Abstract
Shiga-like toxins are ribosome-inactivating proteins (RIP) produced by pathogenic E. coli strains that are responsible for hemorrhagic colitis and hemolytic uremic syndrome. The catalytic A1 chain of Shiga-like toxin 1 (SLT-1), a representative RIP, first docks onto a conserved peptide SD[D/E]DMGFGLFD located at the C-terminus of all three eukaryotic ribosomal stalk proteins and halts protein synthesis through the depurination of an adenine base in the sarcin-ricin loop of 28S rRNA. Here, we report that the A1 chain of SLT-1 rapidly binds to and dissociates from the C-terminal peptide with a monomeric dissociation constant of 13 µM. An alanine scan performed on the conserved peptide revealed that the SLT-1 A1 chain interacts with the anionic tripeptide DDD and the hydrophobic tetrapeptide motif FGLF within its sequence. Based on these 2 peptide motifs, SLT-1 A1 variants were generated that displayed decreased affinities for the stalk protein C-terminus and also correlated with reduced ribosome-inactivating activities in relation to the wild-type A1 chain. The toxin-peptide interaction and subsequent toxicity were shown to be mediated by cationic and hydrophobic docking surfaces on the SLT-1 catalytic domain. These docking surfaces are located on the opposite face of the catalytic cleft and suggest that the docking of the A1 chain to SDDDMGFGLFD may reorient its catalytic domain to face its RNA substrate. More importantly, both the delineated A1 chain ribosomal docking surfaces and the ribosomal peptide itself represent a target and a scaffold, respectively, for the design of generic inhibitors to block the action of RIPs.
Introduction
Shiga toxins such as Shiga-like toxin 1 (SLT-1) are produced by enteropathogenic Escherichia coli strains and represent the major cause of hemorrhagic colitis and hemolytic uremic syndrome [1], [2]. SLT-1 is a type II ribosome-inactivating protein (RIP) composed of a catalytically active A subunit non-covalently associated with a pentamer of B-subunits [3], [4]. This pentamer binds to the glycolipid globotriaosylceramide (CD77,Gb3), an event that leads to its internalization [5], [6], [7]. SLT-1 then traffics in a retrograde manner through the Golgi apparatus where it is proteolytically cleaved into an N-terminal catalytic A1 domain and a C-terminal A2 fragment non-covalently associated with its B-pentamer. Both A chain fragments remain linked by a single disulfide bond which is thought to be reduced in the ER lumen [8], [9], [10]. The A1 domain is then retrotranslocated to the cytosol by virtue of its newly exposed hydrophobic C-terminus, where it eventually docks onto ribosomes and subsequently depurinates a single adenine base (A4324) in the sarcin-ricin loop (SRL) of 28S rRNA [11], [12], [13], [14], [15]. This depurination event creates an apurinic site that prevents elongation factor 1 (EF-1)-dependent amino-acyl tRNA from binding to the ribosome and EF-2-catalysed translocation during elongation, leading to an inhibition of protein synthesis [16], [17], [18].
The protein component of the ribosome was first shown to contribute to the toxicity of RIPs when a 105 fold increase in depurination rate was observed for ricin on native ribosomes when compared to protein-depleted ribosomes [19]. SLT-1 as well as other structurally and functionally related RIPs, require their docking to ribosomal proteins in addition to rRNA to maintain their optimal depurination rate and cytotoxic function [15], [19], [20], [21]. More recently, it has been revealed that the ribosomal protein components required for interacting with either type I (trichosanthin (TCS)) or type II (SLT-1 and ricin) RIPs are the ribosomal proteins RPP0, RPLP1 and RPLP2 (P0, P1, and P2) [15], [20], [22], [23]. These three proteins form the ribosomal stalk which is required for the binding of elongation factors leading to protein translation [24], [25], [26]. The eukaryotic stalk structure is composed of two heterodimers of the P1 and P2 proteins [27], [28], [29], which interact by virtue of the N-terminus of the P1 protein, at two specific locations on the P0 protein [30], [31], [32], [33], which subsequently binds to rRNA [34].
We have previously shown that the A1 chain of SLT-1 interacts with the ribosomal stalk proteins P0, P1, and P2 via a conserved C-terminal peptide (SDXDMGFGLFD, where X = D or E) [15]. In the present study, we demonstrate by yeast-2-hybrid (Y2H) and surface plasmon resonance (SPR) that the A1 chain of SLT-1 interacts with the C-terminal ribosomal stalk peptide with a micromolar dissociation constant. Specifically, the interaction of the A1 chain with the conserved C-terminal peptide SDDDMGFGLFD common to all three ribosomal stalk proteins exhibits a modest binding constant (Kd 13 µM), towards the monovalent peptide, with rapid on and off rates. This transient interaction is mediated by distinct charged and hydrophobic surfaces on the SLT-1 A1 chain, which are also essential for its full catalytic activity. Moreover, alanine-scanning mutagenesis revealed that anionic tripeptide and hydrophobic tetrapeptide motifs within the sequence SDDDMGFGLFD represent key anchor residues recognized by the A1 chain. These findings suggest that the nature of these interactions may play a guiding role in properly orientating RIP catalytic domains towards their substrate, the sarcin-ricin loop, and may represent a scaffold for the generation of RIP-specific antidotes.
Methods
Protein expression and purification
The wild-type SLT-1 was expressed as an N-terminal His8-tagged fusion construct in the E. coli strain JM101 (Agilent Technologies, Mississauga, ON), and purified as an AB5 holotoxin on nickel-NTA resin (Sigma-Aldrich, St. Louis, MO). The A1 chain was further purified from the holotoxin by first treating the purified AB5 variants with the protease furin (New England BioLabs, Ipswich, MA) and reducing the disulfide bond with 10 mM DTT. The A1 chain was then recovered on Nickel-NTA resin in the presence of a guanidine-HCl gradient to remove the untagged A2 chain and B subunits, followed by a re-folding step in PBS.
SLT-1 mutant variants corresponding to those that exhibited a lack of interaction as defined by Y2H screens (R172A, R176A, R179A, R188A, R176/179/188A, V191A, F226A, L233A, and S235A) were created by multi-step PCR using Taq polymerase. The first PCR step consisted of two reactions: (1) using sense primer 2 and one of the mutagenic antisense primers and (2) using an antisense primer 2 and one of the mutagenic sense primers (Table S1). The second step consisted of a single reaction using the previous PCR reactions as templates with sense and antisense primer 2 which allowed for the amplification of the entire mutated sequence including the incorporation of unique restriction endonuclease sites. These mutant gene sequences were digested with the appropriate restriction endonuclease, cloned into the NheI and XhoI sites of the pECHE10a vector (Molecular Templates Inc., Austin, Texas). The resulting SLT-1 A1 mutants were expressed and purified in the same manner as the wild-type SLT-1 A1 chain. The above-mentioned SLT-1 A1 variants used in subsequent experiments were judged by densitometry to be ≥85% pure (Figure S1).
Peptide Synthesis
Synthetic peptides corresponding to the final 17, 11, and 7 residues of the C-terminal domain of ribosomal proteins P1 and P2, a control peptide, as well as all alanine-containing peptide variants of the final 11 residue peptide SDDDMGFGLFD used to measure binding affinities were assembled using the 9-fluorenylmethoxycarbonyl (Fmoc) method and Wang resin on a PS3 Peptide Synthesizer (Protein Technologies Inc., Tucson, AZ). Fmoc-Asp(OMpe)-OH (NovaBiochem, Gibbstown, NJ) and Fmoc-Glu(OBt)-Ser(ΨMe, MePro)-OH (NovaBiochem) dipeptide were used to avoid aspartimide formation. The Fmoc protecting groups were removed using 20% Piperidine/0.1 M HOBt in DMF during synthesis. The N-α-amino group (N-terminus) of the peptides was labeled overnight with biotin using a ten-fold excess of biotin (Molecular Probes, Burlington, ON) in the presence of HCTU/HOBt and DIPEA. Biotinylated peptides were cleaved from their support using 3 mL of a TFA/TIS/EDT/Water (92.5∶2.5∶2.5∶2.5%) mixture for 4 hrs at RT and purified by HPLC on a C18 semi-preparative column using an acetonitrile gradient from 5% to 100% in 20 min. Peptide masses were then confirmed by mass spectrometry.
Yeast-2-Hybrid
Yeast ribosomes are rapidly inactivated by the A chains of RIPs such as SLT-1. A catalytically inactive form of the A1 chain (CIA1) was thus generated by introducing two mutations, namely E167A and R170A, within its catalytic region [11], [15]. These two point mutations decrease the toxicity of the A1 chain by 10,000 fold, therefore enabling yeast to grow during the expression of the A1 chain of SLT-1 [35], [36]. The yeast-2-hybrid (Y2H) technique [37], takes advantage of the GAL4 transcription factor that can be spliced into 2 complementary domains: a DNA binding domain (DNA-BD) and a transcription activation domain (AD). When pairs of bait/prey proteins interact, the GAL4 DNA-BD and transcription AD modules are assembled leading to the activation of survival genes (i.e.: HIS3).
The CIA1 SLT-1 gene sequence was used as a template to construct several charged and hydrophobic single point mutations to alanine by multistep PCR using sets of mutagenic primers (Table S1). The CIA1 as well as the point mutants were cloned into the bait vector pGBKT7 (Clontech, Mountain View, CA) between the NdeI and BamHI sites and expressed as fusions to the C-terminus of the GAL4 DNA-binding domain (GAL4 DNA-BD). The human gene sequence corresponding to RPLP2 (P2) was cloned into the prey vector pGADT7 (Clontech) and expressed as a fusion construct to the GAL4 activation domain (GAL4-AD). The pGBKT7-CIA1 or one of the charged or hydrophobic point mutants were co-transformed with pGADT7-P2 separately into the yeast strain AH109 [MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, URA3::MEL1UAS-MEL1TATA-lacZ, MEL1] (Clontech).
Transformed cells were plated onto SD agar lacking Trp and Leu (−Trp/−Leu), to select for the presence of both plasmids, and incubated for 72 h at 30°C. A single colony of each transformation was then inoculated into SD −Trp/−Leu broth and shaken at 30°C overnight. Overnight cultures were centrifuged at 4000 rpm for five minutes followed by washing and equilibration to OD600 of 1.0 in phosphate-buffered saline (PBS). Samples were then serially diluted 10-fold followed by spotting onto SD agar −Trp/−Leu to select for the presence of both plasmids and SD −Trp/−Leu/−His to select for an interaction between the two proteins. Plates were incubated for 72 h at 30°C.
Surface Plasmon Resonance Measurements
Toxin-peptide binding affinities were assessed by surface plasmon resonance (SPR) using a ProteOnXPR36 array biosensor [38] (Bio-Rad) in PBS buffer at 25°C. A neutravidin (NLC) sensor chip (Bio-Rad) was preconditioned by four pulses of 1 M NaCl, 50 mM NaOH, and 100 mM HCl, which was followed by an equilibration step in PBS. Biotinylated peptides corresponding to the final 17, 11, and 7 residues of the conserved peptide and a control peptide with no sequence homology were immobilized on the NLC sensor chip followed by a wash step in 1 M NaCl, to remove unbound peptide and a PBS wash to equilibrate the chip. Purified SLT-1 wild-type A1 chain was tested at 10 different concentrations in quadruplicate beginning with 30 µM and diluting 2-fold in PBS to 60 nM, followed by an injection of PBS alone. The wild-type A1 chain was exposed to all ribosomal peptide variants. Each concentration of SLT-1 A1 chain variant was exposed to the NLC chip harboring the ribosomal peptides at a flow rate of 50 µl/min for 60 s with a dissociation time of 120 s. A 1 M NaCl wash step was performed for 18 s at a flow rate of 100 µl/min following each protein concentration. The chip was then equilibrated with PBS at a flow rate of 100 µl/min for 120 s.
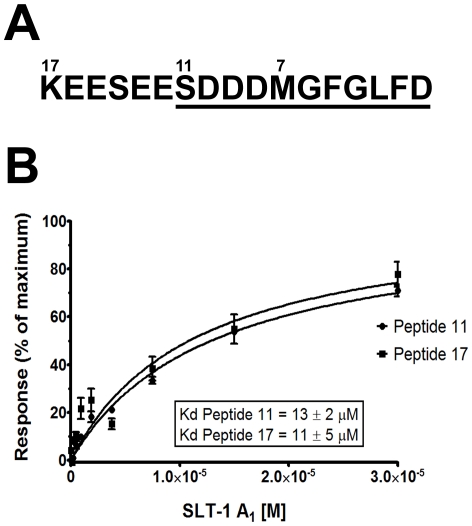
Additional SPR experiments measuring (i) the affinities of charge and hydrophobic A1 chain mutants, (ii) the contributions of electrostatic interactions by increasing salt concentrations, and (iii) to establish the anchor residues within the conserved ribosomal stalk peptide, were performed in triplicate using the same protocol described above. Only the final 11 residues of the conserved peptide KEESEESDDDMGFGLFD were used, as the removal of the six N-terminal residues had no effect on binding and therefore do not contribute significantly to the interaction (Figure 1).
Figure 1. The A1 chain of SLT-1 binds to a conserved C-terminal ribosomal peptide.
(A) Amino acid sequence representing the 17-residue C-terminus common to ribosomal stalk proteins P1 and P2. The last 11 amino acids (underlined) delimit the shortest peptide element shown to interact with the A1 chain [15]. (B) Relative surface plasmon resonance (SPR) signals for the A1 chain of SLT-1 binding to immobilized, biotinylated monomeric synthetic peptides were plotted as a function of SLT-1 A1 chain concentration. The calculated dissociation constants (Kd) suggest that both monomeric peptides have similar affinities for the A1 chain. Each point on the curve represents the average relative SPR signals from experiments performed in quadruplicate.
Equilibrium data was used to calculate peptide binding constants to the A1 chain in light of fast on- and off-rates. In all cases, the response data given from the ribosomal peptides was subtracted from the control peptide as well as PBS alone to eliminate non-specific binding. The resulting data for each concentration was averaged and plotted as percent response units (% RU) versus A1 chain concentration using GraphPad Prism® (Figure 1). In the cases where binding affinities could not be calculated, due to a decrease or loss of binding, raw data was plotted as relative units (RU) compared to the same concentration (15 µM) of wild-type toxin A1 chain in the same buffer (Figures 2B, 2C, 3B, and 3C).
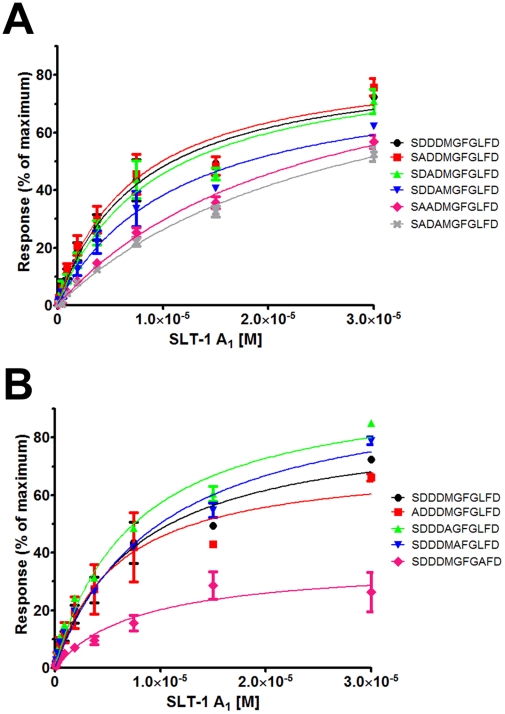
Figure 2. Surface plasmon resonance analysis of alanine-containing peptide variants of the conserved C-terminal ribosomal stalk peptide SDDDMGFGLFD confirms that the interaction with the A1 chain of SLT-1 requires both electrostatic and hydrophobic contacts.
The peptide sequence corresponding to the final 11 residues of the conserved C-terminal peptide (SDDDMGFGLFD) was substituted at each position for an alanine residue. Individual peptides corresponding to a substitution of charged (Panel A) or other residues (Panel B) were biotinylated and immobilized on an NLC SPR sensor chip. Each monomeric peptide was exposed to ten 2-fold serial dilutions of the A1 chain of SLT-1 in triplicate and the responses were subtracted from buffer alone and a control peptide. The SPR responses for the single and double/triple alanine variants were graphed and compared to the control natural peptide. Amino acid substitutions that resulted in a peptide that lacked an interaction with the A1 chain of SLT-1 could not be plotted. Calculated dissociation constants are reported in Table 1.
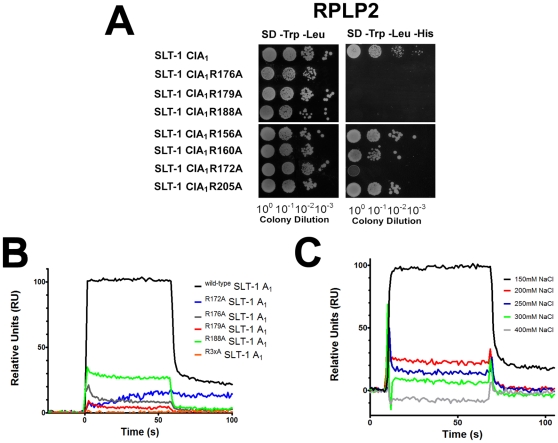
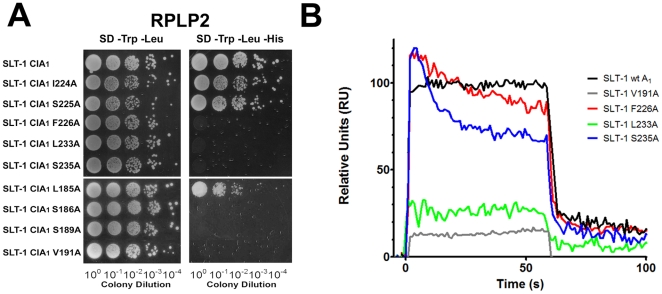
Figure 3. The A1 chain of SLT-1 harbors a cationic surface composed of a cluster of arginine residues that interact with the ribosomal stalk protein P2 and the conserved C-terminal peptide.
(A) A vector expressing a catalytically inactive variant of the SLT-1 A1 domain (CIA1) or one of the arginine-to-alanine point mutants as fusion partners with the GAL4 DNA-BD domain were co-transformed in the yeast strain AH109 with a vector expressing ribosomal protein P2 as a fusion construct to the GAL4-AD. The transformed yeast cells were plated on SD agar −Trp/−Leu. The resulting yeast colonies were grown overnight, and spotted (10 µl) as 10-fold serial dilutions onto SD medium lacking Trp and Leu to select for the presence of each plasmid followed by spotting on SD media lacking Trp, Leu, and His to select for interacting partners leading to colony growth. (B) SPR profiles illustrating the decrease in relative units for the arginine-to-alanine SLT-1 A1 chain variants in relation to the wild-type A1 chain, at a concentration of 15 µM, when presented to the immobilized peptide SDDDMGFGLFD. (C) Increasing salt concentrations led to a decrease or loss of binding of wild-type SLT-1 A1 chain when exposed to the peptide SDDDMGFGLFD. SPR traces were plotted for the wild-type SLT-1 A1 chain (15 µM) as a function of increasing salt concentrations.
Protein Synthesis Assay
T7-coupled transcription-translation (TnT) reticulocyte lysate assays (Promega, Madison, WI) were performed in order to determine if a decrease in ribosomal stalk binding altered the toxicity profiles of SLT-1 A1 chain point mutants when compared to the wild-type toxin. TnT assays were performed, according to the manufacturer's instructions, using eight 10-fold serial dilutions of the wild-type or SLT-1 A1 variants in PBS (starting with 1 µM). Protein synthesis was measured using a luciferase reporter plasmid (500 µg) through the incorporation of [35S]-methionine (10 µCi; GE Healthcare, Piscataway, NJ) after a 90 min incubation at 30°C. Samples (20 µl) were loaded on a gradient (4–12%) SDS PAGE gel, and labeled protein bands revealed using a Storm® Phosphorimager (GE Healthcare). The addition of PBS alone was used as a control.
Results
The catalytic A1 chain of SLT-1 binds to the conserved C-terminal ribosomal peptide
We have previously shown that the A1 chain of SLT-1 binds to the conserved peptide KEESEESD(D/E)DMGFGLFD found at the C-terminus of ribosomal stalk proteins P0, P1 and P2 [15]. However, the molecular details of this interaction are unknown and could provide new evidence surrounding the mechanism of ribosome-inactivation by RIPs. The binding of the conserved C-terminal domain of the ribosomal proteins P1 and P2 to the wild-type SLT-1 A1 chain was thus analyzed by surface plasmon resonance (SPR). Biotinylated peptides corresponding to the final 17, 11 and 7 residues of ribosomal stalk proteins P1 and P2 were immobilized on an sensor chip (NLC; Bio-Rad, Hercules, CA) and each peptide ligand was exposed to increasing concentrations of the wild-type SLT-1 A1 chain. In light of the fast association and dissociation times observed for this binding event, equilibrium data from four independent experiments were instead used to calculate the dissociation constants of peptides binding to the A1 chain. It was determined that the A1 chain of SLT-1 interacted with the C-terminal 17 (KEESEESDDDMGFGLFD) and 11 residues (SDDDMGFGLFD) of P1 and P2 with comparable binding affinities of 11±5 µM and 13±2 µM respectively (Figure 1). The final 7 residues (MGFGLFD) did not bind to wild-type SLT-1 A1 chain (data not shown), as previously reported by pull-down experiments [15].
Delineating key residues within the ribosomal peptide recognized by the SLT-1 A1 chain
The SLT-1 A1 chain docks within the ribosomal stalk by binding to specific residues within the peptide sequence SDDDMGFGLFD. A series of synthetic peptides containing alanine substitutions were generated by solid-phase peptide synthesis, to establish which residues within the peptide motif were important for its interaction with the A1 chain of SLT-1. Specifically, alanine was introduced at each position within the peptide SDDDMGFGLFD. These peptides were modified at their N-terminus with biotin, immobilized on an NLC sensor chip (BioRad), and exposed to graded concentrations of the SLT-1 A1 chain. The SPR equilibrium data was collected, in triplicate, and responses were plotted as a function of A1 chain concentration to calculate dissociation constants. It was determined that mutations of any of the five C-terminal residues to alanine (SDDDMGFGLFD) resulted in a decrease or complete loss of binding to the A1 chain. (Table 1 and Figure 2). Interestingly, individual mutations of the N-terminal, negatively charged aspartyl residues to alanine did not show any significant effect on the affinity suggesting that such interaction with the A1 chain may require the removal of more than one aspartic acid residues within the DDD tripeptide regardless of their position (Figure 2A). Therefore, the contribution of the charged aspartic acid residues within the peptide was further assessed through the generation of double and triple aspartate-to-alanine SLT-1 A1 chain mutants. It was observed that mutations to any two aspartic acid residues to alanines, particularly involving residues at positions 3 and 4 caused at least a two-fold decrease in binding (Table 1 and Figure 2A). This comprehensive binding analysis confirmed that both the charge and hydrophobic elements (Figures 2A and B, respectively) of the conserved peptide SDDDMGFGLFD are required for the optimal binding of the A1 chain of SLT-1 to the ribosomal stalk.
Table 1. Binding of synthetic, alanine-containing peptide variants of the ribosomal stalk peptide SDDDMGFGLFD to the A1 chain of SLT-1 as measured by surface plasmon resonance.
| Peptide | Kd (µM) |
| SDDDMGFGLFD | 10.6±2.4 |
| ADDDMGFGLFD | 12.7±4.3 |
| SADDMGFGLFD | 9.75±2.4 |
| SDADMGFGLFD | 11.6±2.8 |
| SDDAMGFGLFD | 15.9±3.5 |
| SDDDAGFGLFD | 7.5±0.2 |
| SDDDMAFGLFD | 9.9±0.5 |
| SDDDMGAGLFD | ≥50 |
| SDDDMGFALFD | ≥50 |
| SDDDMGFGAFD | 41±11 |
| SDDDMGFGLAD | ≥50 |
| SDDDMGFGLFA | ≥50 |
| SAADMGFGLFD | 23.9±1.6 |
| SADAMGFGLFD | 28.2±1.9 |
| SDAAMGFGLFD | ≥50 |
| SAAAMGFGLFD | ≥50 |
Dissociation constants (Kd) were calculated from curves relating changes in relative surface plasmon resonance signal observed as a function of SLT-1 A1 concentration (Figure 2). Each Kd value was derived from an average of three experiments. A Kd value of ≥50 µM was assigned to peptides displaying no measurable binding to the A1 chain of SLT-1.
The binding between the SLT-1 A1 chain and the conserved peptide SDDDMGFGLFD involves electrostatic interactions
In view of the negatively charged nature of the conserved ribosomal C-terminal peptide (aspartic acid [D] residues), we investigated whether a complementary positively charged surface was present on the A1 chain of SLT-1 that would promote electrostatic interactions with this ribosomal peptide. Surface-exposed arginine residues were mutated to alanine (R-A) in order to assess their importance as anchoring residues for the conserved ribosomal stalk peptide. SLT-1 A1 domain constructs with point mutations at arginine residues were generated by PCR using a catalytically-inactive SLT-1 A1 chain (CIA1) gene for their non-lethal expression in yeast. These R-A mutants were assessed for their ability to bind the full-length P2 protein in a yeast two-hybrid (Y2H) assay where co-transformation of each mutant with P2 were then serially diluted on medium which selected for an interaction between the two proteins (SD −Trp/−Leu/−His). Alanine mutations at residues R172, R176, R179, and R188 in the A1 domain led to a loss of growth, implying that these arginines interact with the ribosomal peptide (Figure 3A). Alanine mutations at other arginine sites within the A1 domain did not affect yeast survival suggesting that they are not involved in ribosomal docking (Figure 3A). Importantly, residues R172, R176, R179, and R188 are clustered on the surface of the A1 chain suggesting the existence of a complementary positively charged surface within the A1 chain.
To further define the effect of charged residues on the binding strength of A1 chain to the peptide SDDDMGFGLFD, we expressed and purified recombinant SLT-1 A1 chain variants, which showed a loss of interaction with the P2 protein by Y2H (R172A, R176A, R179A, R188A, and R176/179/188A), to directly assess their binding affinities towards this peptide by SPR. As expected, the binding affinity of the SLT-1 A1 chain R-A mutants for the peptide (≥50 µM, no measurable binding) was lower than the affinity observed for the wild-type toxin (Kd = 13 µM). The SLT-1 R188A A1 chain mutant was the only variant with a measurable binding constant (approximately 50 µM), while the other SLT-1 A1 chain variants (R172A, R176A, R179A, and R176/179/188A) lacked detectable SPR responses even at high concentrations and were determined to be weaker than 50 µM (Figure 3B), In addition, when the concentration of salt in the running buffer (PBS) was increased, the interaction between the wild-type SLT-1 A1 chain and the peptide SDDDMGFGLFD was decreased, further confirming the importance of electrostatic interactions (Figure 3C).
Hydrophobic interactions also contribute to binding the conserved C-terminal domain of ribosomal stalk proteins
The presence of a hydrophobic tetrapeptide motif (SDDDMGFGLFD) led us to hypothesize that a hydrophobic patch on the A1 chain of SLT-1 may also contribute to its interaction with the ribosomal stalk peptide. To test this hypothesis, several hydrophobic and serine residues namely, L185, V191, I224, S225, F226, L233, and S235 on the surface of the A1 chain that are in the vicinity of the previously defined arginine cluster were mutated to alanine residues. These point mutants were generated by PCR using a catalytically-inactive SLT-1 A1 chain (CIA1) gene serving as a template in order to subsequently express them as non-toxic variants in yeast. Their binding to the ribosomal protein P2 was examined by Y2H (Figure 4A). Yeast transformations were serially diluted on SD-media lacking Trp, Leu, and His, which selects for the interaction between the two proteins. It was determined that residues V191, F226, L233, and S235 are important for the A1 chain – ribosomal peptide interaction, while residues L185, I224 and S225 do not participate in these interactions (Figure 4A). The contribution of the hydrophobic and serine residues located on the surface of the A1 chain was investigated by SPR to confirm the interactions of recombinant A1 chain variants with the C-terminal peptide SDDDMGFGLFD. It was found that the V191A and L233A variants exhibited a large decrease in binding while variants harboring either a F226A or a S235A mutation displayed different binding kinetics and binding affinities of 3 and 5 µM, respectively. These affinities are comparable to that of wild type A1 chain and may be attributed to the altered binding kinetics (Figure 4B and Figure S2).
Figure 4. The interaction of the A1 chain of SLT-1 with the ribosomal stalk protein P2 and the C-terminal peptide SDDDMGFGLFD also involves hydrophobic residues within the A1 chain.
(A) Bait vectors expressing either a catalytically inactive variant of the wild-type SLT-1 A1 domain (CIA1) or one of the hydrophobic mutants were co-transformed in the yeast strain AH109 with a prey vector expressing ribosomal protein P2. The transformed yeast cells were plated on SD agar −Trp/−Leu. The resulting yeast colonies were grown overnight, and spotted (10 µl) as 10-fold serial dilutions onto SD medium lacking Trp and Leu to select for the presence of each plasmid followed by spotting on SD media lacking Trp, Leu, and His to select for interacting partners. (B) SPR profiles (plotted at 15 µM) demonstrate that hydrophobic mutants F226A and S235A in the SLT-1 A1 chain have a minor effect on the binding to the conserved peptide SDDDMGFGLFD and the SLT-1 V191A and L233A A1 chain mutants cause a drastic decrease in binding. Experiments were performed in triplicate.
Altering the cationic or hydrophobic surfaces of the catalytic domain of SLT-1 is sufficient to lower its ability to inhibit protein synthesis
We have shown by both Y2H and SPR that positively charged and hydrophobic residues located on the surface of the A1 chain form cationic and hydrophobic patches allowing for its binding to ribosomal stalk proteins via the C-terminal peptide SDDDMGFGLFD. These interactions were measured as monomeric events, meaning that such interactions and affinity constants reflect complexes involving a single ribosomal peptide binding to a single SLT-1 A1 chain. In reality, the peptide SDDDMGFGLFD is repeated five times within the context of the ribosomal stalk suggesting that even a dissociation constant in the low µM range could be significant due to the higher concentration of low affinity targets that could maintain the docking of the A1 chain to the stalk.
To determine the effects of avidity on the binding and subsequent cytotoxicity of the A1 chain of SLT-1, we further tested whether the charged or hydrophobic residues that are responsible for the interaction with the stalk peptide were also required for its full effect on protein synthesis inhibition. It was thus hypothesized that decreasing the affinity of the A1 chain to the ribosomal stalk proteins by introducing arginine-to-alanine (R-A) or hydrophobic-to-alanine mutations, which have previously been shown to perturb the interaction, would result in a decrease in its ability to inhibit protein synthesis when compared to the wild-type A1 chain. To test this hypothesis, ten-fold serial dilutions of each A1 chain variant (R172A, R176A, R179A, R188A, R176/179/188A, V191A, F226A, L233A, and S235A) were added to a T7-coupled rabbit reticulocyte lysate transcription-translation system to quantify their ability to block the biosynthesis of [35S]-methionine-labeled luciferase. As predicted, A1 chain mutants that exhibited a drastic decrease in affinity for the ribosomal stalk peptide (namely R172A, R176A, R179A, R188A, R176/179/188A, V191A, and L233A) displayed an increase in protein expression as compared to the wild-type toxin (Figure 5). These findings suggest that the binding of the A1 chain to the ribosomal stalk, in the context of a functional eukaryotic ribosome, correlates with its ability to inhibit protein synthesis with all SLT-1 A1 chain mutants able to block protein synthesis at high concentrations. As expected, the S235A mutant did not significantly alter cytotoxicity profiles when compared to the wild type A1 chain. The translation inhibition effects were also smaller for the R188A mutant, which can be attributed to the retention of approximately 30% of the binding compared to the wild-type toxin (Figures 3B and 5). Interestingly, the F226A mutation displayed an increase in catalytic activity as measured by TnT, an observation that is currently under investigation (Figure 5).
Figure 5. Arginine-to-alanine and hydrophobic variants of SLT-1 A1 that bind weakly to the monomeric conserved C-terminal motif display altered ribosome-inactivating activities when compared to the wild-type A1 chain.
Eight ten-fold serial dilutions of the wild-type and each charge and hydrophobic A1 chain variant was dispensed into an in vitro transcription and translation-coupled rabbit reticulocyte lysate system to monitor their ability to block protein synthesis (methods section). The level of in vitro protein synthesis was assessed by measuring the incorporation of [35S]-methionine into the reporter protein luciferase during its synthesis. The expression of radiolabeled luciferase (arrow) was then resolved by SDS-PAGE and quantified using a phosphorimager. The addition of PBS alone (- lane) was used as a control.
Discussion
The catalytic domains of ribosome-inactivating proteins (RIPs) such as SLT-1, ricin, and TCS [15], [20], [22], [23], [39] have been shown to interact with the eukaryotic ribosomal stalk, a heteropentamer composed of two heterodimers of the proteins RPLP1 (P1) and RPLP2 (P2) non-covalently associated to the protein RPP0 (P0) [27], [28], [29], [30], [31], [32], [33]. One known docking site for these catalytic chains on ribosomes is through their binding to a C-terminal peptide KEESEESDXDMGFGLFD (where X is either D or E) encoded by all three stalk proteins P0, P1, and P2. The pentavalent display of this peptide sequence within the stalk provides in theory up to five docking sites for RIP catalytic chains such as the A1 chain of SLT-1 to orient themselves on ribosomes, an event that may facilitate ribosome depurination leading to the inhibition of protein synthesis and apoptosis. The presence of five copies of this peptide motif suggests that valency is a critical factor and that the peptide-RIP interaction may be transient and of low affinity. In the present study, we have established by surface plasmon resonance that the A1 chain of SLT-1 binds to the conserved monomeric 17-residue long C-terminal peptide (KEESEESDDDMGFGLFD) and its truncated 11-residue form (SDDDMGFGLFD) with comparable modest affinities (Kd of 11±5 µM and 13±2 µM, respectively) (Figure 1). Interestingly, the on- and off-rates could not be measured for the A1 chain interacting with the monomeric peptide and such fast association and dissociation rates were expected since the binding of other RIPs namely saporin, restrictocin, and ricin to ribosome also show unusually fast kinetics and catalytic efficiencies [39], [40], [41], [42]. The rapid on- and off-rates are plausible in light of the fact that the interaction between RIPs and the ribosome must be a transient event in order for one ricin molecule to depurinate ∼2000 mammalian ribosomes/min and for a single SLT-1 A1 toxin molecule being required to enter the cytosol to elicit cell death [43], [44].
A previous study by our group had indicated that the C-terminal peptide inhibits the catalytic activity of the SLT-1 A1 chain in blocking protein synthesis in an in vitro transcription-translation assay (McCluskey et al., 2008). This finding suggested that such a peptide may serve as a template in designing a new class of inhibitors able to block the action of RIPs. We thus further analyzed the key components of this peptide involved in its interaction with SLT-1 A1 chain. Specifically, synthetic peptide analogues of this sequence were used to define that the anionic tri-aspartyl sequence DDD and the four C-terminal residues FGLF within the C-terminal peptide (SDDDMGFGLFD) represent the two major anchors that interact with complementary cationic (Figure 2A) and hydrophobic (Figure 2B) surfaces on SLT-1 A1 chain. As in the case of most ER-routed toxins, the A1 chain of SLT-1 contains very few lysines (only 2 lysines located at its N-terminus) [45]. Therefore, arginine residues were projected to contribute to the creation of a positively charged surface on the A1 chain that may interact with the anionic aspartic acid residues of SDDDMGFGLFD. Arginines at positions 172, 176, 179, and 188 were mutated to alanines leading to A1 chain variants that were unable to interact in a yeast-2-hybrid experiment with SDDDMGFGLFD presented in the context of P2 (Figure 3A). These same SLT-1 A1 chain variants were also expressed and the resulting purified recombinant proteins were shown to have a decreased affinity or loss of binding to the conserved peptide SDDDMGFGLFD as determined by SPR (Figure 3B). In addition, increasing salt concentrations inversely correlated with binding of the wild-type A1 chain to the peptide as measured by SPR (Figure 3C). These findings coincide with recent evidence that shows that electrostatic interactions are critical for the binding of ricin to whole ribosomes and for the targeting of restrictocin to the sarcin-ricin loop [39], [40], [46]. Moreover, several surface-exposed hydrophobic or serine residues on the A1 chain in close proximity to the previously defined arginine cluster were mutated to alanine. Residues V191, F226, L233, and S235 were identified by Y2H as critical in maintaining the interaction with the conserved peptide SDDDMGFGLFD, the only docking site on the full length ribosomal protein P2 (Figure 4A). It was confirmed by SPR that residues V191, and L233 serve important roles in the interaction, whereas alanine mutations to residues F226 and S235 had no effect on binding (Figure 4B).
Structural data of a type I RIP, trichosanthin (TCS), and a type III RIP from maize supports our previous hypothesis and experimental data that charged and hydrophobic residues are important for the interaction with the C-terminus to occur [47], [48]. When the structure of SLT-1 A1 chain is compared to that of TCS it reveals the presence of a similar complementary surface-exposed groove theoretically predicted to involve residues R176, R179, and S235 of the A1 chain of SLT-1 [47]. Two of the three residues (R176 and R179) have been identified in this study as being essential for the interaction with the C-terminal peptide. Recent evidence has also been published highlighting the importance of R176 on the depurination activity of SLT-1 [49]. In addition, we have observed that V191, R172, R188, and L233 also contribute to the binding of the A1 chain to the conserved ribosomal peptide. However, the predicted A1 chain-peptide interaction appears to be unique when compared to the known structure of TCS in complex with the conserved peptide [47].
The SLT-1 A1 chain variants harboring R-A or hydrophobic mutations, which display striking decreased affinities for the ribosomal stalk peptide also show a reduction in their abilities to inhibit protein synthesis as measured by in vitro protein translation assays (Figure 5). These variants still retain their catalytic activity at high concentrations suggesting that a single substitution does not affect the overall three-dimensional fold of the A1 chain (Figure 5). These results suggest that depurination may still occur at high concentrations due to rRNA binding or that the pentavalent presentation of the conserved ribosomal peptide in the context of the intact ribosomal stalk still favors A1 chain docking to the stalk. Specifically, it may be sufficient for the A1 chain of SLT-1, and other related RIPs cytotoxic domains, to bind directly to rRNA since the depurination of the sarcin-ricin loop has been observed in protein-depleted ribosomes, although the depurination rate is remarkably reduced [19].
This conserved ribosomal peptide represents a docking site for RIPs as it has been previously shown to interact with the catalytic domains of SLT-1, ricin, TCS, and maize RIP [15], [20], [47], [48]. It is therefore highly likely that SLT-2, an SLT-1 homologue that is produced by clinically more severe bacterial strains [50], would possess similar docking residues as we have shown for SLT-1. Even though the two catalytic domains share only a 55% homology based on amino acid sequence [51], it was observed that most of the surface residues important for the SLT-1-ribosomal peptide interaction were conserved on the surface of SLT-2 when the two toxins were aligned based on their respective tertiary structures (Figure 6). Specifically, five of the six residues shown to be important for the SLT-1 interaction (R172, R176, R179, V191, and L233) were conserved on SLT-2 (R172, R176, R179, Y189, and L232) (Figure 6, panels B and C). The only missing A1 chain interacting residue is arginine 188 of SLT-1, which is absent in SLT-2 (Figure 6, panels B and C). However, the A1 chain variant R188A displayed a catalytic activity comparable to that of wild type A1 chain (Figure 5).
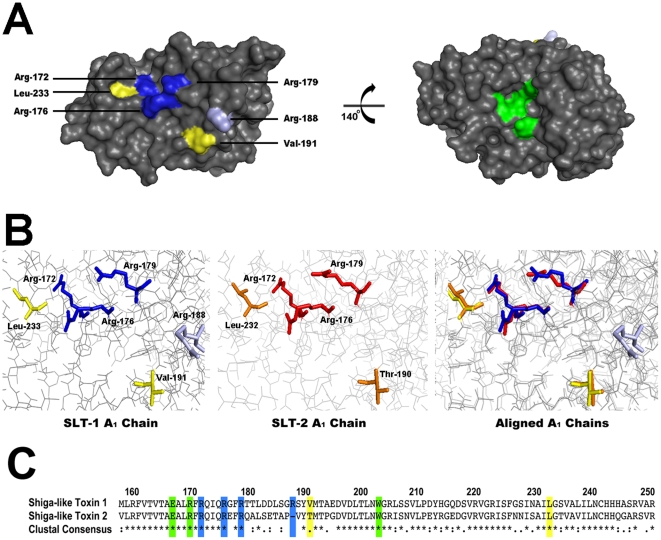
Figure 6. Primary and tertiary structural comparisons between SLT-1 and SLT-2 highlighting the conservation of important ribosomal stalk peptide contact sites.
(A) Left Panel - Surface rendering of the SLT-1 A1 chain (PDB# 1DM0) depicting the cationic (blue) and hydrophobic (yellow) residues essential for optimal binding to the conserved stalk peptide SDDDMGFGLFD as well as Arg-188 (light blue) which has a modest effect on peptide binding. Right Panel – Structure as shown in the left panel rotated by 140°, highlighting the catalytic residues in green. (B) Three-dimensional stick structures of SLT-1 (left panel), SLT-2 (PDB# 1R4P; middle panel), and the structural alignment of the two toxins (right panel). Cationic residues are labeled in blue and red, while hydrophobic residues are labeled in yellow and orange for SLT-1 and SLT-2 respectively. (C) Primary amino acid sequence alignment of SLT-1 and SLT-2 within residues 158 and 250. Catalytic residues are highlighted in green and cationic and hydrophobic residues in blue and yellow, respectively. Surface and stick renderings and alignments were performed using the The PyMOL Molecular Graphics System (Version 1.3, Schrödinger, LLC), whereas amino acid sequences were aligned using BioEdit software [59].
This surface pocket and its affinity for the conserved C-terminal peptide may help explain why different RIP family members have the ability to bind the stalk conserved peptide and also why some RIPs such as pokeweed antiviral protein (PAP) do not require the stalk proteins for ribosome inactivation [52]. For example, the RIP saporin has a homologous tertiary structure to that of TCS, the A chains of SLT-1 and ricin, and was predicted to bind to the C-terminal tail of ribosomal stalk proteins [47]. Yet, we did not observe any measurable interactions between the C-terminal peptide SDDDMGFGLFD and the RIP saporin or the ribonuclease alpha-sarcin, which both target the sarcin-ricin loop, by either pull-down experiments, SPR, or by isothermal titration calorimetry (Figure S3). The A1 chain of SLT-1 was confirmed to interact with the C-terminal peptide by both isothermal calorimetry and pull-down experiments (Figure S3). Although the on and off rates for the toxin-peptide interaction could not be measured by SPR, the HiCaM-tethered peptide was able to interact with the toxin most likely due to the excess (10∶1) of the HiCaM-peptide used in the pull-down experiments in relation to the SLT-1 A1 chain. Thus, the identified surface on the SLT-1 A1 chain may therefore represent only one docking element that may not be generalized to the docking process of all RIPs.
Gastrointestinal disease outbreaks caused by infection with Shiga toxin-producing E. coli strains remain very common and suggest the need for post-infection therapeutic inhibitors [53], [54], [55]. The charged and hydrophobic surfaces mapped in this study provide a binding interface that is distinct from the catalytic site and are required for the full toxicity of SLT-1 in vitro. This structural information can be used for the generation of therapeutic inhibitors for both SLT-1 and SLT-2. For example, it has been shown that small molecule virtual-docking can be exploited to generate chemical compounds directed towards the catalytic domains of ricin, shiga toxins, and Clostridium botulinum neurotoxin [56], [57], [58]. These studies may provide molecular “leads” that may be more suited in terms of cell-permeability, affinities and in vivo stability than ribosomal stalk C-terminal peptide mimics as RIP antidotes.
In summary, the A1 chain of SLT-1 interacts transiently (Kd∼13 µM with rapid on and off rates) with a short 11- amino acid conserved peptide located at the C-terminus of three ribosomal stalk proteins (P0, P1, and P2). The interaction involves both electrostatic and hydrophobic surfaces on both the A1 chain of SLT-1 and the ribosomal peptide SDDDMGFGLFD. Conversely, a cluster of positively charged arginine residues within the A1 chain in spatial proximity to a series of hydrophobic residues were defined as being critical for A1 chain binding to the peptide SDDDMGFGLFD and to inhibit protein synthesis in vitro. Thus, the catalytic A chain of Shiga toxins and other related RIPs may have evolved to interact rapidly and with low affinity to proteins constituting the ribosomal stalk in order to properly orient themselves towards their substrate, the sarcin-ricin loop.
Supporting Information
SDS-PAGE gel showing the relative purities of recombinantly expressed and purified SLT-1 A1 chain mutants. Each SLT-1 variant was expressed and purified as described in the methods section. Purified wild-type SLT-1 A1 and point mutants were analyzed by SDS-PAGE and protein bands visualised by Coomassie blue staining. Numbers below each lane correspond to the purity of the major protein band (as a percentage) in relation to minor contaminating proteins as derived from densitometry measurements using the ImageJ software package.
(TIF)
The SLT-1 A1 chain mutants F226A and S235A bind to the conserved C-terminal ribosomal peptide with similar affinity to wild-type SLT-1 A1. Relative surface plasmon resonance (SPR) signals for the F226A and S235A SLT-1 A1 chain variants binding to immobilized synthetic SDDDMGFGLFD peptide were plotted as a function of SLT-1 A1 chain concentration. The calculated dissociation constants (Kd) suggest that the F226A and S235A mutations in the A1 chain do not affect their affinity for the ribosomal stalk peptide SDDDMGFGLFD. Each point on the curve represents the average relative SPR signals from experiments performed in quadruplicate.
(TIF)
The conserved peptide SDDDMGFGLFD interacts with the A1 chain of SLT-1 but may not be a generic contact site for all ribotoxins. (Top Panel) Phenyl Sepharose bound HiCaM [60] fusion constructs (100 µg) displaying the C-terminal 7 amino acids (Lanes 4–5), 11 amino acids (Lanes 6–7), 17 amino acids (Lanes 8–9) of P1 and P2, or HiCaM alone (Lanes 2–3) were incubated briefly with 10 µg of SLT-1 A1 chain (Lane 1; Panel A), 20 µg saporin (Lane 1; Panel B), or 20 µg sarcin (Lane 1; Panel C) and separated on SDS-PAGE followed by Coomassie blue staining, as described previously [15]. The presence of a protein band in the thrombin cleavage (TC) lanes indicates an interaction and is only seen when the RIP A chain interacts with the final 11 or 17 residues of the conserved peptide. Legend: FT, column flow-through (unbound RIP); TC, thrombin-cleaved peptide. (Lower Panel) Synthetic peptide (starting with 500 µM) was titrated into a sample cell containing a 25 µM solution of degassed recombinant RIP and heat changes were measured using a VP-ITC (MicroCal Inc., Northampton, MA). The resulting calorimetric titration curves, minus the first injection of only 2 µl, were fitted using a single site binding model using the ORIGIN® software.
(TIF)
Primers used to construct expression vectors. Restriction endonuclease sites are underlined and amino acid substitutions are in bold.
(TIF)
Acknowledgments
We are grateful to Dr. Christopher Bachran for supplying saporin used in the SPR experiments as well as Dr. Mohammed Yousef for SPR data analyses.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This project was supported by an operation grant to J.G. from the Canadian Institutes of Health Research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Karmali MA. Infection by verocytotoxin-producing Escherichia coli. Clin Microbiol Rev. 1989;2:15–38. doi: 10.1128/cmr.2.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riley LW. The epidemiologic, clinical, and microbiologic features of hemorrhagic colitis. Annu Rev Microbiol. 1987;41:383–407. doi: 10.1146/annurev.mi.41.100187.002123. [DOI] [PubMed] [Google Scholar]
- 3.Fraser ME, Chernaia MM, Kozlov YV, James MN. Crystal structure of the holotoxin from Shigella dysenteriae at 2.5 A resolution. Nat Struct Biol. 1994;1:59–64. doi: 10.1038/nsb0194-59. [DOI] [PubMed] [Google Scholar]
- 4.Kozlov YV, Chernaia MM, Fraser ME, James MN. Purification and crystallization of Shiga toxin from Shigella dysenteriae. J Mol Biol. 1993;232:704–706. doi: 10.1006/jmbi.1993.1421. [DOI] [PubMed] [Google Scholar]
- 5.Lindberg AA, Brown JE, Stromberg N, Westling-Ryd M, Schultz JE, et al. Identification of the carbohydrate receptor for Shiga toxin produced by Shigella dysenteriae type 1. J Biol Chem. 1987;262:1779–1785. [PubMed] [Google Scholar]
- 6.Lingwood CA, Law H, Richardson S, Petric M, Brunton JL, et al. Glycolipid binding of purified and recombinant Escherichia coli produced verotoxin in vitro. J Biol Chem. 1987;262:8834–8839. [PubMed] [Google Scholar]
- 7.Jacewicz M, Clausen H, Nudelman E, Donohue-Rolfe A, Keusch GT. Pathogenesis of shigella diarrhea. XI. Isolation of a shigella toxin-binding glycolipid from rabbit jejunum and HeLa cells and its identification as globotriaosylceramide. J Exp Med. 1986;163:1391–1404. doi: 10.1084/jem.163.6.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johannes L, Goud B. Surfing on a retrograde wave: how does Shiga toxin reach the endoplasmic reticulum? Trends Cell Biol. 1998;8:158–162. doi: 10.1016/s0962-8924(97)01209-9. [DOI] [PubMed] [Google Scholar]
- 9.Garred O, van Deurs B, Sandvig K. Furin-induced cleavage and activation of Shiga toxin. J Biol Chem. 1995;270:10817–10821. doi: 10.1074/jbc.270.18.10817. [DOI] [PubMed] [Google Scholar]
- 10.Lea N, Lord JM, Roberts LM. Proteolytic cleavage of the A subunit is essential for maximal cytotoxicity of Escherichia coli O157:H7 Shiga-like toxin-1. Microbiology. 1999;145(Pt 5):999–1004. doi: 10.1099/13500872-145-5-999. [DOI] [PubMed] [Google Scholar]
- 11.LaPointe P, Wei X, Gariepy J. A role for the protease-sensitive loop region of Shiga-like toxin 1 in the retrotranslocation of its A1 domain from the endoplasmic reticulum lumen. J Biol Chem. 2005;280:23310–23318. doi: 10.1074/jbc.M414193200. [DOI] [PubMed] [Google Scholar]
- 12.Brigotti M, Carnicelli D, Alvergna P, Mazzaracchio R, Sperti S, et al. The RNA-N-glycosidase activity of Shiga-like toxin I: kinetic parameters of the native and activated toxin. Toxicon. 1997;35:1431–1437. doi: 10.1016/s0041-0101(96)00225-5. [DOI] [PubMed] [Google Scholar]
- 13.Endo Y, Mitsui K, Motizuki M, Tsurugi K. The mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes. The site and the characteristics of the modification in 28 S ribosomal RNA caused by the toxins. J Biol Chem. 1987;262:5908–5912. [PubMed] [Google Scholar]
- 14.Endo Y, Tsurugi K, Yutsudo T, Takeda Y, Ogasawara T, et al. Site of action of a Vero toxin (VT2) from Escherichia coli O157:H7 and of Shiga toxin on eukaryotic ribosomes. RNA N-glycosidase activity of the toxins. Eur J Biochem. 1988;171:45–50. doi: 10.1111/j.1432-1033.1988.tb13756.x. [DOI] [PubMed] [Google Scholar]
- 15.McCluskey AJ, Poon GM, Bolewska-Pedyczak E, Srikumar T, Jeram SM, et al. The catalytic subunit of shiga-like toxin 1 interacts with ribosomal stalk proteins and is inhibited by their conserved C-terminal domain. J Mol Biol. 2008;378:375–386. doi: 10.1016/j.jmb.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 16.Moazed D, Robertson JM, Noller HF. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature. 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- 17.Hausner TP, Atmadja J, Nierhaus KH. Evidence that the G2661 region of 23S rRNA is located at the ribosomal binding sites of both elongation factors. Biochimie. 1987;69:911–923. doi: 10.1016/0300-9084(87)90225-2. [DOI] [PubMed] [Google Scholar]
- 18.Montanaro L, Sperti S, Mattioli A, Testoni G, Stirpe F. Inhibition by ricin of protein synthesis in vitro. Inhibition of the binding of elongation factor 2 and of adenosine diphosphate-ribosylated elongation factor 2 to ribosomes. Biochem J. 1975;146:127–131. doi: 10.1042/bj1460127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Endo Y, Tsurugi K. The RNA N-glycosidase activity of ricin A-chain. The characteristics of the enzymatic activity of ricin A-chain with ribosomes and with rRNA. J Biol Chem. 1988;263:8735–8739. [PubMed] [Google Scholar]
- 20.Chiou JC, Li XP, Remacha M, Ballesta JP, Tumer NE. The ribosomal stalk is required for ribosome binding, depurination of the rRNA and cytotoxicity of ricin A chain in Saccharomyces cerevisiae. Mol Microbiol. 2008;70:1441–1452. doi: 10.1111/j.1365-2958.2008.06492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li XP, Grela P, Krokowski D, Tchorzewski M, Tumer NE. Pentameric organization of the ribosomal stalk accelerates recruitment of ricin a chain to the ribosome for depurination. J Biol Chem. 2010;285:41463–41471. doi: 10.1074/jbc.M110.171793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan DS, Chu LO, Lee KM, Too PH, Ma KW, et al. Interaction between trichosanthin, a ribosome-inactivating protein, and the ribosomal stalk protein P2 by chemical shift perturbation and mutagenesis analyses. Nucleic Acids Res. 2007;35:1660–1672. doi: 10.1093/nar/gkm065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan SH, Hung FS, Chan DS, Shaw PC. Trichosanthin interacts with acidic ribosomal proteins P0 and P1 and mitotic checkpoint protein MAD2B. Eur J Biochem. 2001;268:2107–2112. doi: 10.1046/j.1432-1327.2001.02091.x. [DOI] [PubMed] [Google Scholar]
- 24.Bargis-Surgey P, Lavergne JP, Gonzalo P, Vard C, Filhol-Cochet O, et al. Interaction of elongation factor eEF-2 with ribosomal P proteins. Eur J Biochem. 1999;262:606–611. doi: 10.1046/j.1432-1327.1999.00434.x. [DOI] [PubMed] [Google Scholar]
- 25.Datta PP, Sharma MR, Qi L, Frank J, Agrawal RK. Interaction of the G′ domain of elongation factor G and the C-terminal domain of ribosomal protein L7/L12 during translocation as revealed by cryo-EM. Mol Cell. 2005;20:723–731. doi: 10.1016/j.molcel.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 26.Helgstrand M, Mandava CS, Mulder FA, Liljas A, Sanyal S, et al. The ribosomal stalk binds to translation factors IF2, EF-Tu, EF-G and RF3 via a conserved region of the L12 C-terminal domain. J Mol Biol. 2007;365:468–479. doi: 10.1016/j.jmb.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 27.Grela P, Helgstrand M, Krokowski D, Boguszewska A, Svergun D, et al. Structural characterization of the ribosomal P1A-P2B protein dimer by small-angle X-ray scattering and NMR spectroscopy. Biochemistry. 2007;46:1988–1998. doi: 10.1021/bi0616450. [DOI] [PubMed] [Google Scholar]
- 28.Tchorzewski M, Krokowski D, Boguszewska A, Liljas A, Grankowski N. Structural characterization of yeast acidic ribosomal P proteins forming the P1A-P2B heterocomplex. Biochemistry. 2003;42:3399–3408. doi: 10.1021/bi0206006. [DOI] [PubMed] [Google Scholar]
- 29.Naganuma T, Shiogama K, Uchiumi T. The N-terminal regions of eukaryotic acidic phosphoproteins P1 and P2 are crucial for heterodimerization and assembly into the ribosomal GTPase-associated center. Genes Cells. 2007;12:501–510. doi: 10.1111/j.1365-2443.2007.01067.x. [DOI] [PubMed] [Google Scholar]
- 30.Krokowski D, Boguszewska A, Abramczyk D, Liljas A, Tchorzewski M, et al. Yeast ribosomal P0 protein has two separate binding sites for P1/P2 proteins. Mol Microbiol. 2006;60:386–400. doi: 10.1111/j.1365-2958.2006.05117.x. [DOI] [PubMed] [Google Scholar]
- 31.Perez-Fernandez J, Remacha M, Ballesta JP. The acidic protein binding site is partially hidden in the free Saccharomyces cerevisiae ribosomal stalk protein P0. Biochemistry. 2005;44:5532–5540. doi: 10.1021/bi047332r. [DOI] [PubMed] [Google Scholar]
- 32.Hagiya A, Naganuma T, Maki Y, Ohta J, Tohkairin Y, et al. A mode of assembly of P0, P1, and P2 proteins at the GTPase-associated center in animal ribosome: in vitro analyses with P0 truncation mutants. J Biol Chem. 2005;280:39193–39199. doi: 10.1074/jbc.M506050200. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalo P, Lavergne JP, Reboud JP. Pivotal role of the P1 N-terminal domain in the assembly of the mammalian ribosomal stalk and in the proteosynthetic activity. J Biol Chem. 2001;276:19762–19769. doi: 10.1074/jbc.M101398200. [DOI] [PubMed] [Google Scholar]
- 34.Santos C, Ballesta JP. Characterization of the 26S rRNA-binding domain in Saccharomyces cerevisiae ribosomal stalk phosphoprotein P0. Mol Microbiol. 2005;58:217–226. doi: 10.1111/j.1365-2958.2005.04816.x. [DOI] [PubMed] [Google Scholar]
- 35.Yamasaki S, Furutani M, Ito K, Igarashi K, Nishibuchi M, et al. Importance of arginine at position 170 of the A subunit of Vero toxin 1 produced by enterohemorrhagic Escherichia coli for toxin activity. Microb Pathog. 1991;11:1–9. doi: 10.1016/0882-4010(91)90088-r. [DOI] [PubMed] [Google Scholar]
- 36.Hovde CJ, Calderwood SB, Mekalanos JJ, Collier RJ. Evidence that glutamic acid 167 is an active-site residue of Shiga-like toxin I. Proc Natl Acad Sci U S A. 1988;85:2568–2572. doi: 10.1073/pnas.85.8.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 38.Bravman T, Bronner V, Lavie K, Notcovich A, Papalia GA, et al. Exploring “one-shot” kinetics and small molecule analysis using the ProteOn XPR36 array biosensor. Anal Biochem. 2006;358:281–288. doi: 10.1016/j.ab.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 39.Li XP, Chiou JC, Remacha M, Ballesta JP, Tumer NE. A two-step binding model proposed for the electrostatic interactions of ricin a chain with ribosomes. Biochemistry. 2009;48:3853–3863. doi: 10.1021/bi802371h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin S, Zhou HX. Dissection of the high rate constant for the binding of a ribotoxin to the ribosome. Proc Natl Acad Sci U S A. 2009;106:6974–6979. doi: 10.1073/pnas.0900291106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korennykh AV, Correll CC, Piccirilli JA. Evidence for the importance of electrostatics in the function of two distinct families of ribosome inactivating toxins. Rna. 2007;13:1391–1396. doi: 10.1261/rna.619707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sturm MB, Schramm VL. Detecting ricin: sensitive luminescent assay for ricin A-chain ribosome depurination kinetics. Anal Chem. 2009;81:2847–2853. doi: 10.1021/ac8026433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Endo Y, Tsurugi K. The RNA N-glycosidase activity of ricin A-chain. Nucleic Acids Symp Ser. 1988:139–142. [PubMed] [Google Scholar]
- 44.Tam PJ, Lingwood CA. Membrane cytosolic translocation of verotoxin A1 subunit in target cells. Microbiology. 2007;153:2700–2710. doi: 10.1099/mic.0.2007/006858-0. [DOI] [PubMed] [Google Scholar]
- 45.Hazes B, Read RJ. Accumulating evidence suggests that several AB-toxins subvert the endoplasmic reticulum-associated protein degradation pathway to enter target cells. Biochemistry. 1997;36:11051–11054. doi: 10.1021/bi971383p. [DOI] [PubMed] [Google Scholar]
- 46.Korennykh AV, Piccirilli JA, Correll CC. The electrostatic character of the ribosomal surface enables extraordinarily rapid target location by ribotoxins. Nat Struct Mol Biol. 2006;13:436–443. doi: 10.1038/nsmb1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Too PH, Ma MK, Mak AN, Wong YT, Tung CK, et al. The C-terminal fragment of the ribosomal P protein complexed to trichosanthin reveals the interaction between the ribosome-inactivating protein and the ribosome. Nucleic Acids Res. 2009;37:602–610. doi: 10.1093/nar/gkn922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y, Mak AN, Shaw PC, Sze KH. Solution structure of an active mutant of maize ribosome-inactivating protein (MOD) and its interaction with the ribosomal stalk protein P2. J Mol Biol. 2010;395:897–907. doi: 10.1016/j.jmb.2009.10.051. [DOI] [PubMed] [Google Scholar]
- 49.Di R, Kyu E, Shete V, Saidasan H, Kahn PC, et al. Identification of amino acids critical for the cytotoxicity of Shiga toxin 1 and 2 in Saccharomyces cerevisiae. Toxicon. 2011;57:525–539. doi: 10.1016/j.toxicon.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noris M, Remuzzi G. Hemolytic uremic syndrome. J Am Soc Nephrol. 2005;16:1035–1050. doi: 10.1681/ASN.2004100861. [DOI] [PubMed] [Google Scholar]
- 51.Jackson MP, Neill RJ, O'Brien AD, Holmes RK, Newland JW. Nucleotide sequence analysis and comparison of the structural genes for Shiga-like toxin I and Shiga-like toxin II encoded by bacteriophages from Escherichia coli 933. J FEMS Microbiology Letters. 1987;44:109–114. doi: 10.1016/0882-4010(87)90106-9. [DOI] [PubMed] [Google Scholar]
- 52.Ayub MJ, Smulski CR, Ma KW, Levin MJ, Shaw PC, et al. The C-terminal end of P proteins mediates ribosome inactivation by trichosanthin but does not affect the pokeweed antiviral protein activity. Biochem Biophys Res Commun. 2008;369:314–319. doi: 10.1016/j.bbrc.2008.01.170. [DOI] [PubMed] [Google Scholar]
- 53.Frank C, Werber D, Cramer JP, Askar M, Faber M, et al. Epidemic Profile of Shiga-Toxin-Producing Escherichia coli O104:H4 Outbreak in Germany - Preliminary Report. N Engl J Med. 2011 doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- 54.Kupferschmidt K. Infectious diseases. As E. coli outbreak recedes, new questions come to the fore. Science. 2011;333:27. doi: 10.1126/science.333.6038.27. [DOI] [PubMed] [Google Scholar]
- 55.Voelker R. Rare E. coli strain races through Europe; high rate of kidney failure reported. Jama. 2011;306:29. doi: 10.1001/jama.2011.889. [DOI] [PubMed] [Google Scholar]
- 56.Bai Y, Watt B, Wahome PG, Mantis NJ, Robertus JD. Identification of new classes of ricin toxin inhibitors by virtual screening. Toxicon. 2010;56:526–534. doi: 10.1016/j.toxicon.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pang YP, Park JG, Wang S, Vummenthala A, Mishra RK, et al. Small-molecule inhibitor leads of ribosome-inactivating proteins developed using the doorstop approach. PLoS One. 2011;6:e17883. doi: 10.1371/journal.pone.0017883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roxas-Duncan V, Enyedy I, Montgomery VA, Eccard VS, Carrington MA, et al. Identification and biochemical characterization of small-molecule inhibitors of Clostridium botulinum neurotoxin serotype A. Antimicrob Agents Chemother. 2009;53:3478–3486. doi: 10.1128/AAC.00141-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tchorzewski M, Boldyreff B, Issinger O, Grankowski N. Analysis of the protein-protein interactions between the human acidic ribosomal P-proteins: evaluation by the two hybrid system. Int J Biochem Cell Biol. 2000;32:737–746. doi: 10.1016/s1357-2725(00)00017-0. [DOI] [PubMed] [Google Scholar]
- 60.McCluskey AJ, Poon GM, Gariepy J. A rapid and universal tandem-purification strategy for recombinant proteins. Protein Sci. 2007;16:2726–2732. doi: 10.1110/ps.072894407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SDS-PAGE gel showing the relative purities of recombinantly expressed and purified SLT-1 A1 chain mutants. Each SLT-1 variant was expressed and purified as described in the methods section. Purified wild-type SLT-1 A1 and point mutants were analyzed by SDS-PAGE and protein bands visualised by Coomassie blue staining. Numbers below each lane correspond to the purity of the major protein band (as a percentage) in relation to minor contaminating proteins as derived from densitometry measurements using the ImageJ software package.
(TIF)
The SLT-1 A1 chain mutants F226A and S235A bind to the conserved C-terminal ribosomal peptide with similar affinity to wild-type SLT-1 A1. Relative surface plasmon resonance (SPR) signals for the F226A and S235A SLT-1 A1 chain variants binding to immobilized synthetic SDDDMGFGLFD peptide were plotted as a function of SLT-1 A1 chain concentration. The calculated dissociation constants (Kd) suggest that the F226A and S235A mutations in the A1 chain do not affect their affinity for the ribosomal stalk peptide SDDDMGFGLFD. Each point on the curve represents the average relative SPR signals from experiments performed in quadruplicate.
(TIF)
The conserved peptide SDDDMGFGLFD interacts with the A1 chain of SLT-1 but may not be a generic contact site for all ribotoxins. (Top Panel) Phenyl Sepharose bound HiCaM [60] fusion constructs (100 µg) displaying the C-terminal 7 amino acids (Lanes 4–5), 11 amino acids (Lanes 6–7), 17 amino acids (Lanes 8–9) of P1 and P2, or HiCaM alone (Lanes 2–3) were incubated briefly with 10 µg of SLT-1 A1 chain (Lane 1; Panel A), 20 µg saporin (Lane 1; Panel B), or 20 µg sarcin (Lane 1; Panel C) and separated on SDS-PAGE followed by Coomassie blue staining, as described previously [15]. The presence of a protein band in the thrombin cleavage (TC) lanes indicates an interaction and is only seen when the RIP A chain interacts with the final 11 or 17 residues of the conserved peptide. Legend: FT, column flow-through (unbound RIP); TC, thrombin-cleaved peptide. (Lower Panel) Synthetic peptide (starting with 500 µM) was titrated into a sample cell containing a 25 µM solution of degassed recombinant RIP and heat changes were measured using a VP-ITC (MicroCal Inc., Northampton, MA). The resulting calorimetric titration curves, minus the first injection of only 2 µl, were fitted using a single site binding model using the ORIGIN® software.
(TIF)
Primers used to construct expression vectors. Restriction endonuclease sites are underlined and amino acid substitutions are in bold.
(TIF)