Abstract
BIX-01294 and its analogs were originally identified and subsequently designed as potent inhibitors against histone H3 lysine 9 (H3K9) methyltransferases G9a and G9a-like protein (GLP). Here we show BIX-01294 and its analog E67 can also inhibit H3K9 Jumonji demethylase KIAA1718 with half-maximal inhibitory concentrations in low micro-molar range. Crystallographic analysis of KIAA1718 Jumonji domain in complex with E67 indicated the benzylated six-membered piperidine ring was disordered and exposed to solvent. Removing the moiety (generating compound E67-2) has no effect on the potency against KIAA1718, but unexpectedly lost inhibition against GLP by a factor of 1500. Furthermore, E67 and E67-2 have no effect on the activity against histone H3 lysine 4 (H3K4) demethylase JARID1C. Thus our study provides a new avenue for designing and improving the potency and selectivity of inhibitors against H3K9 Jumonji demethylases over H3K9 methyltransferases as well as H3K4 demethylases.
Keywords: Epigenetics, Histone lysine demethylation, Enzymatic inhibition, BIX analogs
The Jumonji domain containing protein (histone) lysine demethylases are members of the cupin family of α-ketoglutarate (α-KG) and Fe2+ dependent dioxygenases 1. There are nearly 30 Jumonji containing proteins with known or suggested lysine demethylation function in human proteome 2. Abnormal activities of many of these enzymes have been implicated in various pathological conditions like cancer, developmental and neurological disorders 3; 4. Thus, these enzymes are novel targets for therapeutics 5; 6; 7.
Currently known inhibitors of Jumonji demethylases resemble the chemical structure of α-ketoglutarate, the cofactor, or succinate, which is the byproduct generated during enzymatic decarboxylation of α-ketoglutarate 8; 9; 10. Consequently, the cofactor-mimic compounds could inhibit a large numbers of α-ketoglutarate dependent dioxygenases by competing for the cofactor-binding sites.
BIX-01294 (a diazepin-quinazoline-amine derivative; Fig. 1a) inhibits activities of G9a 11 and G9a-like protein (GLP) lysine methyltransferase by mimicking the bound conformation of histone H3 Lys4 to Arg8, residues N-terminal to the target lysine 9 of histone H3, in the substrate peptide-binding groove 12. Addition of a lysine or methyl-lysine mimic to BIX-01294 enhances its potency of inhibition of G9a and GLP due to the insertion of the mimics into the target lysine-binding channel 13; 14; 15; 16. It is interesting to note that both lysine methyltransferases and demethylases recognize lysines in methylated and unmethylated states, whether they are substrates or reaction products. We therefore explore whether BIX analogs can function as selective inhibitors of human KIAA1718 (a H3K9me2 demethylase, also known as JHDM1D), in comparison with their inhibitions of human GLP (a H3K9 methyltransferase, also known as EHMT1) - whose loss-of-function mutations cause the 9q34 subtelomeric deletion syndrome 17 - and human JARID1C (a H3K4me3 demethylase, also known as SMCX) - mutations of which affect its demethylase activity are found in X-linked mental retardation patients 18; 19.
Fig. 1.
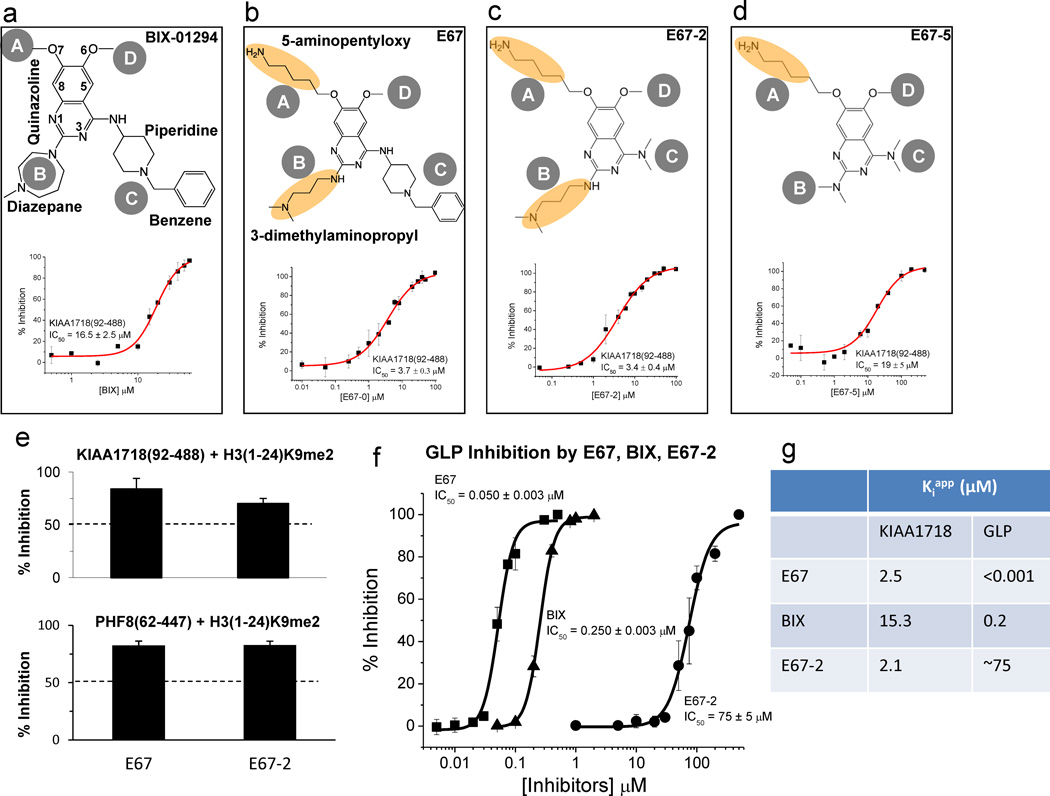
Chemical structures for (a) BIX-01294, (b) E67, (c) E67-2 and (d) E67-5. The inhibition (IC50 values) of compounds against demethylation reaction by KIAA1718 (Jumonji domain) is plotted against various concentrations of inhibitor [I], under the conditions of 2.5 µM enzyme [E] and 5 µM H3(1-24)K9me2 peptide substrate [S] in 50 mM HEPES pH 7.0, 1 mM α-KG, 50 µM Fe(NH4)2(SO4)2 and 2 mM ascorbic acid. The enzyme and the respective inhibitor were preincubated for 10 min at room temperature (approximately 21 °C). Adding substrate H3 peptide initiated reaction for 10 min at 37 °C and subjected to mass-spectrometry-based inhibition assay as previously described 12.
(e) Percentage of inhibition of the H3K9me2 demethylation activities of KIAA1718 Jumonji domain (top panel) and PHF8 Jumonji domain (bottom panel), under the conditions of 2.5 µM [E], 5 µM [S] and 10 µM [I]. Enzymes were pre-incubated with the inhibitors for 10 min at room temperature (approximately 21 °C) prior to the addition of substrate peptide. The reactions were then incubated at 37 °C for 10 min (KIAA1718) or 60 min (PHF8).
(f) The inhibition (IC50 values) of E67 and E67-2 against methylation reaction by GLP (SET domain) is plotted against various concentrations of compound, under the conditions of 0.1 µM [E] and 5 µM H3 peptide (residues 1–15) [S] in 20 mM Tris 8.0, 5 mM dithiothreitol (DTT) and 100 µM S-adenosyl-l-methionine (AdoMet). GLP was pre-incubated with varying concentrations of E67 and E67-2 for 5 min at room temperature (approximately 21 °C). Adding substrate H3 peptide initiated reaction for 5 min at 30° C.
(g) Comparison of Kiapp values, calculated based on three zones of enzyme-inhibitor interactions defined by Straus and Goldstein (1943)31: Kiapp=IC50-0.5[E] (if Kiapp/[E] in the range of 10–0.01) or Kiapp<0.01[E] (if IC50 ~ 0.5[E]) or Kiapp ~ IC50 (if Kiapp/[E] > 10).
KIAA1718 belongs to a small family of Jumonji proteins with three members (PHF2, PHF8 and KIAA1718) 2. Each of these proteins harbors two domains in its respective N-terminal half (Supplementary Fig. S1a): a PHD domain that binds tri-methylated histone H3 lysine 4 (H3K4me3) - a modification associated with transcriptional activation – and their linked Jumonji domains which remove methyl marks (H3K9me2, H3K27me2, or H4K20me1) that are associated with transcriptional repression 20; 21; 22. KIAA1718 removes methyl marks from dimethylated H3 lysine 9 (H3K9me2) and/or lysine 27 (H3K27me2) 20; 23; 24, which shares an ARKS sequence. Here, we focus on residues 92–488 from KIAA1718 (Jumonji) and 1–488 (containing PHD and Jumonji) (Supplementary Fig. S1b).
Inhibition of KIAA1718 by BIX and its analog E67
First, we found BIX-01294 inhibits the demethylation activity of KIAA1718 Jumonji domain (residues 92–488) with an IC50 (half-maximal inhibitory concentration) value of approximately 16.5 µM, using a histone H3K9me2 peptide (residues 1–24) as substrate, by a mass spectrometry-based demethylation assay 20 (Fig. 1a). Next, we tested the BIX analog E67 (Fig. 1b), which was originally generated with improved inhibition for GLP 14, with the replacement of the O7-methoxy group with a 5-aminopentyloxy substituent at site A and the diazepane ring with a 3-dimethylaminopropyl group at site B. The cumulative effect of these changes in E67 decreases its IC50 by a factor of approximately 4.5 in comparison to that of BIX.
Structure of KIAA1718 Jumonji domain bound with E67
We co-crystallized KIAA1718 Jumonji domain with E67, which was dissolved in aqueous buffer, in the presence of α-ketoglutarate and Ni2+ ion (Supplementary Fig. S1c). The structure was solved at a resolution of 2.8 Å (Table 1). The first 22 residues (amino acids 92–113) and the last 9 residues (amino acids 480–488) of KIAA1718 were not observed. The protein component of the structure is highly similar to that of KIAA1718 in the absence of bound peptide substrate (Fig. 2a; PDB 3KVA) and that of PHF8 in the presence of bound H3 peptide substrate (Fig. 2c; PDB 3KV4), with a root mean squared deviation (rmsd) of approximately 0.3 Å and 1.3 Å, respectively, when comparing 366 pairs of Cα atoms (residues 114–479).
Table 1.
Crystallographic data collection and refinement statistics.
| KIAA1718 (92-488) | |
|---|---|
| Data Collection | |
| Co-factors | Ni2+, α-ketoglutarate |
| Inhibitor | E67 |
| Space Group | P6122 |
| Cell dimensions | α = β = 90°, γ = 120° |
| a, b, c (Å) | 77.5, 77.5, 290.1 |
| Resolution (Å) | 26.63 – 2.79 |
| Rmerge | 0.124 (0.708) |
| I/σI | 25.5 (5.2) |
| Completeness (%) | 99.9 (100) |
| Redundancy | 18.7 (19.4) |
| Refinement | |
| Resolution (Å) | 2.79 |
| No. of Reflections | 13299 |
| Rwork/Rfree | 0.188/0.256 |
| No. of Atoms | |
| Proteins | 2930 |
| Heterogen | 69 |
| Water | 43 |
| B-factors (Å2) | |
| Protein | 48.5 |
| Inhibitor (E67) | 81.8 |
| Water | 41.2 |
| α-ketoglutarate | 45.9 |
| R.m.s. deviations | |
| Bond Lengths (Å) | 0.008 |
| Bond angles (°) | 1.18 |
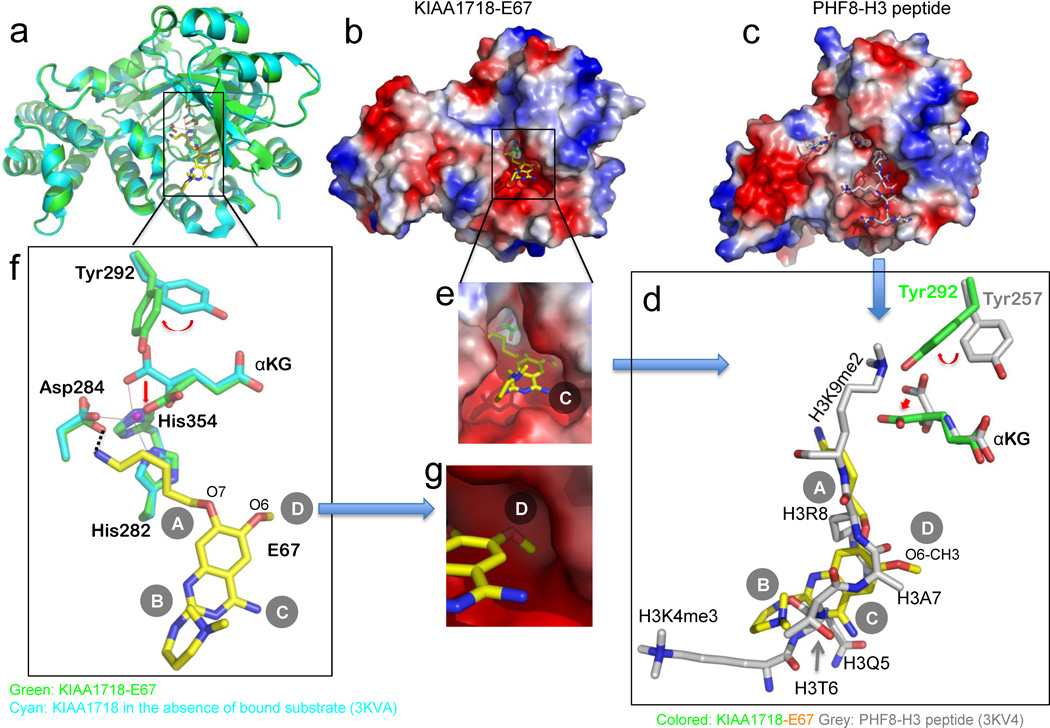
Fig. 2. Structure of KIAA1718-E67 complex.
(a) Superimposition of KIAA1718 Jumonji domain structures with bound E67 (colored in green) and in the absence of bound substrate (in cyan; PDB ID: 3KVA).
(b,c) Surface representation of the KIAA1718 Jumonji domain with E67 bound in the acidic substrate peptide binding groove (b), which is occupied by a histone peptide in the closely related PHF8 structure (c; PDB ID: 3KV4). The surface charge at neutral pH is displayed as red for negative, blue for positive, and white for neutral.
(d) Superimposition of H3 peptide (grey; taken from PDB ID: 3KV4) and E67 (yellow). The inhibitor occupies the space of bound histone H3 peptide from Lys4 (main chain) to Lys9 (main chain). The corresponding tyrosine (Tyr292 in KIAA1718 and Tyr257 in PHF8) adopts different rotamer conformation resulting in a different mode of binding of α-ketoglutarate (αKG).
(e) Enlarged view of E67 binding site with α-ketoglutarate (green) in the bottom of the pocket.
(f) Network of interactions centered on the amino group of 5-aminopentyloxy moiety at site A of E67. The metal ion (in small ball) is coordinated by side chains of His282, Asp284, His354 and two oxygen atoms of α-ketoglutarate.
(g) The methyl group of the O6 methoxy at site D of E67, mimics the side chain of histone H3 Ala7 (see panel d), is bound in a shallow surface pocket of KIAA1718.
E67 lies in a location occupied by the histone H3 peptide in the PHF8-peptide complex (compare Fig. 2b with Fig. 2c). E67 occupies approximately the same space as that by histone H3 lysine 4 (main chain) to lysine 9 (Fig. 2d), with a total of buried interface area of 422 Å2. The acidic binding pocket is wide open and the α-ketoglutarate can be seen at the bottom of the pocket (Fig. 2e). The 5-aminopentyloxy moiety at site A is extended near the metal cofactor in the active site, with the aliphatic chain stacking with the planar surface of His282 and the terminal amino group forming a weak hydrogen bond with one of the carboxyl oxygen atoms of Asp284 (3.4 Å) (Fig. 2f). The side chains of His282 and Asp284, together with His354, coordinate the binding of the metal ion. The N,N-dimethylpropylamine side chain at site B is near the acidic entrance of the substrate binding groove on the protein surface (Fig. 2e) and the terminal dimethylated amino group folds back and stacks with the planar surface of the quinazoline ring (Fig. 2f).
The methyl group of the O6 methoxy at site D is inserted into a shallow surface pocket of an inner wall formed by Gln200 and Thr279 (Fig. 2g), mimicking the side chain of histone H3 Ala7 (H3A7; Fig. 2d). The O6 methyoxy-H3A7 mimic has also been observed in the interactions of E67 as well as BIX with GLP methyltransferase 12; 14. Thus, we suggest the O6 methyoxy at site D is an important discriminator of H3K9 enzymes (methylase and demethylases) vs. H3K4 enzyme(s) (see below). The N-benzyl-4-amino-piperidine moiety at site C is not observed in the structure. We could however model the moiety in the current structure without steric clashes with the protein by pointing the moiety to the solvent (Supplementary Fig. S1d).
Generation of compound E67-2
Based on the structural information of KIAA1718-E67 complex, we thus generated a smaller compound (E67-2) without the N-benzyl-4-amino-piperidine moiety. E67-2 inhibits KIAA1718 Jumonji domain with an IC50 value of 3.4 µM (Fig. 1c), the same as that of E67 (Fig. 1b). Removal of both of the side chains at sites B and C (compound E67-5) increases the IC50 value by a factor of approximately 5 (Fig. 1d). Compounds E67 and E67-2 retain the inhibitory potency towards related PHF8 Jumonji domain (Fig. 1e).
Compound E67 was originally designed to improve the potency against H3K9 methyltransferase activities of G9a and GLP (with an IC50 value of 50 nM; Fig. 1f) 14. Unexpectedly, the new E67 derivative (E67-2) reduced inhibitory effect against GLP by a factor of approximately 1500, resulting in an IC50 of 75 µM (Fig. 1f).
Effect of PHD domain on the activity of Jumonji domain
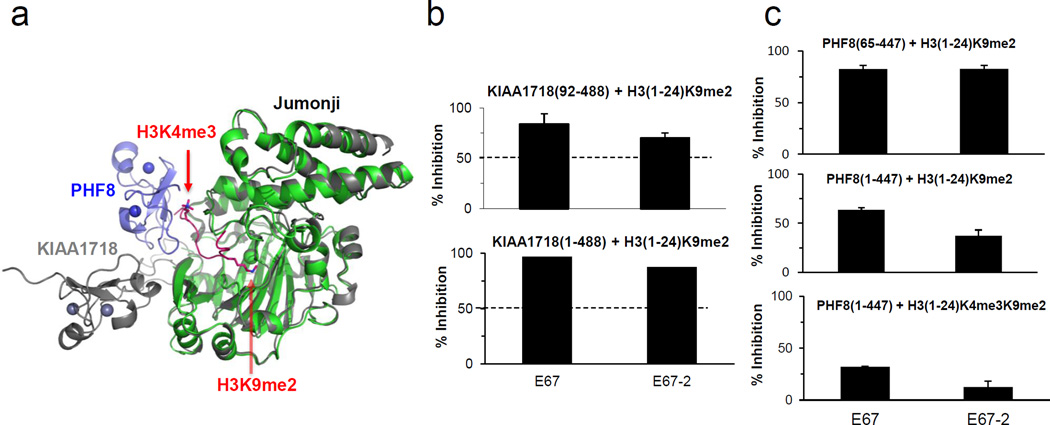
KIAA1718 and PHF8 have a N-terminal H3K4me3-binding PHD domain linked to their respective catalytic Jumonji domain (Fig. 3a) {Horton, 2010 #919}. The inhibition (under the same assay conditions) for the KIAA1718 (residues 1–488), including both the PHD and Jumonji domains, is comparable to those measured for the Jumonji domain only (residues 92–488) (Fig. 3b), suggesting that the PHD domain of KIAA1718 has minor influence on the inhibitory properties of these compounds that target the Jumonji active site. On the other hand, while retaining the inhibitory potency towards related PHF8 Jumonji domain (Fig. 1e and Fig. 3c, top panel), compounds E67 and E67-2 have much reduced inhibition on PHF8 (residues 1–447) on the doubly methylated H3(1-24)K4me3K9me2 peptide substrate (Fig. 3c, bottom panel). This can be explained by the structural observation that KIAA1718 has an extended conformation between the two domains that allows the compound to be diffused into the active site, whereas PHF8 has a closed conformation, which limits the access to the Jumonji active site when its PHD domain engages H3K4me3 (Fig. 3a). Removal of the N-terminal PHD domain from the protein (Fig. 3c, top panel) or the H3K4me3 mark from the substrate (Fig. 3c, middle panel) renders these inhibitors more effective in inhibiting H3K9me2 demethylation activity of PHF8, indicating that binding of the H3K4me3 mark by the PHD domain impairs the inhibitor binding to the active site (Fig. 3c, bottom panel).
Fig. 3. Effect of PHD domain on the activity of Jumonji domain.
(a) Superimposition of KIAA1718 (PDB 3KV5; colored grey) and PHF8 (PDB 3KV4; colored blue for PHD and green for Jumonji) with a bound H3 peptide containing K4me3 and K9me2 (colored red).
(b) Percentage of inhibition for KIAA1718 (residues 1–488) (bottom panel) is comparable to those measured for the Jumonji domain only (residues 92–488) (top panel).
(c) Percentage of inhibition of the H3K9me2 demethylation activities of PHF8 Jumonji domain (residues 65–447) on H3(1-24)K9me2 (top panel), PHF8 (residues 1–447) on H3(1-24)K9me2 (middle panel), and PHF8 (residues 1–447) on the doubly methylated H3(1-24)K4me3K9me2 peptide substrate (bottom panel), under the conditions of 10 µM [I], 5 µM [S] and 2.5 µM [E] in 50 mM HEPES pH 7.0, 1 mM αKG, 50 µM Fe(NH4)2(SO4)2, and 2 mM ascorbic acid. For PHF8(1-447) activity on H3(1-24)K4me3K9me2 substrate (bottom panel), reaction mixtures were incubated for 10 minutes at 37 °C. For PHF8(1-447) and PHF8(65-447) on H3(1-24)K9me2 substrate, mixtures were incubated for 60 minutes at 37 °C. These time points were chosen to access the linear range of the product formation curve by these enzymes without completely exhausting the initial substrate.
Selectivity over JARID1C, an H3K4me3 demethylase
We next tested whether the current inhibitors (E67 and E67-2) against KIAA1718 and PHF8 (two related H3K9me2 demethylases) can also inhibit H3K4me3 demethylation activity of JARID1C enzyme (Supplementary Fig. S2a). We purified the N-terminal half of JARID1C (residues 1–839) containing the catalytic Jumonji domain (Supplementary Fig. S2b). The JARID1C is an active H3K4me3 demethylase, with an optimum pH at 6.5–6.8 (Supplementary Fig. S2c) and maximal activity at 50 mM NaCl (Supplementary Fig. S2d). The demethylation activity is also dependent on substrate H3 peptide length and is active on peptide longer than 12 amino acids (Supplementary Fig. S2e).
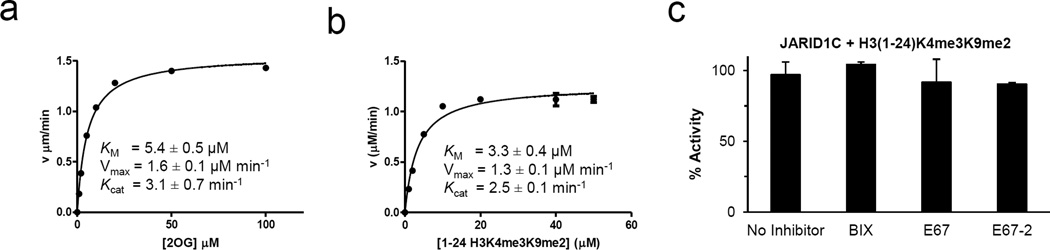
We used a fluorescence-based demethylation assay based on a formaldehyde dehydrogenase-coupled continuous reaction 20 to determine the kinetic characteristics of JARID1C. We determined the KM for α-ketoglutarate to be approximately 5 µM (Fig. 4a), and KM for peptide substrate approximately 3 µM (under the conditions of α-ketoglutarate concentration being 200 µM, at which reaction velocity is well saturated) (Fig. 4b). The turnover rate (Kcat) of JARID1C is approximately 2.5 ~ 3 min−1, which is significantly faster than most known Jumonji histone lysine demethylases (comparing Kcat ~ 0.1 min−1 for PHF8 and KIAA1718 20 and Kcat ~ 0.01–0.07 min−1 for JMJD2A and JMJD2D 25).
Fig. 4. Kinetic parameters and inhibition of JARID1C.
Michaelis-Menten plots of JARID1C (residues 1–839) for α-ketoglutarate (αKG) (a) and substrate H3 peptide (b). Initial velocity was plotted against αKG or peptide concentration and fitted with the Michaelis-Menten equation. Different concentrations of formaldehyde standards were made fresh and used to calibrate fluorescence intensities to determine initial velocities. The turnover number of FDH is fast enough to ensure the clearance of formaldehyde from JARID1C catalysis is not delayed by FDH conversion. Reactions were performed in 40 µl volume under the conditions of 20 µM H3(1-24)K4me3K9me2 [S], 0.5 µM JARID1C [E], 2 mM ascorbic acid, 1 mM αKG, 0.1 mM Fe(II), 20 mM MES pH 6.5, 50 mM NaCl, 0.6 mM APAD+ and 3 µM FDH at 37 °C for 10 min monitored by a BioTek Synergy™ 4 Hybrid Microplate Reader. All elements were pre-incubated at 37 °C for 15 min followed by addition of peptide and APAD+ to initiate the reaction. Experiments were done in triplicates and initial velocities were then plotted against peptide or α-KG (also know as 2-oxoglutarate or 2OG) concentrations and fit with the Michaelis-Menten equation using GraphPad Prism 5.0.
(c) H3K4me3 demethylation activity of JARID1C(1-839) on H3(1-24)K4me3K9me2 substrate, under the conditions of 0.25 µM enzyme [E], 10 µM peptide substrate [S] and 10 µM inhibitor [I] in 50 mM MES pH 6.8. Enzyme was pre-incubated with inhibitor at room temperature (~ 21 °C) prior to the addition of substrate peptide and the reactions were lasted for 10 min at 37 °C.
As evident from the activity data (Fig. 4c), neither BIX nor its analogs (E67 and E67-2) can significantly inhibit JARID1C demethylation activity on the H3K4me3 peptide substrates. This is consistent with the fact that recognition of H3K4 and H3K9 methylation marks involves different sets of backbone and side chain interactions and the BIX analogs tested in this work are originally designed to mimic H3K9 binding.
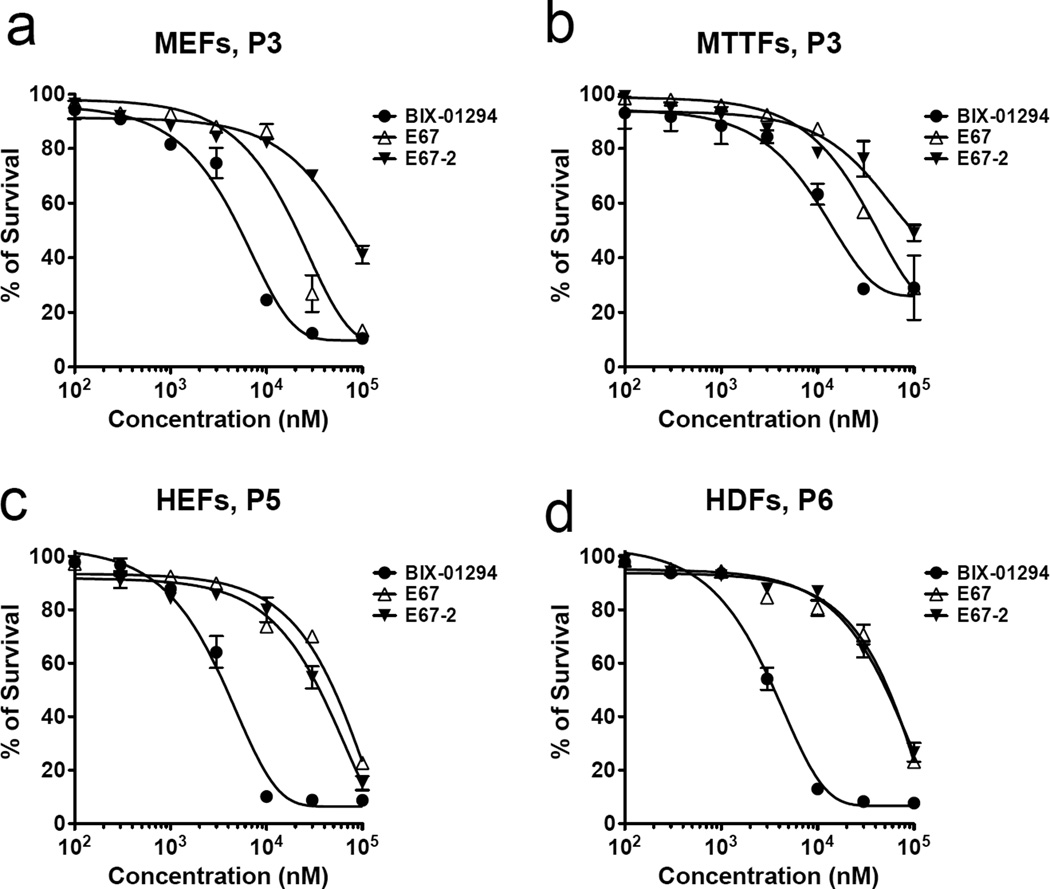
Finally, we tested the cell toxicity of the three compounds in mouse and human primary fibroblasts. Like E67 14, E67-2 has significantly reduced cell toxicity in vivo (Fig. 5), compared with BIX, in all four types of mouse and human primary fibroblasts treated. For E67-2, the LC50 (half-maximal lethal concentration) values are 45.8 µM in mouse cells and 52.1 µM in human cells, respectively, comparable to that of E67 (24.1 µM for mouse cells and 95.6 µM for human cells) but much higher than that of BIX-01294 (7.4 µM for mouse cells and 3.2 µM human cells).
Fig. 5. Cytotoxicity of E67-2 compounds to primary fibroblasts.
Mouse and human primary fibroblasts were treated with various concentrations of BIX-01294, E67, and E67-2. Primary mouse embryonic fibroblasts (MEFs) and mouse tail-tip fibroblasts (MTTFs) were isolated from E13.5 embryo and tail, respectively, from 129S2/SvPasCrl inbred mouse (Charels River, Wilmington, MA, USA). Human embryonic fibroblasts (HEFs), HE-19, was purchased from RIKEN Cell Bank (Ibaraki, Japan). Human dermal fibroblasts (HDFs) were purchased from American Type Culture Collection (ATCC, Washington, DC, USA). All primary cells (100 µl) were cultured in Dulbecco’s modified Eagle’s Media (DMEM, Invitrogen, Carlsbad, CA, USA) supplement with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA, USA), 1x Antibiotic-Antimycotic (Invitrogen, Carlsbad, CA, USA), 2 mM L-glutamine (GlutaMAX, Invitrogen, Carlsbad, CA, USA), 100 µM Minimum Essential Media Non-Essential Amino Acids (MEM NEAA, Invitrogen, Carlsbad, CA, USA) and 100 µM β-mercaptoethanol (Sigma, St. Louis, MO, USA). Cell cytotoxicity was measured with the cell counting kit-8 (CCK-8, Dojindo, Kumamoto, Japan). Five thousand cells in 100 µl were seeded per well. After incubation for 24 h at 37°C, 5% CO2, cells were treated with BIX-01294, E67, and E67-2 at 100, 300, 1,000, 3,000, 10,000, 30,000, and 100,000 nM and incubated another 24 h. A 10 µl of CCK-8 solution was added to each well and the plate was incubated for another 4 h. Cell viability was determined with colorimetric assays for measuring the activity of dehydrogenase enzyme from live cells that reduce a chemical dye, WST-8, to an orange-color formazan dye which can be detected at a specific wavelength of light. The absorbance of formazan dye at 450 nm is directly proportional to the number of living cells. The absorbance was read at 450 nm using Synergy 2 microplate reader (BioTek, Winooski, VT, USA).
Discussion
Here we show that BIX-01294, a H3K9 peptide mimic small molecule compound, can inhibit both H3K9 methyltransferase (GLP: IC50 = 0.25 µM; Fig. 1f) and demethylase (KIAA1718: IC50 =16.5 µM; Fig. 1a). Because the IC50 values are enzyme concentration dependent, we thus compared Kiapp Copeland (2005) 26 for the two H3K9 modifying enzymes (Fig. 1g). Both BIX and E67 inhibit GLP better than KIAA1718 by a factor of approximately 76 (comparing Kiapp values of 0.2 vs. 15.3 µM) and 2500 (0.001 vs. 2.5 µM), respectively. By deleting the benzylated six-membered piperidine ring moiety, as suggested by the co-crystal structure of KIAA1718-E67 complex, E67-2 is more selective against KIAA1718 than GLP by a factor of 35 (comparing Kiapp values of 2.1 vs. 75 µM). This reversal of selectivity is only due to the severe loss in activity against GLP without affecting inhibition against KIAA1718. Therefore, the current structure of KIAA1718–E67 complex provides new avenues for further improving the potency and selectivity of the inhibitor by strengthening KIAA1718-E67 interactions. For example, the N,N-dimethylpropylamine side chain at site B has little contact with the protein and replacement with group(s) that maximize the interaction near the entrance of the substrate binding groove may increase the potency against KIAA1718. Iterative cycles of crystallography, synthesis, and bioassay will ultimately aid successful design of selective and potent epigenetic inhibitors of histone lysine demethylases.
Synthesis of compounds
E67 has been prepared as previously reported 14. For E67 derivatives (Supplementary Fig. S3), the anthranilic nitrile 2 was prepared by alkylation with benzyl bromide under basic conditions of the commercially available 4-hydroxy-3-methoxybenzonitrile 1 followed by nitration under standard conditions, and subsequent reduction of the nitro intermediate with iron dust in an acidic medium. The intermediate 2 was then converted into the dichloroquinazoline 3 through a reaction with methyl chloroformate followed by cyclization with hydrogen peroxide under basic conditions, and final halogenation by means of POCl3 under reflux conditions. Treatment of 3 with dimethylamine at room temperature provided the 4-substituted intermediate 4. This latter was converted into 5 by the mean of a further treatment with dimethylamine at 110 °C in a sealed tube. The intermediate 5 was then converted into the final compound E67-5 by debenzylation through catalytic hydrogenation, followed by Mitsunobu reaction with the Boc-protected 5-aminopentan-1-ol, and final deprotection and salification with HCl. The intermediate 6 was prepared starting from 4 by debenzylation with trifluoroacetic acid under reflux conditions and subsequent Mitsunobu reaction with the Boc-protected 5-aminopentan-1-ol. The chloroquinazoline 6 was then converted into the final compound E67-2 by reaction with the commercially available N1,N1-dimethylpropane-1,3-diamine at 110 °C in a sealed tube and final deprotection and salification with HCl.
Crystallography
The X-ray diffraction data (Table 1) were collected at the Advanced Photon Source, Argonne National Laboratory, SERCAT beamline (22-BM). Diffraction data were processed and scaled by HKL2000 27. We used PHENIX 28 for molecular replacement, using a previously solved KIAA1718 (residues 92–488) structure (PDB 3KVA) 20 as the initial model, and refinement. Coot 29 was used for graphic model building. PRODRG server 30 generated the initial model of E67 compound. The ReadySet software integrated in PHENIX generated the structural restraints for the inhibitor molecule. PyMOL (DeLano Scientific) was used to prepare the structural figures.
Highlights.
BIX-01294 inhibits both H3K9 methyltransferase and H3K9 Jumonji demethylase
E67-2 selectively inhibits H3K9 Jumonji demethylase over H3K9 methyltransferase as well as H3K4 demethylase
E67-2 has low cytotoxicity
Supplementary Material
Acknowledgements
We thank Dr. John R. Horton for maintaining local X-ray facility and help with X-ray data collection in synchrotron, Dr. Justin E. Jones for comments and Dr. Robert A. Copeland for suggestion of calculating Kiapp values. The Department of Biochemistry at the Emory University School of Medicine supported the use of the SER-CAT synchrotron beamline at the Advanced Photon Source of Argonne National Laboratory, local X-ray facility and MALDI-TOF mass spectrometry. This work was supported by NIH grants (GM068680-07 to X.C and GM092035-01 to X.C. and Y.Y.), by PRIN 2009PX2T2E and by the FP7 Project BLUEPRINT/282510 to A.M. X.C. is a Georgia Research Alliance Eminent Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession number
Protein Data Bank: The coordinates and structure factors have been deposited with accession numbers 3U78 for KIAA1718 Jumonji domain in complex with E67.
Author Contributions
A.K.U. performed KIAA1718 and PHF8 purifications, crystallographic analysis of E67-bound KIAA1718 Jumonji domain structure that leads to the designing of E67 analogs and mass spectrometry-based inhibition assays including the inhibition studies on GLP activity with E67 analogs; R.H. performed purification of JARID1C and fluorescence-based kinetic measurements; Y.C. purified GLP enzyme; D.R., D.L. and A.M. performed chemical synthesis and wrote the method of chemical synthesis; J.W.H. and Y.Y. performed cytotoxicity assay and wrote the methods thereof; X.Z. guided JARID1C experiments and X.C. organized and designed the scope of the study; A.K.U. and X.C. wrote the manuscript and all were involved in analyzing data and helped in writing and revising the manuscript.
Supporting Information
Supplementary data associated with this article can be found, in the online version at doi:
References
- 1.Clissold PM, Ponting CP. JmjC: cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2beta. Trends Biochem Sci. 2001;26:7–9. doi: 10.1016/s0968-0004(00)01700-x. [DOI] [PubMed] [Google Scholar]
- 2.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 3.Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- 4.Pedersen MT, Helin K. Histone demethylases in development and disease. Trends Cell Biol. 2010;20:662–671. doi: 10.1016/j.tcb.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Copeland RA, Olhava EJ, Scott MP. Targeting epigenetic enzymes for drug discovery. Curr Opin Chem Biol. 2010;14:505–510. doi: 10.1016/j.cbpa.2010.06.174. [DOI] [PubMed] [Google Scholar]
- 6.Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010;28:1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rotili D, Mai A. Targeting Histone Demethylases: A New Avenue for the Fight against Cancer. Genes & Cancer. 2011;2:663–679. doi: 10.1177/1947601911417976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King ON, Li XS, Sakurai M, Kawamura A, Rose NR, Ng SS, Quinn AM, Rai G, Mott BT, Beswick P, Klose RJ, Oppermann U, Jadhav A, Heightman TD, Maloney DJ, Schofield CJ, Simeonov A. Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors. PLoS ONE. 2010;5:e15535. doi: 10.1371/journal.pone.0015535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rose NR, Woon EC, Kingham GL, King ON, Mecinovic J, Clifton IJ, Ng SS, Talib-Hardy J, Oppermann U, McDonough MA, Schofield CJ. Selective inhibitors of the JMJD2 histone demethylases: combined nondenaturing mass spectrometric screening and crystallographic approaches. J Med Chem. 2010;53:1810–1818. doi: 10.1021/jm901680b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamada S, Suzuki T, Mino K, Koseki K, Oehme F, Flamme I, Ozasa H, Itoh Y, Ogasawara D, Komaarashi H, Kato A, Tsumoto H, Nakagawa H, Hasegawa M, Sasaki R, Mizukami T, Miyata N. Design, synthesis, enzyme-inhibitory activity, and effect on human cancer cells of a novel series of jumonji domain-containing protein 2 histone demethylase inhibitors. J Med Chem. 2010;53:5629–5638. doi: 10.1021/jm1003655. [DOI] [PubMed] [Google Scholar]
- 11.Kubicek S, O'Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, Kelly TA, Jenuwein T. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 12.Chang Y, Zhang X, Horton JR, Upadhyay AK, Spannhoff A, Liu J, Snyder JP, Bedford MT, Cheng X. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nat Struct Mol Biol. 2009;16:312–317. doi: 10.1038/nsmb.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu F, Chen X, Allali-Hassani A, Quinn AM, Wasney GA, Dong A, Barsyte D, Kozieradzki I, Senisterra G, Chau I, Siarheyeva A, Kireev DB, Jadhav A, Herold JM, Frye SV, Arrowsmith CH, Brown PJ, Simeonov A, Vedadi M, Jin J. Discovery of a 2,4-diamino-7-aminoalkoxyquinazoline as a potent and selective inhibitor of histone lysine methyltransferase G9a. J Med Chem. 2009;52:7950–7953. doi: 10.1021/jm901543m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang Y, Ganesh T, Horton JR, Spannhoff A, Liu J, Sun A, Zhang X, Bedford MT, Shinkai Y, Snyder JP, Cheng X. Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases. J Mol Biol. 2010;400:1–7. doi: 10.1016/j.jmb.2010.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu F, Barsyte-Lovejoy D, Allali-Hassani A, He Y, Herold JM, Chen X, Yates CM, Frye SV, Brown PJ, Huang J, Vedadi M, Arrowsmith CH, Jin J. Optimization of Cellular Activity of G9a Inhibitors 7-Aminoalkoxy-quinazolines. J Med Chem. 2011;54:6139–6150. doi: 10.1021/jm200903z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vedadi M, Barsyte-Lovejoy D, Liu F, Rival-Gervier S, Allali-Hassani A, Labrie V, Wigle TJ, Dimaggio PA, Wasney GA, Siarheyeva A, Dong A, Tempel W, Wang SC, Chen X, Chau I, Mangano TJ, Huang XP, Simpson CD, Pattenden SG, Norris JL, Kireev DB, Tripathy A, Edwards A, Roth BL, Janzen WP, Garcia BA, Petronis A, Ellis J, Brown PJ, Frye SV, Arrowsmith CH, Jin J. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat Chem Biol. 2011;7:566–574. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleefstra T, Brunner HG, Amiel J, Oudakker AR, Nillesen WM, Magee A, Genevieve D, Cormier-Daire V, van Esch H, Fryns JP, Hamel BC, Sistermans EA, de Vries BB, van Bokhoven H. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am J Hum Genet. 2006;79:370–377. doi: 10.1086/505693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 19.Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, Shi Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- 20.Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol. 2010;17:38–43. doi: 10.1038/nsmb.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, Desai A, Dorrestein PC, Glass CK, Rosenfeld MG. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466:508–512. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, Yaghi NK, Lim H, Garcia BA, Brizuela L, Zhao K, Roberts TM, Shi Y. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010;466:503–507. doi: 10.1038/nature09261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang C, Xiang Y, Wang Y, Li X, Xu L, Zhu Z, Zhang T, Zhu Q, Zhang K, Jing N, Chen CD. Dual-specificity histone demethylase KIAA1718 (KDM7A) regulates neural differentiation through FGF4. Cell Res. 2010;20:154–165. doi: 10.1038/cr.2010.5. [DOI] [PubMed] [Google Scholar]
- 24.Yokoyama A, Okuno Y, Chikanishi T, Hashiba W, Sekine H, Fujiki R, Kato S. KIAA1718 is a histone demethylase that erases repressive histone methyl marks. Genes Cells. 2010;15:867–873. doi: 10.1111/j.1365-2443.2010.01424.x. [DOI] [PubMed] [Google Scholar]
- 25.Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat Struct Mol Biol. 2007;14:689–695. doi: 10.1038/nsmb1273. [DOI] [PubMed] [Google Scholar]
- 26.Copeland RA. Evaluation of enzyme inhibitors in drug discovery: a guide for medicinal chemists and pharmacologists. Hoboken, New Jersey: John Wiley & Sons, Inc.; 2005. p. 182. [PubMed] [Google Scholar]
- 27.Otwinowski Z, Borek D, Majewski W, Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr A. 2003;59:228–234. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- 28.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 30.Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 31.Straus OH, Goldstein A. Zone behavior of enzymes illustrated by the effect of dissociation constant and dilution on the system cholinesterase-physostigmine. J. Gen. Physiol. 1943;26:559–585. doi: 10.1085/jgp.26.6.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.