Abstract
Background
The 16S rRNA gene is the gold standard in molecular surveys of bacterial and archaeal diversity, but it has the disadvantages that it is often multiple-copy, has little resolution below the species level and cannot be readily interpreted in an evolutionary framework. We compared the 16S rRNA marker with the single-copy, protein-coding rpoB marker by amplifying and sequencing both from a single soil sample. Because the higher genetic resolution of the rpoB gene prohibits its use as a universal marker, we employed consensus-degenerate primers targeting the Proteobacteria.
Methodology/Principal Findings
Pyrosequencing can be problematic because of the poor resolution of homopolymer runs. As these erroneous runs disrupt the reading frame of protein-coding sequences, removal of sequences containing nonsense mutations was found to be a valuable filter in addition to flowgram-based denoising. Although both markers gave similar estimates of total diversity, the rpoB marker revealed more species, requiring an order of magnitude fewer reads to obtain 90% of the true diversity. The application of population genetic methods was demonstrated on a particularly abundant sequence cluster.
Conclusions/Significance
The rpoB marker can be a complement to the 16S rRNA marker for high throughput microbial diversity studies focusing on specific taxonomic groups. Additional error filtering is possible and tests for recombination or selection can be employed.
Introduction
The small subunit ribosomal RNA gene is the recognized gold standard for estimating the phylogenetic diversity in microbial communities (e.g. [1], [2], [3]). This marker gene is universally present and has the advantage of containing both highly conserved fragments, facilitating the design of PCR primers targeting all members of a community, and more variable regions that allow for the discrimination of different microbial taxa. Furthermore, the identity of 16S rRNA gene sequences collected from the environment can be related to the taxonomic identity of sequences obtained from cultivated, characterized strains. With the introduction of high throughput (pyro)sequencing methods in studies of microbial diversity, large datasets are rapidly accruing that allow patterns of sequence recovery to be examined in depth across multiple habitats and samples [4]. However, the 16S rRNA gene is not without potential drawbacks, and the use of alternative markers has been proposed, including the beta subunit of DNA polymerase, rpoB [5], [6], [7], [8], [9], [10].
The use of the rpoB gene offers various potential advantages over standard 16S rRNA gene-based approaches. First, since most bacterial genomes contain multiple copies of the 16S rRNA gene, and copy number varies per species, extrapolation of relative abundances from gene recovery frequencies is seriously impaired. This is further complicated by the fact that sequence variation between the different 16S rRNA gene copies present exists in some genomes [11], [12]). rpoB typically occurs in a single copy [9]. Second, the high level of conservation across 16S rRNA genes can obscure most intraspecific, and sometimes interspecific (e.g. [13]) variation. In contrast, the higher resolution rpoB marker is capable of revealing molecular variation down to the population level [6]. Third, genetic divergence of rpoB correlates better with overall genomic divergence and provides better bootstrap support for phylogenetic reconstruction [6]. Fourth, given the fact that rpoB is a protein-encoding gene, the data generated from this marker is more readily interpreted in an evolutionary framework. Fifth, (pyro)sequencing error is an important confounding factor in studies of microbial diversity using 16S rRNA gene sequences [14], [15], [16]. Given that rpoB is single-copy, essential protein-encoding gene, sequence errors can be readily identified and removed if they introduce disruptions in reading frame.
Here, we test the performance of the rpoB gene as a marker for pyrosequencing-based assessments of bacterial diversity in soil, and compare results using this marker to those obtained by the conventional 16S rRNA gene marker. Due to the lower degree of conservation within the rpoB gene and our desire to provide deep sampling, we chose to restrict the current study to a single, important bacterial phylum, namely the Proteobacteria. To this effect, consensus-degenerate PCR primers were developed for the amplification of proteobacterial rpoB gene fragments for direct 454-based pyrosequencing. After the resulting sequences were subjected to denoising [15], [17], it was examined how diversity estimation was improved by additional reading-frame correction. We examined the performance of both markers with respect to quantifying diversity of the proteobacterial fraction of the community. Lastly, we explored the potential of using rpoB gene sequences to yield insights into population-level processes.
Materials and Methods
Sample Collection and DNA Extraction
A single soil sample was collected from the experimental grassland field ‘De Ossekampen’ in Wageningen, The Netherlands (51°58′14″N; 5°38′19″E) and stored at −20°C for 6 days before DNA extraction. After manually removing plant material, DNA was extracted from twelve 0.25 g subsamples using the Mo-Bio PowerSoil DNA Isolation kit (Mo-Bio Laboratories) after which the DNA was pooled. DNA extraction was performed according to the manufacturer's instructions, with the only modification being that the soil suspension was heated to 65°C for 5 minutes followed by 10 minutes horizontal shaking (Retsch mixer mill, 30 Hz).
PCR Amplification and Pyrosequencing
The V4 region of the 16S rRNA gene was amplified using forward primer 515f (5-GCCTTGCCAGCCCGCTCAGGTGTGCCAGCMGCCGCGGTAA-3) containing the 454 Life Sciences primer B, the broadly conserved bacterial primer 515F, and a 2-base linker sequence ‘TC’, and using reverse primer 806r (5-GCCTCCCTCGCGCCATCAGGGGGACTACVSGGGTATCTAAT-3) containing the 454 Life Sciences primer A and the bacterial primer 806R [18].
A fragment of the rpoB gene (region in-between primers binding sites corresponding to E. coli K-12 nucleotide positions 1671-2049) was amplified using forward primer 1f (5-GCCTTGCCAGCCCGCTCAGTCCGTGCACCCCACCcaytayggnmg-3) containing 454 Life Sciences primer B, a 2-base linker sequence ‘TC’ and consensus-degenerate primer CGTGCACCCCACCcaytayggnmg, and using reverse primer 1r (5- GCCTCCCTCGCGCCATCAGCCCACGGCCTGCckytgcatrtt-3) containing 454 Life Sciences primer A, a 2-base linker sequence ‘CC’ and consensus-degenerate primer CACGGCCTGCckytgcatrtt. The consensus degenerate primers were designed using the program iCODEHOP [19] on the basis of an alignment of 39 diverse Proteobacterial species (Table S1). This approach combines a 3′ fully degenerate primer spanning three to four codons with a longer 5′ consensus clamp containing most probable codons for that region. The pyrosequencing primer was actually not used during 454 sequencing because another primer was ligated by the sequencing company (rendering our primers longer than necessary for PCR).
PCRs contained 0.5 µl (30 pmol/µl) of each forward and reverse primer, 1 µl template DNA, 2.5 µl 10× PCR Buffer, 2.5 µl dNTP's (2 mM), 17.8 µl ddH2O water and 0.2 µl of FastStart Expand TAQ DNA polymerase (5 units/µl) (Roche). Samples were initially denatured at 95°C for 5 min, then amplified using 20 cycles for the 16S RNA gene and 30 cycles for the rpoB gene (30 s at 95°C, 45 s at 55°C and 90 s at 72°C plus a final extension of 15 mins. at 72°C to ensure complete amplification of the target region). 96 and 192 PCR amplifications were performed for the 16S rRNA- and rpoB genes, respectively, after which samples were pooled. Using many separate PCRs instead of one reaction, allowed us to minimize the number of PCR cycles and therefore potential PCR-induced bias and error.
Pooled samples were precipitated by adding 2 µl of 3 M NaOAc (pH 5.2), 1 µl of 100 mM Na2EDTA (pH 8.0) and 1 µl 20 µg/µl glycogen to 20 µl of PCR reaction and 2 volumes (96×23 µl and 192×23 µl respectively) of ice-cold 96% ethanol, followed by vortexing. Precipitated DNA was centrifuged at 3838×g for 10 min at 4°C, and the resulting pellet was washed in ice-cold 70% ethanol followed by another round of centrifugation. After removal of the supernatant, the pellet was dried in a speedvac (Thermoscientific) and resuspended in 125 µl of PCR grade ddH2O.
Because a second, low quantity amplicon was present in the PCRs targeting the rpoB gene, these samples were run on a 2% agarose gel and the desired band excised and purified using the Qiaquick Gel Extraction kit (Qiagen). The 16S rRNA gene PCR products were purified using the Qiaquick PCR Purification kit (Qiagen). Both samples were subjected to pyrosequencing on a 454 Life Sciences Genome Sequencer FLX (Roche) machine at ¼ plate capacity each by Macrogen Inc. (Seoul, Korea). All sequences were submitted in the MG-RAST database: http://metagenomics.anl.gov/linkin.cgi?metagenome=4476877.3 (rpoB) and http://metagenomics.anl.gov/linkin.cgi?metagenome=4476876.3 (16S rRNA).
PCR and Sequencing Error Correction
Raw 16S rRNA and rpoB 454 reads were filtered for reads associated with errors by removing all sequences that did not have a perfect match to the degenerate primer. Reads where the first noisy flow (0.5–0.7) occurred before position 360 out of 800 were also removed. The reads were then truncated to a length of 720 flows. The filtered reads were subsequently denoised using the AmpliconNoise-Perseus pipeline [17]: 454 errors were removed by the flowgram clustering program PyroNoise [15], forward primers were removed from forward reads, the reverse primer was removed from the reverse reads and PCR point mutations were removed by the sequence clustering program SeqNoise. The chimera classifier Perseus was then used to identify the probability of each read being chimeric through rigorous search against possible parents. Any read with a 50% or greater probability of being chimeric was removed. Finally, reverse primers were removed from forward reads and vice versa.
In addition to filtering and denoising, a further level of error correction was employed for the rpoB marker by translating nucleotide sequences into amino acids and removing any sequences possessing nonsense mutations. As a last check, any sequences containing obviously aberrant stretches of amino acids were manually removed after visual inspection of colour-coded alignments in MEGA 4 [20] (http://www.megasoftware.net/mega4/mega.html). Lastly, the reading frame correction method was employed on the undenoised dataset to allow comparison of the two methods.
OTU Construction and Taxonomy
OTUs (Operational Taxonomic Units) were constructed from distances calculated following exact pairwise alignment by the Needleman-Wunsch algorithm [21]. These pairwise distances were used in a complete linkage hierarchical clustering to generate OTUs at different nucleotide sequence difference cut-offs. OTUs were constructed for both the 16S rRNA and rpoB forward and reverse cleaned sequences. In addition, OTUs were calculated for the two genes using both forward and reverse sequences; the pairwise alignments and resulting distances ignore terminal gaps, allowing composite OTUs to be produced. The most frequent sequence in an OTU was then identified for every cluster. The 16S rRNA and rpoB sequence reads were classified by employing the same database (1477 prokaryote genomes; NCBI Complete Microbial Genomes, February 2011), BLAST type (nucleotide) and classification algorithm (LCA; using the default parameters in MEGAN [22]).
Estimation of Species Diversity
Both markers can be compared at the level of total diversity quantified by each, but, since the rpoB marker was specifically designed to target Proteobacteria, the comparison is more meaningfully made when restricted to this phylum. Following OTU construction at 2.3% sequence difference for rpoB and 1% for the 16S rRNA gene (see Results for a justification of OTU cut-off values), we compared OTU diversity for reads classified as Proteobacteria. Rarefaction curves were calculated to show the effect of sample size on observed diversity. To estimate the total diversity in the community, a Bayesian method was used to fit taxa abundance distributions to the observed OTU frequencies [23]. As predictions have previously been found to be sensitive to the choice of distribution [23], both the log-normal and Sichel distributions were used.
Population Genetic Analyses
Partial rpoB gene sequences classified by MEGAN as belonging to the same species were selected for population-level analyses. These DNA sequences were then aligned at the amino acid sequence level using the Clustal algorithm in MEGA 4 [20]. A Minimum Spanning Tree based on nucleotide differences was subsequently constructed using HapStar (http://www.fo.am/hapstar) [24]. A NeighborNet network analysis was constructed using SplitsTree4 (http://www.splitstree.org) [25]. The PHI test of homologous recombination [26] was also conducted using SplitsTree4.
Results
Sequence Error Correction
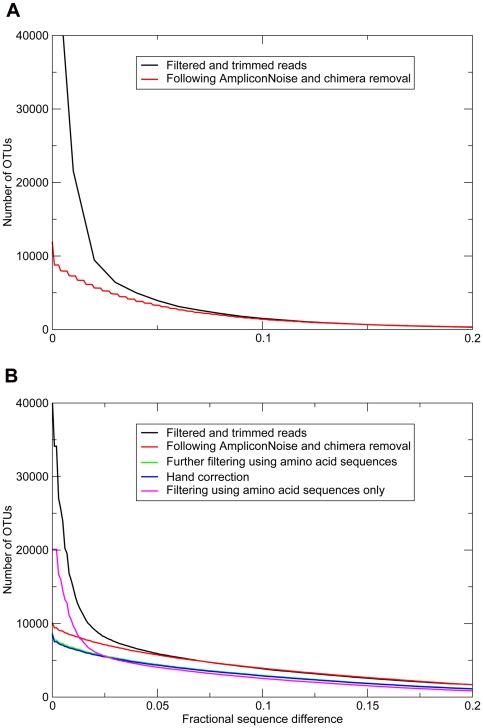
As has been demonstrated before [17], noise removal by AmpliconNoise and Perseus significantly reduced observed OTU richness. This is clearly displayed in Fig. 1, where the number of unique Operational Taxonomic Units (OTUs) for denoised and undenoised 16S rRNA and rpoB marker genes are plotted as a function of OTU cut-off value. Subsequent to denoising, rpoB nucleotide sequences were translated into amino acid sequences and any sequences containing nonsense mutations (i.e. containing stop codons or frame shifts) were removed, followed by back translation. After this automated step, 382 sequences out of 31,684 were manually removed as they were visually very dissimilar (Fig. 1B). At the level of unique sequences (0% OTU cut-off), this reading frame correction lowered observed denoised diversity with an additional 1.2%. Although this effect is small compared to the effect of denoising (which removed 27.6% of unique OTUs), the resulting data were largely freed of artefactual sequences that could negatively affect downstream population genetic analyses. When reading frame correction was applied to undenoised sequences, an excess of OTUs was observed above the 2.3% species cut-off compared to the combined use of denoising and reading-frame correction, but OTU richness was nearly identical for both approaches below this cut-off (Fig. 1B).
Figure 1. Number of OTUs as a function of fractional sequence difference (OTU cut-off) for the 16S rRNA marker gene (A) and the rpoB marker gene (B).
OTU number is plotted for filtered and trimmed sequences (undenoised), denoised sequences and denoised- and reading frame corrected sequences (automated and manual correction). The latter treatments were only applicable to rpoB.
Observed and Estimated Diversity
Our soil sample displayed a taxonomic composition characteristic for temperate soils [27] with abundant Proteobacteria, Acidobacteria and Actinobacteria as determined from the 16S rRNA gene sequences (Fig. S1). In order to compare the overlap in sequence target of the rpoB and 16S rRNA primers, we compared taxonomies using the same database and classifier (Fig. S1). It was obvious that the rpoB primers did not exclusively target Proteobacteria, with for instance large numbers of the unrelated Actinobacteria also amplified. The distributions among the proteobacterial classes also exhibited differences, which is to be expected as primer sets will inevitably have distinct biases (Fig. S1).
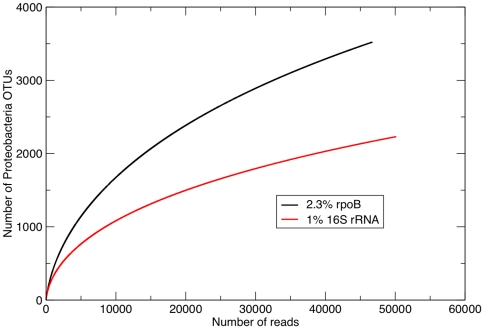
Proteobacterial diversity was quantified at the OTU cut-off level that corresponded to an average DNA-DNA Hybridization (DDH) value of 70% used to delineate bacterial species [6], [7]. This value is 1% for the 16S rRNA gene [28] (although wider cut-offs are commonly employed for this gene as well). A value of 2.3% rpoB divergence has been shown to correspond best to an average 70% DDH value [7] (this also held true for the fragment of the rpoB gene utilized here, data not shown). Figure 2 shows an accumulation curve plotting the number of proteobacterial OTUs as a function of sampled reads, using the 16S rRNA and rpoB species definition OTU cut-offs. It is immediately apparent that the curves are very different, with the 16S rRNA marker indicating a distribution skewed towards more frequently occurring species (the slower accumulation of OTU richness being caused by repeated sampling of identical OTUs). A considerably higher number of proteobacterial species were found using the rpoB marker (Table 1).
Figure 2. Rarefaction curves showing mean expected OTU number for Proteobacteria as a function of sample size.
The 1% and 2.3% cut-offs for 16S rRNA and rpoB are chosen to reflect species definitions (see text).
Table 1. Estimation of species richness for sequences classified as Proteobacteria.
| marker (OTU cut-off) | reads | observed n species | estimated n specieslog-normal distribution* | estimated n species Sichel distribution* | estimated n species Chao estimator |
| rpoB (2.3%) | 46,704 | 3521 | 15,501 : 19,897 : 25,829 | 7299 : 8166 : 9401 | 5646 |
| 16S rRNA (1%) | 50,145 | 2231 | 12,384 : 18,900 : 42,824 | 5036 : 5978 : 7699 | 3490 |
* = The parametric total diversity estimates are given as lower 95% confidence interval : median : upper 95% confidence interval.
The total proteobacterial species richness for both markers was estimated using fitting of the Sichel and lognormal distributions, as well as using the more conservative, non-parametric Chao estimator (Table 1). Both the log-normal and Sichel distributions yielded a greater total richness for rpoB sequences as compared to 16S rRNA gene sequences, but since confidence intervals of the two estimators overlap, this difference was not significant. The log-normal distribution was a marginally, but not significantly, better fit than the Sichel distribution for the rpoB data, but the converse was true for the 16S rRNA gene.
The proteobacterial OTU frequency-abundance distributions for the rpoB and 16S rRNA gene markers were highly skewed and well described by the fitted log-normal and Sichel functions (Fig. S2 A and B). The curve for the 16S rRNA gene was flatter than that for the rpoB gene, indicating a more skewed distribution. This effect was subtle, but it is reinforced by calculating the amount of sampling required to observe 90% of the true diversity [23] (Table 2). The choice of distribution has a large impact on required sampling effort, but for both distributions the median number of reads required is at least an order of magnitude larger for the 16S rRNA than for the rpoB gene. This result is significant in that the predictions for the two genes for the same choice of distribution lie outside of each other's confidence intervals (Table 2).
Table 2. 90% sampling effort, defined as number of reads required to observe 90% of the true diversity, for proteobacterial species.
| marker (OTU cut-off) | 90% sampling effortlog-normal distribution* | 90% sampling effortSichel distribution* |
| rpoB (2.3%) | 2.46e+07 : 7.15e+07 : 2.21e+08 | 4.27e+05 : 5.86e+05 : 8.65e+05 |
| 16S rRNA (1%) | 5.36e+08 : 3.67e+09 : 1.63e+11 | 2.55e+06 : 4.16e+06 : 8.36e+06 |
* = The parametric total diversity estimates are given as lower 95% confidence interval : median : upper 95% confidence interval.
Population Genetic Analyses
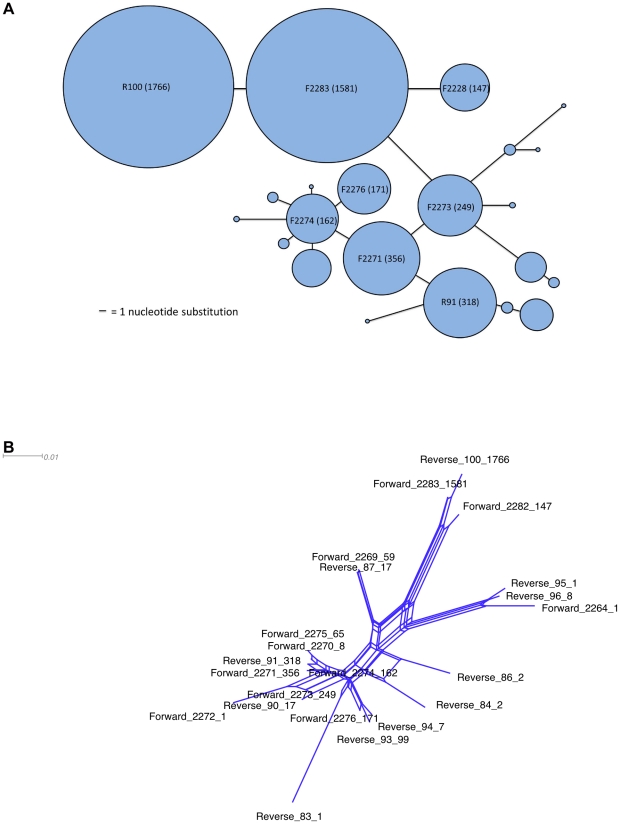
In order to demonstrate the suitability of pyrosequencing data from the given marker gene for analyses within a population-level framework, all sequences with more than 94% similarity to a particularly abundant sequence (1766 identical individuals, 82% nucleotide identity to both Anaeromyxobacter dehalogenans 2CP-1 and Anaeromyxobacter sp. K) were examined in more detail. First, a Minimum Spanning Tree was created (Fig. 3A). MST analysis is a useful tool for visualizing evolutionary relationships at the population level, where ancestral and evolved genotypes coexist (unlike phylogenetic trees, where extant genotypes are represented by the tips of branches and ancestors are represented by internal nodes). Such ‘population snapshots’ have been primarily generated with allelic MLST data using the eBurst algorithm [29], [30] (http://eburst.mlst.net/) but have also been applied to single-gene nucleotide data obtained from isolates (e.g. [31]).
Figure 3. Population-level analyses.
A: a Minimum Spanning Tree for all sequences more than 94% similar to abundant Anaeromyxobacter sequence R100. Circle size equates with the number of sequences, bar length equates with the number of nucleotide substitutions between sequences. B: a NeighbourNet phylogenetic network based on the same sequences as in A.
The shape of the MST (Fig. 3A) does not point to an epidemic population structure, where a common, presumably well-adapted genotype is connected to multiple, less frequent ‘offshoot’ genotypes [32]. Instead, multiple common genotypes are all connected; such a ‘straggly’ appearance has been shown to be the result of high homologous recombination rates in eBurst-generated population snapshots [33]. It remains to be investigated whether recombination influences the MST algorithm used here in the same way, but a PHI test indicated highly significant levels of recombination in this dataset (p<0.0001). The influence of recombination is also indicated by the reticulated structure of the associated phylogenetic network (Fig. 3B).
Since proteobacterial diversity is so very high, most sequences in this dataset were separated by many substitutions. In order to truly zoom in on population-level processes, a much greater amount of sequencing would be needed and/or more specific primers would need to be employed. This will give better coverage below the species-level and allow for more options with respect to tests of recombination and selection. When well-characterized functional genes are chosen, it should also be possible to survey mutations of known effect on protein function. This was actually also possible for the fragment of rpoB sequenced here, as it covers a region (cluster II) where mutations conferring rifampicin resistance have been found in various species [34]. None of these mutations were found to be present in our dataset.
Discussion
In this study, we sought to compare the merit of the protein-coding rpoB marker gene with that of the standard 16S rRNA marker gene for assessing bacterial community diversity by amplifying and pyrosequencing both genes from the same soil sample. Single-copy protein-coding genes essential for cell functioning have an ‘internal check’ in that any disruption of the reading frame found must be the result of experimental error. Roche 454 pyrosequencing has the advantage of yielding a large number of sequences, but it is also vulnerable to miscalls of homopolymer runs that cause frame shifts [15]. Denoising algorithms can mitigate this effect to a large extent, as this and previous studies [15], [17] have shown, but reading frame correction was found to provide a clear additional benefit (Fig. 1). Reading frame correction in rpoB was found to perform equally well as the combined use of denoising and reading frame correction for taxa above the species-level (Fig. 1). This correction step might prove especially useful for sequencing protein-coding genes using the Illumina method, for which no denoising program yet exists.
In any ecosystem survey, it is important to quantify diversity in such a way that it can be related to the basic unit of community diversity, the species, as unambiguously as possible. Current microbial ecology studies use 16S rRNA gene sequences as a proxy for species diversity. However, the divergence of rpoB correlates better to overall genomic divergence [7], allowing for the selection of a more reliable OTU cut-off. The observed richness (Table 1) and diversity (Fig. 2) of proteobacterial species was markedly higher when quantified using rpoB. This phenomenon is likely to be at least partly due to the effect of copy number, where individual bacteria add multiple identical (or highly similar) copies of 16S rRNA genes to the sequenced pool. Remarkably, however, the total estimated number of proteobacterial species did not differ between the two markers (Table 1). This implies that rpoB can be a more efficient marker than the 16S rRNA gene when a subset of total diversity is targeted, revealing more species for any population sample and requiring an order of magnitude fewer reads to obtain 90% of the true diversity (Table 2).
Although its lower level of conservation makes it less suited for high-level classification, rpoB offers greater resolution than the 16S rRNA gene, making it in theory a better marker to distinguish between strains and species. The greater resolution of protein-encoding housekeeping genes compared to ribosomal genes has been appreciated for some time, and many isolation-based studies exist employing the rpoB gene (using this gene either exclusively, e.g. [35], [36], [37], or using this gene in combination with other housekeeping genes in Multilocus Sequence Typing (MLST) studies, e.g. [38], [39], [40]). In addition, the rpoB gene and similarly conserved single-copy markers have been extracted from metagenomic datasets for specific analyses [41], [42].
Our study is one of the first to apply PCR-based pyrosequencing to a protein-coding gene from the environment (see also [43], [44]). Three main disadvantages are associated with the use of rpoB (or any other protein-coding marker). First, it is not conserved enough to be of use as a universal marker and only a subset of the microbial community can be targeted. Second, assigning taxonomy to the sequences is problematic because no appropriate databases and classifiers are available. Third, experiments using complex but defined communities are necessary to rigorously test primer bias (although this holds true for the 16S rRNA marker also). However, in comparison with the 16S marker, single-copy protein-coding genes offer several advantages: there is a possibility for additional error-correction, sampling efficiency could be higher, insights can be provided into population-level processes such as homologous recombination rate and mutations of known effect can be surveyed. Finally, with more advanced computing options becoming available, such data can be analyzed not only on the level of OTUs, but also directly at the sequence level, for instance in neutral models developed in population genetics [45].
Supporting Information
MEGAN classification of 16S RNA (red) and rpoB (blue) genes up to the Class level (normalized read frequencies).
(TIF)
The frequency of 16S rRNA 1% Proteobacteria OTUs (A) and rpoB 2.3% Proteobacteria OTUs (B) with a given abundance. The axes are log scaled and data points have been aggregated to reduce observed noise. We also show fits of the log-normal and Sichel distributions to this data.
(TIF)
Strains used to design the rpoB primers.
(DOCX)
Acknowledgments
We thank Rob Geerts at Plant Research International, Wageningen, The Netherlands for access to the Ossekampen field site and the provision of metadata (made available in the MG-RAST sequence deposition). This is Publication No. 5185 of the Netherlands Institute of Ecology (NIOO-KNAW).
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by a Netherlands Organisation for Scientific Research (NWO) Vici grant to Dr. Kowalchuk (http://www.nwo.nl/nwohome.nsf/pages/sppd_5r2qe7_eng). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hugenholtz P, Goebel BM, Pace NR. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, et al. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward DM, Weller R, Bateson MM. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;345:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- 4.Huse SM, Dethlefsen L, Huber JA, Mark Welch D, Relman DA, et al. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Case RJ, Boucher Y, Dahllof I, Holmstrom C, Doolittle WF, et al. Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl Environ Microbiol. 2007;73:278–288. doi: 10.1128/AEM.01177-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adekambi T, Drancourt M, Raoult D. The rpoB gene as a tool for clinical microbiologists. Trends Microbiol. 2009;17:37–45. doi: 10.1016/j.tim.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Adekambi T, Shinnick TM, Raoult D, Drancourt M. Complete rpoB gene sequencing as a suitable supplement to DNA-DNA hybridization for bacterial species and genus delineation. Int J Syst Evol Microbiol. 2008;58:1807–1814. doi: 10.1099/ijs.0.65440-0. [DOI] [PubMed] [Google Scholar]
- 8.Mollet C, Drancourt M, Raoult D. rpoB sequence analysis as a novel basis for bacterial identification. Mol Microbiol. 1997;26:1005–1011. doi: 10.1046/j.1365-2958.1997.6382009.x. [DOI] [PubMed] [Google Scholar]
- 9.Santos SR, Ochman H. Identification and phylogenetic sorting of bacterial lineages with universally conserved genes and proteins. Environ Microbiol. 2004;6:754–759. doi: 10.1111/j.1462-2920.2004.00617.x. [DOI] [PubMed] [Google Scholar]
- 10.Wooley JC, Godzik A, Friedberg I. A primer on metagenomics. PLoS Comput Biol. 2010;6:e1000667. doi: 10.1371/journal.pcbi.1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pei AY, Oberdorf WE, Nossa CW, Agarwal A, Chokshi P, et al. Diversity of 16S rRNA genes within individual prokaryotic genomes. Appl Environ Microbiol. 2010;76:3886–3897. doi: 10.1128/AEM.02953-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh DA, Bapteste E, Kamekura M, Doolittle WF. Evolution of the RNA polymerase B′ subunit gene (rpoB′) in Halobacteriales: a complementary molecular marker to the SSU rRNA gene. Mol Biol Evol. 2004;21:2340–2351. doi: 10.1093/molbev/msh248. [DOI] [PubMed] [Google Scholar]
- 13.Fox GE, Wisotzkey JD, Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 14.Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2010;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- 15.Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- 16.Reeder J, Knight R. The ‘rare biosphere’: a reality check. Nat Methods. 2009;6:636–637. doi: 10.1038/nmeth0909-636. [DOI] [PubMed] [Google Scholar]
- 17.Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing Noise From Pyrosequenced Amplicons. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci U S A. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rose T, Henikoff J, Henikoff S. CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucl Acids Res. 2003;31:3763–3766. doi: 10.1093/nar/gkg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 21.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quince C, Curtis TP, Sloan WT. The rational exploration of microbial diversity. ISME J. 2008;2:997–1006. doi: 10.1038/ismej.2008.69. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 24.Teacher AG, Griffiths DJ. HapStar: automated haplotype network layout and visualization. Mol Ecol Resour. 2011;11:151–153. doi: 10.1111/j.1755-0998.2010.02890.x. [DOI] [PubMed] [Google Scholar]
- 25.Huson DH, Bryant D. Application of Phylogenetic Networks in Evolutionary Studies. Mol Biol Evol. 2005;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 26.Bruen TC, Philippe H, Bryant D. A simple and robust statistical test to detect the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janssen PH. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol. 2006;72:1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- 29.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spratt BG, Hanage WP, Li B, Aanensen DM, Feil EJ. Displaying the relatedness among isolates of bacterial species – the eBURST approach. FEMS Microbiol Lett. 2004;241:129–134. doi: 10.1016/j.femsle.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 31.Zamborsky DJ, Nishiguchi MK. Phylogeographical patterns among Mediterranean sepiolid squids and their Vibrio symbionts: environment drives specificity among sympatric species. Appl Environ Microbiol. 2011;77:642–649. doi: 10.1128/AEM.02105-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feil EJ. Small change: keeping pace with microevolution. Nat Rev Microbiol. 2004;2:483–495. doi: 10.1038/nrmicro904. [DOI] [PubMed] [Google Scholar]
- 33.Turner KM, Hanage WP, Fraser C, Connor TR, Spratt BG. Assessing the reliability of eBURST using simulated populations with known ancestry. BMC Microbiol. 2007;7:30. doi: 10.1186/1471-2180-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, et al. Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 35.Ben Salah I, Adekambi T, Raoult D, Drancourt M. rpoB sequence-based identification of Mycobacterium avium complex species. Microbiol. 2008;154:3715–3723. doi: 10.1099/mic.0.2008/020164-0. [DOI] [PubMed] [Google Scholar]
- 36.Gaget V, Gribaldo S, Tandeau de Marsac N. A rpoB signature sequence provides unique resolution for the molecular typing of cyanobacteria. Int J Syst Evol Microbiol. 2011;61:170–183. doi: 10.1099/ijs.0.019018-0. [DOI] [PubMed] [Google Scholar]
- 37.Minegishi H, Kamekura M, Itoh T, Echigo A, Usami R, et al. Further refinement of the phylogeny of the Halobacteriaceae based on the full-length RNA polymerase subunit B′ (rpoB′) gene. Int J Syst Evol Microbiol. 2010;60:2398–2408. doi: 10.1099/ijs.0.017160-0. [DOI] [PubMed] [Google Scholar]
- 38.Olvera A, Cerda-Cuellar M, Aragon V. Study of the population structure of Haemophilus parasuis by multilocus sequence typing. Microbiol. 2006;152:3683–3690. doi: 10.1099/mic.0.29254-0. [DOI] [PubMed] [Google Scholar]
- 39.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salerno A, Deletoile A, Lefevre M, Ciznar I, Krovacek K, et al. Recombining population structure of the Enterobacteriaceae Plesiomonas shigelloides revealed by multilocus sequence typing. J Bacteriol. 2007;189:7808–7818. doi: 10.1128/JB.00796-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu M, Eisen JA. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 2008;9:R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu D, Wu M, Halpern A, Rusch DB, Yooseph S, et al. Stalking the fourth domain in metagenomic data: searching for, discovering, and interpreting novel, deep branches in marker gene phylogenetic trees. PLoS ONE. 2011;6:e18011. doi: 10.1371/journal.pone.0018011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hou XL, Cao QY, Jia HY, Chen Z. Pyrosequencing analysis of the gyrB gene to differentiate bacteria responsible for diarrheal diseases. Eur J Clin Microbiol Infect Dis. 2008;27:587–596. doi: 10.1007/s10096-008-0477-7. [DOI] [PubMed] [Google Scholar]
- 44.Oakley BB, Carbonero F, Dowd SE, Hawkins RJ, Purdy KJ. Contrasting patterns of niche partitioning between two anaerobic terminal oxidizers of organic matter. ISME J. 2011 doi: 10.1038/ismej.2011.165. doi: 10.1038/ismej.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuhner MK. Coalescent genealogy samplers: windows into population history. Trends Ecol Evol. 2009;24:86–93. doi: 10.1016/j.tree.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MEGAN classification of 16S RNA (red) and rpoB (blue) genes up to the Class level (normalized read frequencies).
(TIF)
The frequency of 16S rRNA 1% Proteobacteria OTUs (A) and rpoB 2.3% Proteobacteria OTUs (B) with a given abundance. The axes are log scaled and data points have been aggregated to reduce observed noise. We also show fits of the log-normal and Sichel distributions to this data.
(TIF)
Strains used to design the rpoB primers.
(DOCX)