Abstract
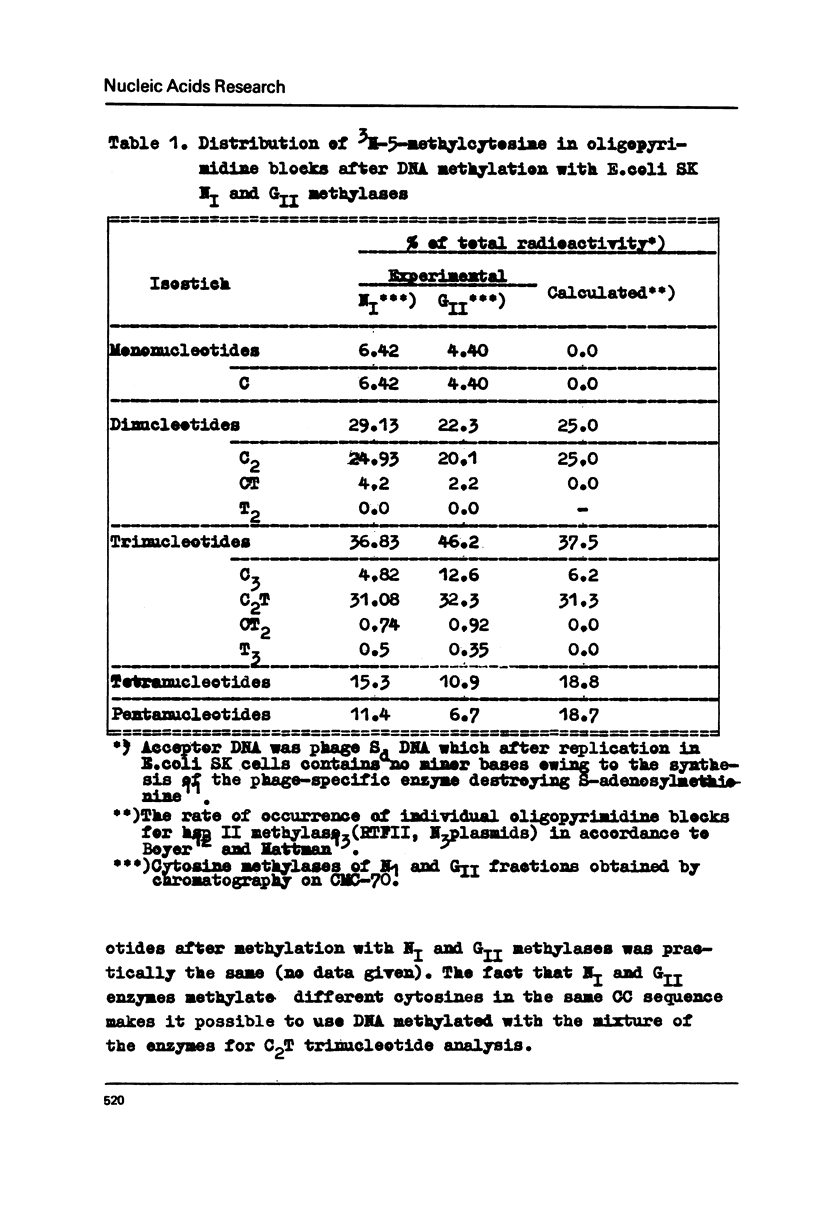
Two different cytosine DNA-methylases, NI and GII, are present in Escherichia coli SK. The GII methylase recognizes the five-member symmetric sequence: 5'...NpCpCpApGpGpN...3'. This sequence is identical with the recognition site of the hsp II type determined by RII plasmid but, in contrast to RII methylase, the GII enzyme methylates cytosine located on the 5' side of the site. By analogy with the isoshizomery of the restricting endonucleases, RII and GII DNA methylaeses may be called isomethymers which recognize the same site but methylate different bases. Since the phage of the SK and hsp II phenotypes is effectively restricted in respective cells it may be assumed that the isomethymeric modification does not provide any protection against the corresponding restrictases. NI methylase recognizes the five-member symmetric site which represents an inverted sequence of the GII site: 5'...NpGpGpApCpCpN...3'. In this case cytosine at the 3'-end of the recognition site is methylated.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arber W., Linn S. DNA modification and restriction. Annu Rev Biochem. 1969;38:467–500. doi: 10.1146/annurev.bi.38.070169.002343. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Chow L. T., Dugaiczyk A., Hedgpeth J., Goodman H. M. DNA substrate site for the EcoRII restriction endonuclease and modification methylase. Nat New Biol. 1973 Jul 11;244(132):40–43. doi: 10.1038/newbio244040a0. [DOI] [PubMed] [Google Scholar]
- Gefter M., Hausmann R., Gold M., Hurwitz J. The enzymatic methylation of ribonucleic acid and deoxyribonucleic acid. X. Bacteriophage T3-induced S-adenosylmethionine cleavage. J Biol Chem. 1966 May 10;241(9):1995–2006. [PubMed] [Google Scholar]
- Nikol'skaia I. I., Aleksandrova S. S., Lopatina N. G., Debov S. S. Fraktsionirovanie i ochistka metilaz DNK i spetsificheskikh endonukleaz iz kletok Escherichia coli SK. Biokhimiia. 1977 Apr;42(4):598–608. [PubMed] [Google Scholar]
- Nikolskaya I. I., Lopatina N. G., Chaplygina N. M., Debov S. S. The host specificity system in Escherichia coli SK. Mol Cell Biochem. 1976 Nov 30;13(2):79–87. doi: 10.1007/BF01837057. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Restriction endonucleases. CRC Crit Rev Biochem. 1976 Nov;4(2):123–164. doi: 10.3109/10409237609105456. [DOI] [PubMed] [Google Scholar]


