Abstract
RecQ helicases feature multiple domains in their structure, of which the helicase domain, the RecQ-Ct domain and the HRDC domains are well conserved among the SF2 helicases. The helicase domain and the RecQ-Ct domain constitute the catalytic core of the enzyme. The domain interfaces are the DNA binding sites which display significant conformational changes in our molecular dynamics simulation studies. The preferred conformational states of the DNA bound and unbound forms of RecQ appear to be quite different from each other. DNA binding induces inter-domain flexibility leading to hinge mobility between the domains. The divergence in the dynamics of the two structures is caused by changes in the interactions at the domain interface, which seems to propagate along the whole protein structure. This could be essential in ssDNA binding after strand separation, as well as aiding translocation of the RecQ protein like an inch-worm.
Keywords: Helicase mechanism, Homology modeling, Molecular Dynamics simulations, unwinding, translocation
Background:
RecQ helicase is a member of the helicase superfamily 2 (SF2) [1– 2]; it translocates in the 3' to 5' direction and contains the conserved DEAH box motif [3]. The enzyme plays an important role in DNA damage response, chromosomal stability maintenance and has a vital role in maintaining genome homeostasis [4– 6]. RecQ is found to be conserved among both prokaryotes and eukaryotes, among higher organisms, multiple paralogs of the enzyme have been observed. In humans, five members of this family are currently known out of which mutations in three have been found to be linked to enhanced sister chromatid exchange and hereditary diseases (Bloom syndrome, Werner syndrome, and Rothmund-Thomson syndrome), which display clinical symptoms of premature aging and predisposition to cancers [7– 9].Three conserved sequence elements are commonly found in RecQ helicases, namely the Helicase domain, the RecQ-C-terminal (RecQ-Ct) and Helicase-and-RNaseD-like-C-terminal (HRDC) domains [10]. The helicase and RecQ-Ct domains together constitute the catalytic core of RecQ. In addition to these elements, eukaryotic RecQ proteins often contain N - and C - terminal extensions that confer additional functions like exonuclease domain in WRN, nuclear localization signals in BLM [9, 10] and the Zn finger and winged helix motifs in the RecQ-Ct domain [11].The complete structure of RecQ helicases has so far been resistant to attempts of crystallisation, so the X-ray structure is available for only two domains of the E.coli RecQ protein. We have adopted the homology modeling technique to construct the structure of the enzyme and carried out molecular dynamics (MD) simulations of the RecQ model and its DNA-docked complex to understand the mechanism of action of the protein. Although complete conformational sampling for a multi-domain protein requires an MD trajectory of very long time scales, snapshots of domain motions that are viable can be investigated by sampling small time segments. Studies on related members of the family suggest that DNA binding enhances the inherent flexibility in the structure [12, 13]. Our molecular dynamics (MD) studies predict that the preferred conformational states of the DNARecQ complex and RecQ are distinct from each other. Besides, the DNA binding seems to augment domain flexibility and coordinate domain movements in the structure that may eventually facilitate ssDNA binding, culminating in strand separation.
Methodology
MODELLER9v5 [14] was used to generate multiple models of RecQ (GenPept ID: YP_001465308) using three template structures (PDB ID: 1WUD, 1OYW, 1N4A). The best model [Figure 2 (a)] was selected on the basis of DOPE assessment score, and the MODELLER objective function. Quality of the best model selected was improved further by performing loop refinement using MODELLER. After loop refinement the best model selected on the basis of the objective function was then subjected to 500 steps of steepest descent and 200 steps of conjugate gradient energy minimization methods using GROMACS 3.3-2[15]. The energy minimization process converged for both the methods and the energy minimized structure thus generated was evaluated using the server PROCHECK [16].
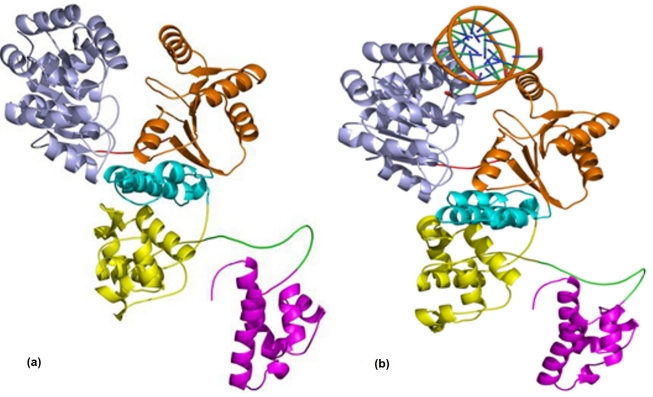
Figure 2.
The RecQ Model generated from (a) Modeller; (b) the DNA-bound RecQ complex. The helicase subdomains are shown in light blue and orange colour, RecQ-Ct domain is shown in cyan and yellow colour and HRDC domain in magenta colour.
Experimental studies show that E.coli RecQ helicase has strong strand separation activity for double stranded blunt ended DNA [1], so a blunt ended dsDNA with one complete turn was used for docking the RecQ protein. The best model generated for E.coli RecQ from MODELLER9v5 was docked with double stranded (ds) blunt end DNA (NDB entry BD0026) [Figure 2 (b)] using PATCHDOCK [17]. The interaction surface of the RecQ was predicted based on its similarity with its homologue and, presence of positively charged residues on the predicted binding surface. Molecular dynamics simulation of the E.coli RecQ was carried out to confirm that the predicted model is stable and also to understand the dynamics of the system and to explore the possible conformational states of the system. MD simulations were also carried out for E.coli RecQ and its DNA complex; the results provided insight into the mechanism by which the enzyme may function.
Discussion:
Failure to crystallize the whole E.coli RecQ protein suggests that the highly flexible loop connecting the HRDC domain and the catalytic core domain may be detrimental to it. Bernstein et al. [18] suggested that the HRDC domain preferentially binds single stranded DNA after strand separation and then moves closer to the catalytic core domain, facilitated by the flexible loop. There is no experimental evidence to suggest any interaction between the catalytic core and HRDC domain. As a long DNA is the natural substrate of the enzyme, the HRDC domain need not necessarily move to the catalytic site for the domain to be able to hold single stranded DNA and for the protein to be stable. Therefore, the extended structure [Figure 1] predicted by MODELLER9v5 posed to be a reasonable model for further investigation which was supported by structure validation based on quality assessment. During the 5 ns MD simulation, the structure of the RecQ model remained stable and there were no major changes in the conformation, where as the structure of the DNA-RecQ complex, changed conformation and showed significant dynamics. Predominant domain movements were observed while sampling the structures as the dynamics evolved. Using single-linkage method of cluster formation [15] the structures at different time steps during the simulation were clustered to find similar conformational states attained during the simulation. The trajectories of the RecQ structure displayed three major clusters; one centered about 840ps structure, the other two about 1440ps and 4760ps structures but, the representative structures of these clusters were not very different from each other [Figure 3 (a)]. While, in the DNA-RecQ complex, there were two major clusters; one centered about 800ps structure and the other one about 3.7 ns structure and, the representative structures were quite different from each other [Figure 3 (b)]. These observations suggest that in the absence of DNA the domains are in a stable conformation, which is clearly not the case for the RecQ-DNA complex. The helicase domain of RecQ has two subdomains and, analysis of the RecQ structure at different time steps suggested that on binding with DNA a hinge movement was induced between the subdomains. The helicase subdomain2 drifted away from the subdomain1 carrying with it the RecQ-Ct and HRDC domain.
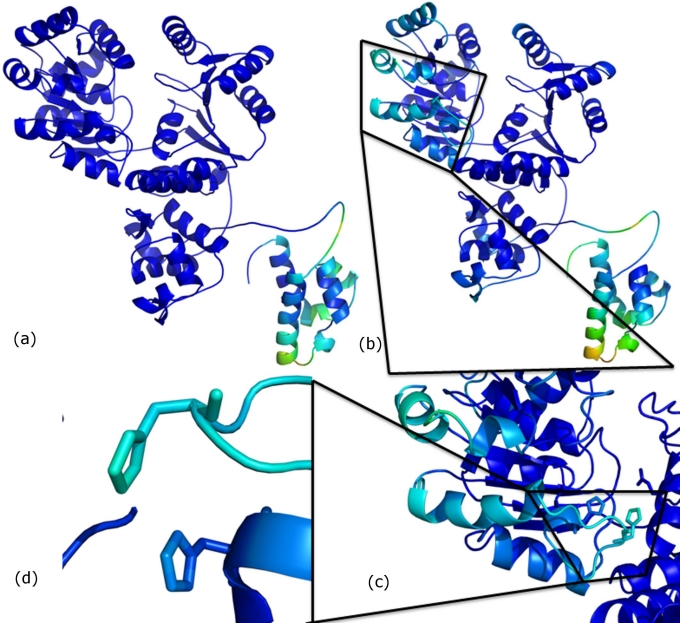
Figure 1.
The B-factor calculations for (a) the RecQ simulation and (b) for the RecQ-DNA complex. The HRDC domain appears in a lighter shade indicating higher flexibility in the region (c) some residues in helicase domain show fluctuation on DNA binding (d) these residues include His149 and His156.
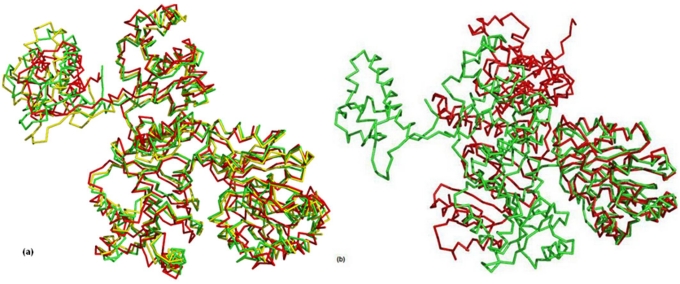
Figure 3.
(a) Representative structures of major clusters for RecQ at 840ps (green), 1440ps (yellow) and 4760ps (red) time steps, respectively; (b) Representative structure of major clusters for DNA-RecQ complex at 800ps (green) and 3700ps (red) time steps, respectively. Hinge motion moves the helicase subdomain2 away from the helicase subdomain1.
The root mean square deviation of the domains at different time step with reference to the starting structure showed fluctuations which were quite distinct for the 3 major domains. The RMSD in the helicase domain was more for the RecQ structure (˜2.7Å) compared to the DNA-RecQ complex (˜2.2Å) while, the RMSD of the RecQ-Ct domain was more (3.5Å) in the DNA-RecQ complex as compared to the RecQ structure (˜2.9Å). Similarly, the HRDC domain deviated less (˜2.3Å) in the RecQ structure compared to the complex (˜2.7Å). The increase in RMSD of the RecQ-Ct domain and the HRDC domain may be attributed to the presence of DNA binding motifs e.g., the winged helix and helix turn helix motif, which fluctuate more in the DNA bound form which could enhance DNA binding affinity [19] whereas, stability of helicase domain in the complex form may be explained by the non-covalent interactions between the enzyme and its substrate.
To find out the average fluctuation of each residue in the RecQ protein and in the DNA - RecQ complex, RMSF of each residue was calculated after fitting to a reference frame and then converted to B-factor values [15]. For the RecQ only structure, regions other than the HRDC domain did not have much fluctuation [Figure 1 (a)]. The HRDC region was expected to be comparatively more flexible than the rest of the structure as it was connected by a flexible loop. However, the fluctuation in the HRDC domain was larger in the DNA-RecQ complex, where the HRDC domain showed intrinsic fluctuation of amino acid residues in addition to whole domain motion [Figure 1 (b)]. The fluctuation was more for residues on helix 1 of HRDC domain when DNA was bound, these residues are known to be crucial for single strand DNA binding [19– 20]. It appears that the ssDNA binding positively charged residues on the surface of helix 1 of HRDC domain were induced by fluctuations in the winged helix region of RecQ-Ct domain. Fluctuation was observed in some residues of helicase subdomain1 [Figure 1 (c)] including two histidine residues (His149 and His156) [Figure 1 (d)] which are proposed to be involved in the interaction between the two subomains [11]. This observation suggests that these residues serve as a messenger for the subsequent domains so that, the changes required in conformation of the protein can be induced on substrate binding.
Conclusion:
DNA binding at the helicase domain induces fluctuation in the subsequent domains. The HRDC domain known to bind ssDNA, helps to tether the separated ssDNA and prevents reannealing. The HRDC domain contains positively charged residues on helix 1 which are crucial for its DNA binding affinity. In other words, upon DNA binding at the interface of the helicase subdomains, the whole RecQ helicase undergoes a series of induced motions which are co-ordinated. Comparison of the behavior of RecQ helicase with its homologue RepA [12] and PcrA [13] helicase display similarility in domain motions. The subdomains in the structures alternately bind the double stranded and single stranded DNA and inch forward along the strand in the 3‘ to 5’ direction and there are periods in the cycle of motion when the helicase is attached to both the single stranded and double stranded parts of the substrate [13]. We perceived analogous dynamics in the RecQ helicase too from our MD simulation. The dynamic behaviour of the E.coli RecQ helicase has been revealed from the simulations for the first time. The existence of preferred conformational states for the RecQ protein in the free state and DNA bound state are quite distinct from each other which gives a molecular basis to change in shape upon interaction with DNA. Substrate binding at the interface of the subdomains triggers co-ordinated domain motions which ordain it the inchworm mechanism of action.
Acknowledgments
The MD calculations have been performed mostly in the computer server of the Center for Computational Natural Sciences and Bioinformatics, IIIT-Hyderabad, INDIA, as part of the M.S. thesis of NP, SG was faculty here before shifting to her current position. The authors thank Dr. B. Gopalakrishnan (A.T.C., TCS, Hyderabad), Dr. B. Prabhakar (IIIT- Hyderabad) and Dr. Deva Priyakumar (IIIT- Hyderabad) for their valuable discussions and support.
Footnotes
Citation:Pandey et al, Bioinformation 7(8): 371-374(2011)
References
- 1.P Janscak, et al. J Mol Biol. 2003;330:29. [Google Scholar]
- 2.DA Bernstein, JL Keck. Nucleic Acids Research. 2003;31:2778. doi: 10.1093/nar/gkg376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.ME Fairman-Williams, et al. Curr Opin Struct Biol. 2010;20:313. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WK Chu, ID Hickson. Nat Rev Cancer. 2009;9:644. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 5.L Bjergbaek, et al. Swiss Med Wkly. 2002;132:433. [Google Scholar]
- 6.JA Cobb, et al. FEBS Lett. 2002;529:43. [Google Scholar]
- 7.ID Hickson. Nat Rev Cancer. 2003;3:169. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 8.CJ Epstein, et al. Medicine. 1966;45:177. [Google Scholar]
- 9.S Kitao, et al. Genomics. 1999;61:268. [Google Scholar]
- 10.A Vindigni, ID Hickson. HFSP J. 2009;3:153. doi: 10.2976/1.3079540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MP Killoran, JL Keck. Nucleic Acids Res. 2006;34:4098. doi: 10.1093/nar/gkl538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.S Korolev, et al. Cell. 1997;90:635. [Google Scholar]
- 13.SS Velankar, et al. Cell. 1999;97:75. [Google Scholar]
- 14.A Sali, TL Blundell. J Mol Biol. 1993;234:779. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 15.E Lindahl, et al. J Mol Mod. 2001;7:306. [Google Scholar]
- 16.RA Laskowski, et al. J Appl Cryst. 1993;23:283. [Google Scholar]
- 17.D Schneidman- Duhovny, et al. Nucleic Acids Res. 2005;33:W363. [Google Scholar]
- 18.DA Bernstein, et al. EMBO J. 2003;22:4910. doi: 10.1093/emboj/cdg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DA Bernstein, JL Keck. Structure. 2005;13:1173. [Google Scholar]
- 20.Z Liu, et al. Structure. 1999;7:1557. [Google Scholar]