao AG, et al. Cloning two P5CS genes from bioenergy sorghum and their expression profiles under abiotic stresses and MeJA treatment. Plant Sci. 2011;181:652–659. doi: 10.1016/j.plantsci.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 45.Oguchi K, Tanaka N, Komatsu S, Akao S. Methylmalonate-semialdehyde dehydrogenase is induced in auxin-stimulated and zinc-stimulated root formation in rice. Plant Cell Rep. 2004;22:848–858. doi: 10.1007/s00299-004-0778-y. [DOI] [PubMed] [Google Scholar]
- 46.Chinnusamy V, Zhu J, Zhu JK. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007;12:444–451. doi: 10.1016/j.tplants.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Ling KS, Zhu HY, Gonsalves D. Complete nucleotide sequence and genome organization of Grapevine leafroll-associated virus 3, type member of the genus Ampelovirus. J Gen Virol. 2004;85:2099–2012. doi: 10.1099/vir.0.80007-0. [DOI] [PubMed] [Google Scholar]
- 48.Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, et al. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 49.Huang D, Wu W, Abrams SR, Cutler AJ. The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J Exp Bot. 2008;59:2991–3007. doi: 10.1093/jxb/ern155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riera M, Valon C, Fenzi F, Giraudat J, Leung J. The genetics of adaptive responses to drought stress: abscisic acid-dependent and abscisic acid-independent signalling components. Physiol Plantarum. 2005;123:111–119. [Google Scholar]
- 51.Zenoni S, Ferrarini A, Giacomelli E, Xumerle L, Fasoli M, et al. Characterization of transcriptional complexity during berry development in Vitis vinifera using RNA-Seq. Plant Physiol. 2010;152:1787–1795. doi: 10.1104/pp.109.149716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Searle I, Coupland G. Induction of flowering by seasonal changes in photoperiod. EMBO J. 2004;23:1217–1222. doi: 10.1038/sj.emboj.7600117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, et al. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:D192–196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 56.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 57.Rambaldi D, Ciccarelli FD. FancyGene: dynamic visualization of gene structures and protein domain architectures on genomic loci. Bioinformatics. 2009;25:2281–2282. doi: 10.1093/bioinformatics/btp381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang H, Wang X, Bowers JE, Ming R, Alam M, et al. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 2008;18:1944–1954. doi: 10.1101/gr.080978.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parkinson H, Kapushesky M, Kolesnikov N, Rustici G, Shojatalab M, et al. ArrayExpress update—from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37:D868–872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wise R, Caldo R, Hong L, Shen L, Cannon E, et al. BarleyBase/PLEXdb: a unified expression profiling database for plants and plant pathogens. Methods Mol Biol. 2007;406:347–363. doi: 10.1007/978-1-59745-535-0_17. [DOI] [PubMed] [Google Scholar]
- 61.Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99:909–917. [Google Scholar]
- 62.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 63.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 64.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 65.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Nat Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alternatively spliced transcripts of VvALDH2B4 . (A) Exon-intron structure of alternatively spliced transcripts of VvALDH2B4; (B) Alignment of the 5′-open reading frame (ORF) sequences of the three alternative splice variants of VvALDH2B4. Translational initiation sites are marked with blue boxes. The 113 bp retained intron of VvALDH2B4_v2 causes a frame shift in translation and a different translational initiation site.
(PDF)
Details of publicly available grape array datasets and grape ALDH expression profiles.
(XLS)