ABSTRACT
Many Gram-negative bacteria utilize specialized secretion systems to inject proteins (effectors) directly into host cells. Little is known regarding how bacteria ensure that only small subsets of the thousands of proteins they encode are recognized as substrates of the secretion systems, limiting their identification through bioinformatic analyses. Many of these proteins require chaperones to direct their secretion. Here, using the newly described protein interaction platform assay, we demonstrate that type 3 secretion system class IB chaperones from one bacterium directly bind their own effectors as well as those from other species. In addition, we observe that expression of class IB homologs from seven species, including pathogens and endosymbionts, mediate the translocation of effectors from Shigella directly into host cells, demonstrating that class IB chaperones are often functionally interchangeable. Notably, class IB chaperones bind numerous effectors. However, as previously proposed, they are not promiscuous; rather they recognize a defined sequence that we designate the conserved chaperone-binding domain (CCBD) sequence [(LMIF)1XXX(IV)5XX(IV)8X(N)10]. This sequence is the first defined amino acid sequence to be identified for any interspecies bacterial secretion system, i.e., a system that delivers proteins directly into eukaryotic cells. This sequence provides a new means to identify substrates of type III secretion systems. Indeed, using a pattern search algorithm for the CCBD sequence, we have identified the first two probable effectors from an endosymbiont, Sodalis glossinidius.
IMPORTANCE
Many Gram-negative pathogens utilize type 3 secretion systems to deliver tens of effectors into host cells. In order to understand the diverse ways that these organisms cause disease, it is necessary to identify their effectors, many of which require chaperones to be secreted. Here we establish that class IB chaperones are not promiscuous, as previously proposed, but rather recognize a conserved effector sequence. We demonstrate that pattern search algorithms based on this defined sequence can be used to identify previously unknown effectors. Furthermore, we observe that class IB chaperones from at least seven bacterial species are functionally interchangeable. Not only do they bind and mediate the delivery of their own set of effectors into host cells but they also bind to type 3 substrates from other bacteria, suggesting that inhibitors that block chaperone-effector interactions could provide a novel means to effectively treat infections due to Gram-negative pathogens, including organisms resistant to currently available antibiotics.
Introduction
Over 30 species of Gram-negative bacteria, both pathogens and endosymbionts, utilize type 3 secretion systems (T3SSs) to deliver tens of proteins, referred to as effectors, directly into host cells (1). Type 3 effectors target a variety of host cellular processes to promote bacterial spread and survival. While the protein components of these complex secretion machines are highly conserved, each bacterial species delivers its own unique repertoire of effectors into host cells. Although many effectors require chaperones to be secreted, little is known regarding how they are defined as type 3 secreted substrates, limiting their identification through bioinformatic analyses.
Extensive work mapping the regions of effectors required for their secretion has established that two domains play a role, the N-terminal secretion sequence and, in many cases, a downstream chaperone-binding domain (2, 3). The N-terminal secretion sequence is not a specific sequence, but rather, as recent experimental data suggest, an intrinsically structurally disordered region (4). Machine learning algorithms can identify known effectors based on the nature of their N-terminal residues. However, the full utility of these algorithms in identifying new effectors is still unknown (5–7).
Two distinct but structurally related classes of chaperones mediate the secretion of type 3 effectors (8, 9). Class IA chaperones are almost always located within operons adjacent to the genes that encode their one or two cognate effectors. In contrast, class IB chaperones are encoded within large operons surrounded by components of the type 3 secretion machinery rather than effectors. On the basis of the results of structural analyses (10–13), it is hypothesized that effectors interact with class IA and class IB chaperones via a conserved structural motif, the β-strand motif (11), and that it is likely that the chaperone-effector complex is the signal recognized by the type III secretion apparatus (10, 11). The two best-characterized class IB chaperones are Spa15 from Shigella flexneri and InvB from Salmonella enterica serovar Typhimurium SPI1 (Salmonella pathogenicity island 1) T3SSs. Each has been established to mediate the secretion of multiple effectors, nine in the case of Shigella (14–16) and four in the case of Salmonella (18, 23, 24). Given their ability to interact with numerous effectors, Spa15 and InvB have been proposed to be promiscuous in their recognition of effectors (19).
Here, we present evidence that class IB chaperones from seven different bacterial species, including pathogens and endosymbionts, are functionally interchangeable. Specifically, class IB chaperones from one species can bind and mediate the type 3 secretion-dependent translocation of effectors from another. These class IB chaperones are not promiscuous, as previously proposed, but rather recognize a defined amino acid sequence motif, which we designate the conserved chaperone-binding domain (CCBD). The CCBD overlaps the previously identified structural β-strand motif, providing evidence that residues of the CCBD sequence directly bind to chaperones. However, the CCBD demonstrates that class IB chaperones recognize a conserved amino acid pattern. The CCBD sequence is the first identified defined amino acid sequence that is common to effectors from any interspecies bacterial secretion system, i.e., one used by bacteria to deliver proteins into eukaryotic cells. Uncovering this sequence not only refines our understanding of how interactions between chaperones and effectors are defined but also provides a new means to identify type 3 substrates from bacteria that encode class IB. Indeed, based on the results of a pattern search algorithm of the Sodalis glossinidius genome for proteins that contain the CCBD sequence, we identified the first two likely effectors from an endosymbiont.
RESULTS
Conserved recognition of effectors by Shigella and Salmonella class IB chaperones.
It is well established that class IA chaperone-dependent effectors are recognized as substrates of heterologous T3SSs when their cognate chaperone is also present (20, 21). In contrast, at the start of this study, little was known regarding the behavior of class IB chaperone-dependent effectors in heterologous systems. For example, it was not known whether “promiscuous” chaperones from one system could bind and mediate the secretion of effectors from another. To investigate this possibility, we tested whether class IB chaperones from Shigella and Salmonella could bind each other’s effectors using the Saccharomyces cerevisiae yeast-based protein interaction platform (PIP) assay, an assay previously established to be more sensitive than the yeast two-hybrid assay in detecting chaperone-effector interactions (16).
The PIP assay is a visualization system for identifying interacting proteins in living cells. In this assay, one protein is fused to μNS, a reoviral protein that forms inclusions (platforms) when expressed in eukaryotic cells; a second protein is fused to a fluorescent protein (16). When coexpressed in yeast, if the two proteins interact, the fluorescent fusion protein is recruited to the platforms and fluorescent foci are observed. Using the PIP assay, we observed interactions between Salmonella InvB and 10 of 23 Shigella effectors, the same 10 that interact with Shigella Spa15 in the PIP assay (see Fig. S1A in the supplemental material) (16). In all but one case, the majority of yeast cells visualized exhibited fluorescent foci. The exception was green fluorescent protein (GFP)-IpgB1, where the percentage of yeast cells that displayed fluorescent foci with expression of μNS-InvB and GFP-IpgB1 was decreased compared to those expressing μNS-Spa15 and GFP-IpgB1 (34 versus 68%). This observation suggests that IpgB1 interacts more weakly with InvB than Spa15, as recent studies demonstrate that the percentage of yeast displaying fluorescent foci reflect the relative in vitro binding affinities of the two proteins for each other (22).
In a complementary set of PIP assays, Salmonella InvB and Shigella Spa15 each interacted with three Salmonella effectors that require InvB for their efficient secretion by the Salmonella T3SS (SopA, SopE1, and SopE2) (see Fig. S1B in the supplemental material) (18, 23, 24). However, the fourth Salmonella InvB-dependent effector, SipA, bound only InvB. Neither InvB nor Spa15 interacted with any of the 13 Shigella effectors previously established to not require Spa15 for their secretion, including those that bind class IA chaperones, demonstrating that these class IB chaperones exhibit some specificity in their interactions (Fig. S1C) (16).
Complementation of secretion of Shigella Spa15-dependent effectors by Salmonella InvB.
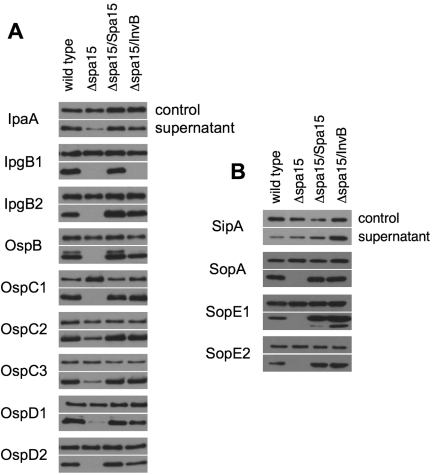
To determine whether the interactions detected in the PIP assay were physiologically relevant, we tested whether expression of the Salmonella InvB chaperone would restore secretion of Shigella Spa15-dependent effectors from a strain lacking Spa15 (Shigella ∆spa15 mutant). Complementation with InvB restored secretion to essentially wild-type levels for eight of the nine Shigella Spa15-dependent effectors (Fig. 1A), all but IpgB1, the effector that exhibited decreased interaction with InvB in the PIP assay. Similarly, secretion of the three Salmonella InvB-dependent effectors that bind Spa15 (SopA, SopE1, and SopE2) from the Shigella ∆spa15 mutant was only observed with complementation by either Spa15 or InvB (Fig. 1B). SipA, the only Salmonella InvB effector that interacted exclusively with InvB in the PIP assay, was secreted at relatively low levels from both the wild-type Shigella and the Shigella ∆spa15 mutant, suggesting that it can be secreted in the absence of a chaperone. Complementation of the Shigella ∆spa15 mutant strain with Salmonella InvB, but not Shigella Spa15, results in increased levels of secreted SipA (Fig. 1B). Together, the results of these studies demonstrate that the class IB chaperones from Salmonella and Shigella are nearly functionally interchangeable. These are the first class I chaperones yet to be identified to bind and promote the secretion of each other’s effectors.
FIG 1 .
Shigella and Salmonella class IB chaperones are functionally interchangeable. S. flexneri strains (wild type, Δspa15 mutant, and the Δspa15 mutant complemented with Shigella Spa15 or Salmonella InvB) and expressing the designated FLAG-tagged Shigella (A) or Salmonella (B) effectors were grown under conditions that induce T3SS. The supernatant proteins were precipitated with TCA, separated by SDS-PAGE, and immunoblotted with anti-FLAG and anti-IcsA antibodies. IcsA is an autotransporter and secreted by a mechanism other than type 3 secretion and serves as a loading control. The blots shown are representative of at least three experiments.
For these experiments, epitope-tagged versions of each effector were expressed under the control of a weakened version of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible trc promoter from a low-copy-number plasmid (pBR ori) (25). This allowed us to ensure that the effectors were expressed at close to physiologic levels when type 3 secretion was induced (16), since at least in the case of Shigella OspF, this system results in similar levels of expression as observed under native conditions (data not shown). Similarly, for the complementation experiments, Spa15 and InvB were expressed under the control of a weakened trc promoter from another low-copy-number plasmid (pSC101 ori). We observed no differences in the levels of secretion of each of the 9 Spa15-dependent effectors from wild-type Shigella versus Shigella ∆spa15 mutant complemented with a plasmid expressing Spa15, suggesting that the phenotypes observed are not due to Spa15 or InvB overexpression. Thus, Salmonella InvB and Shigella Spa15 are able to recognize and mediate the secretion of a subset of each other’s effectors.
Identification of a conserved CCBD sequence.
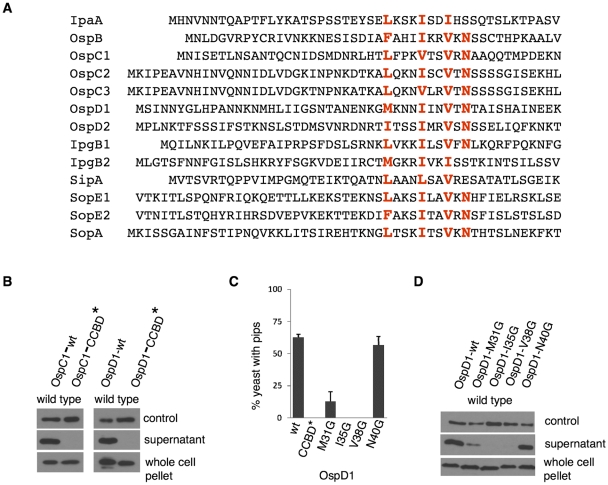
The observation that Spa15 and InvB recognize the same subset of effectors suggested that these chaperones are discriminate in their recognition of effectors and led us to search for a conserved chaperone-binding domain (CCBD) sequence. Previous studies had failed to recognize a sequence common to all type 3 effectors. However, when we restricted our alignment studies to the N-terminal 70 residues of the 13 Shigella and Salmonella effectors that interact with Spa15 and/or InvB, we identified a shared sequence, (LMIF)1XXX(IV)5XX(IV)8X(N)10 (Fig. 2A). Interestingly, the only effector that does not contain isoleucine or valine at position 5 is Salmonella SipA, the effector that interacts exclusively with Salmonella InvB. This consensus sequence is not found in any of the 11 Shigella effectors that do not require Spa15 for their secretion (16) or in any Salmonella effectors, other than those that interact with InvB. This sequence is the first defined amino acid sequence that has been identified that is shared by secreted effectors of any interspecies bacterial secretion system, and we designate it the conserved chaperone-binding domain sequence.
FIG 2 .
Identification of the conserved chaperone-binding domain, a sequence common to Shigella and Salmonella class IB-dependent effectors. (A) Multiple-sequence alignment of effectors whose secretion is promoted by InvB or Spa15. The CCBD sequence is shown in large red letters. (B to D) Wild-type Shigella expressing epitope-tagged alleles of wild-type (wt) and mutant alleles of OspC1 and OspD1 were grown under conditions that induce T3SS. The residues at positions 1, 5, 8, and 10 in CCBD* were changed to alanines. The supernatant and pellet fractions were immunoblotted with anti-FLAG antibody to assess for secretion and protein stability, respectively. The supernatant fractions were also immunoblotted with anti-IcsA antibody as a loading control. All blots shown are representative of at least three experiments. (C) Yeast coexpressing μNS-Spa15 and the designated GFP-OspD1 alleles were visualized 4 to 5 hours after induction of each protein in the PIP assay. The percentage of yeast expressing fluorescent foci (pips) was determined by counting 100 cells. The values shown are representative of two independent experiments done in triplicate.
Site-directed mutagenesis studies confirmed a role for the CCBD sequence in mediating chaperone-effector interactions. The conversion of all four of the conserved residues (positions 1, 5, 8, and 10) of the CCBD sequence of Shigella OspC1 or OspD1 to alanines completely abolished their secretion via the Shigella T3SS (Fig. 2B) and resulted in loss of interaction of OspD1 and Spa15 in the PIP (Fig. 2C) and yeast two-hybrid (Y2H) protein-protein interaction assays (see Table S1 in the supplemental material). We also examined whether single mutations at position 1, 5, 8, or 10 of the CCBD sequence would disrupt secretion and/or chaperone binding. For these studies, each residue was mutated to glycine to disrupt the hydrophobic contacts predicted to mediate chaperone-effector interaction in accordance with the corresponding positions in the SipA-InvB crystal structure. Point mutations at positions 1 and 10, the less conserved residues, resulted in a mild-to-moderate decrease in secretion, while mutation of either position 5 or 8, positions invariantly present as isoleucines or valines, essentially disrupted all secretion (Fig. 2D). In each case, the single point mutations disrupted interactions between Spa15 and OspD1 in the PIP and Y2H assays (Table S1) with the magnitude of loss paralleling the observed loss in secretion (Fig. 2C). Since decreased interactions in the PIP and Y2H assays are both associated with decreased binding affinities determined in vitro (22, 26), these observations suggest that these residues of the CCBD sequence directly bind class IB chaperones, as observed in the SipA-InvB cocrystal structure (11).
Interactions between the CCBD and class IB chaperones determine substrate specificity.
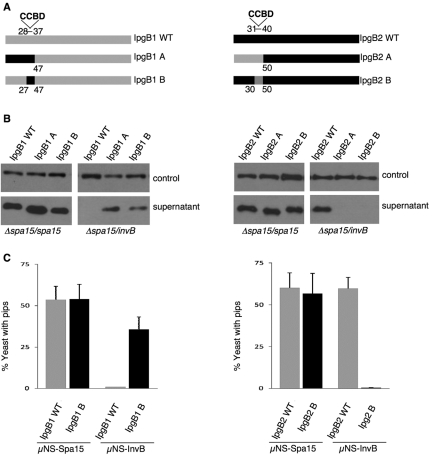
We next investigated whether interactions between the CCBD and class IB chaperones are sufficient to define chaperone substrate specificity. To address this question, we exploited our earlier observations that Salmonella InvB binds and complements the secretion of all of the Shigella CCBD-containing effectors except for IpgB1 (Fig. 1). IpgB1 shares a high degree of structural homology with IpgB2 (27), another Shigella CCBD-containing effector, suggesting that the unstructured N-terminal regions of these proteins dictate whether InvB recognizes them as effectors. Indeed, by swapping the first 47 residues of IpgB1 with the first 50 residues of IpgB2 (the respective regions of the effectors located upstream of their conserved structural domains), we switched their substrate specificity, as InvB now recognizes IpgB1, but not IpgB2, as a type 3 substrate (Fig. 3A and B). This substrate specificity is not determined by the N-terminal secretion signal, as additional swaps established that substrate specificity maps to regions that encompass the CCBD sequences of the two proteins (Fig. 3A and B). The switch in recognition of the IpgB1/IpgB2 hybrids likely represents differences in their binding affinities, as the ability of InvB to complement the secretion of these proteins from the Shigella Δspa15 mutant correlates with its capacity to bind the proteins in the PIP assay (Fig. 3C). All of the hybrid effectors were secreted by the Shigella Δspa15 mutant in the presence of Spa15 (Fig. 3B). These observations suggest that the CCBD is a major determinant in defining interactions between effectors and class IB chaperones.
FIG 3 .
The CCBD sequence defines chaperone-effector interactions. (A) Schematic of IpgB1/IpgB2 hybrid proteins used in this study. (B) Shigella Δspa15 mutant complemented with Spa15 or InvB and expressing epitope-tagged alleles of the designated IpgB1 or IpgB2 hybrid proteins grown under conditions that induce T3SS. The supernatant fractions were immunoblotted with anti-FLAG antibody to assess for secretion. The supernatant fractions were also immunoblotted with anti-IcsA antibody as a loading control. (C) Yeast coexpressing μNS-Spa15 or μNS-InvB and the designated IpgB1/IpgB2 alleles were visualized 4 to 5 hours after induction of each protein in the PIP assay. The percent of yeast expressing fluorescent foci (pips) was determined by counting 100 cells. The values shown are representative of two independent experiments done in triplicate.
The CCBD sequence is unique to effectors that bind class IB chaperones.
Notably, at least in the case of SipA, whose crystal structure in complex with InvB has been solved, the CCBD domain overlaps with the β-strand motif, a region characterized by three hydrophobic residues (ψ) separated by 1 to 4 variable residues (ψX2–4ψX1–2ψ) (11). This structural motif is found in effectors that bind class IA and IB chaperones. Unlike the CCBD sequence, there is no sequence conservation of residues in the β-strand motif other than their hydrophobic nature. Over 35 type 3 effectors have been proposed to contain the structural β-strand motif, including those dependent on Spa15 or InvB for their secretion (11). We randomly selected and tested whether nine of these effectors (Salmonella SifA, SifB, SigD, SptP, and SseJ, Escherichia coli EspH, Map, and Tir, and Yersinia pseudotuberculosis YopH) interact with Spa15 or InvB in the PIP assay (see Fig. S2A in the supplemental material). This set included six effectors (SigD, SptP, EspH, Map, Tir, and YopH) previously shown to bind class IA chaperones. None of these effectors interacted with Spa15 or InvB. Similarly, we observed no interactions in the PIP assay between the nine Shigella CCBD-containing effectors and two atypical class IA chaperones that have been established to bind multiple effectors, CesT from enterohemorrhagic E. coli and SrcA from the Salmonella SPI2 T3SS (Fig. S2C) (28, 29). Therefore, we hypothesize that the amino acid sequence of the CCBD defines the specificity of interactions of class IB chaperones with their cognate effectors.
Conserved recognition of CCBD-containing effectors by class IB chaperones in distantly related species.
We next investigated whether the recognition of CCBD-containing effectors is restricted to closely related pathogens like Shigella and Salmonella or a feature common to all class IB chaperones. Towards this goal, we tested the ability of seven additional class IB chaperones, four from human pathogens (Proteus mirabilis [30]), Burkholderia dolosa, Burkholderia mallei [31] and Yersinia enterocolitica [32]) and three from endosymbionts (Sodalis glossinidius [33, 34] and Hamiltonella defensa [35]) to bind to Shigella CCBD-containing effectors. Sodalis glossinidius, a tsetse fly endosymbiont, contains three T3SSs, two of which (SSR1 and SSR2) encode InvB homologs. Similarly, Y. enterocolitica encodes two T3SSs, the well-studied Ysc T3SS and the relatively poorly characterized Ysa T3SS. Only the latter system encodes a class IB chaperone.
To increase the throughput of our protein interaction studies, we developed an automated microscopy-based version of the PIP assay. Automating the PIP assay decreased the time needed for image acquisition and analysis allowing for the rapid analysis of hundreds of yeast cells for the formation of fluorescent foci (pips) (Fig. 4A). One issue we encountered with the automated PIP assay was that the segmentation software used had difficulty identifying fluorescent foci in yeast that express weakly fluorescent GFP fusion proteins, specifically GFP-OspB, GFP-OspC3, and GFP-OspD2. To account for differences in GFP intensities (Fig. 4B), for each effector protein, we normalized the number of yeast that exhibited fluorescent foci with each homolog to those observed with Shigella Spa15.
FIG 4 .
Summary of interactions of class IB chaperones with CCBD-containing effectors obtained by automated microscopy. (A) Images of yeast expressing GFP-OspD1 in the presence of either μNS (negative control) or μNS-Spa15 (positive control) obtained via automated microscopy. The images in the left column are unprocessed, while the images in the right column represent pip-positive (green) and pip-negative (red) yeast as determined by the IN Cell Workstation segmentation software. (B) The heatmap shown (45) represents relative number of yeast with fluorescent foci (pips) visualized with coexpression of the designated μNS-class IB chaperone and GFP-effector fusion protein pairs. For each GFP fusion protein, the absolute percentage of yeast expressing fluorescent foci was normalized to the absolute percentage of yeast expressing fluorescent foci when the GFP-effector fusion protein was coexpressed with μNS-Spa15. The data are representative of 3 independent experiments done in duplicate. At least 6,000 cells were quantified per condition.
As summarized in Fig. 4B (raw data shown in Fig. S3A in the supplemental material), all of the class IB chaperones, except those from the Sodalis SSR1 and B. mallei T3SSs, interacted with Shigella CCBD-containing effectors. The lack of detection of interactions of the Sodalis SSR1 and B. mallei homologs with effectors was not due to their inability to form platforms in yeast, as fluorescent foci were observed when each of these chaperones was simultaneously fused to a cyan fluorescent protein-μNS fusion protein (Fig. S3B).
Functionally interchangeability as a general feature of class IB chaperones.
To confirm that the chaperone-effector interactions detected in the PIP assay were physiologically relevant, we investigated whether each of the class IB homologs could complement translocation of the nine Shigella CCBD-containing effectors using the well-established TEM-1 β-lactamase reporter assay (36). Briefly, in this assay, HeLa cells are preloaded with CCF4/AM, a fluorescence resonance energy transfer (FRET)-based dye, that emits green fluorescence. When translocated into host cells, the TEM-1 β-lactamase fusion proteins cleave the substrate, disrupting FRET and resulting in cells that emit blue fluorescence (Fig. 5A). The translocation efficiency of each effector corresponds to the percentage of cells that fluoresce blue.
FIG 5 .
Functional interchangeability of class IB chaperones in a type 3 translocation assay. (A) Images of HeLa cells preloaded with CCF4/AM infected with wild-type (WT) Shigella or Shigella Δspa15 mutant expressing OspD1–TEM-1. (B) The heatmap shown (45) is representative of the percent blue cells observed when the HeLa cells were infected with Shigella Δspa15 mutant strain that carries plasmids that express the designated class IB chaperone with each of the effector–TEM-1 fusion proteins. Translocation was quantified by measuring the percentage of cells that fluoresce blue (cleaved CCF4/AM). Data are representative of two independent experiments done in triplicate and at least 600 cells were counted for each sample.
To facilitate our studies, we adapted the TEM-1 β-lactamase reporter assay to monitor the translocation of Shigella effectors into HeLa cells in a 96-well format. Effector–TEM-1 fusions were expressed under the control a weakened trc IPTG-inducible promoter from a low-copy-number plasmid (pBR ori). After the expression of effectors was induced, HeLa cells were infected with Shigella at a multiplicity of infection (MOI) of 100 for 2 h. The cells were examined and the numbers of green and blue cells were counted. These conditions were used for our high-throughput studies given that at the 2-h time point we observed similar levels of translocation of all nine effectors into host cells with wild-type Shigella and the Shigella Δspa15 mutant complemented with Spa15 (Fig. S4A in the supplemental material).
As summarized in Fig. 6B and shown in Fig. S4A in the supplemental material, all of the class IB chaperones, except those from the Sodalis SSR1 and B. mallei T3SSs, complemented the translocation of CCBD-containing effectors from the Shigella Δspa15 mutant into host cells. Lack of translocation by the Sodalis SSR1 and B. mallei chaperones was not due to lack of expression or decreased stability of these proteins, as both effectors are present at levels similar to Spa15 in Shigella (Fig. S4B). Rather, as neither interact with CCBD-containing effectors in the PIP assay, it is likely that these homologs do not complement secretion because they do not bind the effectors. Interestingly, Sodalis SSR2, like Salmonella InvB, poorly complemented the translocation of IpgB1. Consistent with this observation, the Sodalis SSR2 homolog, like Salmonella InvB, complements the secretion into the media of all the Shigella CCBD-containing effectors, except for IpgB1 (Fig. S5). Taken together, the PIP and translocation assay results suggest that, with the exception of homologs from the Sodalis SSR1 and B. mallei T3SSs, the majority of class IB chaperones are functionally interchangeable and recognize a conserved amino acid domain, the CCBD sequence.
FIG 6 .
SG0567 and SG0764 are the first identified effectors from an endosymbiont. (A) Wild-type Shigella or the Shigella Δspa15 mutant complemented with the Spa15 homologs from the Sodalis SSR1 or SSR2 T3SSs and expressing FLAG-tagged Sodalis candidate effectors were grown under conditions that induce T3SS. The supernatant proteins were immunoblotted with anti-FLAG and anti-IcsA antibodies. The blots shown are representative of at least three experiments. (B) HeLa cells preloaded with CCF4/AM were infected with wild-type Shigella or the Shigella Δspa15 mutant expressing the designated protein fused to TEM-1 for 2 hours. The cells were fixed, and the percentage of blue cells were determined. Values are representative of three independent experiments done in triplicate, and at least 600 cells were counted for each sample (means plus standard errors of the means [SEM] [error bars] are shown).
Identification of the first effectors from an endosymbiont based on a pattern search algorithm for the CCBD sequence.
While the proteins that comprise T3SSs, including class IB chaperones, are highly conserved, each bacterial species delivers its own unique set of effectors into host cells. Although class IB chaperones can be identified based on homology searches, with the exceptions of Shigella Spa15 and Salmonella InvB, none of their cognate effectors are known. Since most of the homologs bind and mediate the translocation of Shigella CCBD-containing effectors, we reasoned that we could identify their native effectors by searching their respective genomes for open reading frames (ORFs) that contain the CCBD sequence. We focused our initial studies on Sodalis glossinidius, particularly given that no effectors from an endosymbiont have yet been identified. Using the Bioinformatics Toolkit pattern search algorithm (37) for the CCBD sequence [(LMIF)1XXX(IV)5XX(IV)8X(N)10] in the first 25 to 45 N-terminal residues of annotated ORFs (a region chosen based on the location of the CCBD sequence in all 13 Shigella and Salmonella Spa15/InvB-dependent effectors), we identified 13 Sodalis proteins, 9 of which are annotated as housekeeping proteins. We focused our studies on the remaining proteins that were annotated as hypothetical ORFs (see Fig. S6A in the supplemental material).
Two of these four Sodalis glossinidius proteins, SG0576 and SG0764, were stably expressed in Shigella, and both of these were secreted into the media by wild-type Shigella under conditions that induce type 3 secretion (Fig. 6A). The secretion of both was impaired or absent from the Shigella ∆spa15 mutant, and as predicted based on the results of binding and translocation studies, expression of the class IB homolog from the Sodalis SSR2, but not the SSR1 T3SS, restored secretion of both proteins from the Shigella ∆spa15 mutant (Fig. 6A). Neither protein was secreted from a Shigella strain that does not form a functional T3SS due to loss of expression of the structural protein MxiM (Shigella ∆mxiM mutant) (see Fig. S6B in the supplemental material) (38). Last, SG0576 and SG0764 were both translocated into host cells via the Shigella T3SS in a class IB chaperone-dependent manner (Fig. 6B). Together, these observations strongly suggest that these proteins are substrates of the Sodalis SSR2 T3SS. Thus, we have identified the first type 3 effectors from an endosymbiont.
DISCUSSION
Many Gram-negative bacteria utilize T3SSs to deliver tens of proteins directly into host cells during the course of an infection. The identification of effectors remains challenging, even in the postgenomics era, in part given their lack of a defined searchable conserved secretion signal. Here, we demonstrate that class IB chaperones are not promiscuous, as previously proposed, but rather recognize a defined amino acid sequence, which we designate the conserved chaperone-binding domain. This CCBD sequence is recognized by class IB chaperones from both endosymbionts and animal pathogens. Seven of the nine class IB homologs studied here not only bound CCBD-containing effectors but were functionally interchangeable in the context of the Shigella T3SS. Notably, by using a pattern search algorithm to screen for proteins that contain the CCBD sequence, we successfully identified the first putative effectors from an endosymbiont, Sodalis glossinidius.
Candidate effectors have traditionally been identified from bacterial genome sequences by searching for proteins that share sequence similarity with type 3 effectors or mammalian proteins as well as by focusing on those encoded by genes within pathogenicity islands or exhibit a distinctive GC content. Recently, several groups have developed machine learning algorithms to identify candidate effectors based on their N-terminal 15 to 20 residues, the secretion signal. Curiously, using available Web-based tools for two of these algorithms, we found that the SIEVE (39) and Effective T3 (5) algorithms identify only about half of the 15 Shigella, Salmonella, and Sodalis CCBD-containing effectors, suggesting that CCBD-based searches will lead to the identification of effectors missed by the machine learning algorithms.
While the conserved residues of the CCBD are clearly important, they are not sufficient to define effectors, as this sequence is found in bacterial housekeeping proteins, which are unlikely to be secreted. This is not surprising, as additional determinants play a role in defining effectors. For example, our pattern search algorithms for the CCBD sequence motif do not select for proteins that encode an N-terminal secretion signal. In addition, it is possible, that the variable residues of the CCBD sequence, the residues indicated as X in the consensus sequence, play a role in defining an effector. Curiously, these residues in the verified effectors are enriched for nucleophilic and charged residues. This is a particularly interesting finding, as recent work suggests that the three-dimensional structure of the chaperone-effector complex defines the secretion signal (10, 40). Last, binding to the chaperone does not ensure that a protein is secreted via the T3SS, as prior to being secreted, the effectors are unfolded by an T3SS-associated ATPase that presumably enables them to be transported through a ~20-angstrom channel into host cells. Indeed, heterologous proteins with high intrinsic stability that have been engineered to encode amino-terminal secretion signals are not secreted (41). Future experimental work and bioinformatic analyses designed to address the possibilities outlined above will likely result in refinements that improve the specificity of pattern search algorithms for the detection of effectors.
In summary, our current work demonstrates that by exploiting the functional interchangeability of class IB chaperones and their conserved recognition of the CCBD sequence motif we have developed a new experimental genome-mining means for the identification of previously unknown effectors. Notably, while it is relatively straightforward to monitor the secretion and translocation of Shigella effectors, with the exception of the Salmonella SPI T3SS, in vitro conditions that induce other T3SS genes that encode class IB chaperones are currently unknown. However, as demonstrated with Sodalis CCBD-containing effectors, candidate effectors from bacteria that encode functionally interchangeable class IB chaperones can be screened for those that are recognized as substrates of the Shigella T3SS. Once likely effectors of these relatively poorly characterized T3SSs are identified, these proteins can be used as a readout to identify in vitro conditions that induce their respective native T3SSs. Furthermore, our discovery that class IB chaperones recognize a conserved sequence provides a new means to pursue the development of novel antimicrobial agents using rationale drug design approaches.
MATERIALS AND METHODS
Plasmids.
All of the plasmids involving S. flexneri and many of the S. Typhimurium ORFs have been previously described (16, 42). The remaining bacterial and yeast expression plasmids were created via Gateway (Invitrogen) site-specific recombination (16).
Basically, the ORF encoding each effector and each chaperone was amplified from genomic DNA (gDNA) by nested PCR such that it is flanked by attB sites. In addition to an attB site, a Shine-Dalgarno sequence was introduced upstream of each ORF. We created open (lacking a stop codon) and closed versions of each ORF when carboxy fusions were desired. Mutant alleles of the effectors were created by sewing PCR. The amplified genes were then introduced into pDNR223 and/or pDNR221 to create Gateway entry vectors via BP reactions (Invitrogen). Each insert was sequence verified and subsequently transferred to a variety of Gateway destination vectors via LR reactions (Invitrogen).
For the μNS fusion proteins in the PIP assays, the chaperones and effectors were introduced into pAG416GAL-μNS-ccdB, pAG415GAL-μNS-ccdB, and/or pAG415GAL-CFP-μNS-ccdB. For the GFP fusion proteins, the chaperones and the Salmonella effectors, with the exception of SipA, were introduced into pAG413GAL-GFP-ccdB and pBY011 (43). Conventional cloning was used created GFP-SipA (amino acids 1 to 254) in pRS313.
To generate FLAG tag fusions for the secretion assays, the wild-type, mutant, and hybrid effectors were introduced into pDSW206-FLAG-ccdB (ColE1 ori, ampicillin resistance) (16, 25), and the chaperones were introduced into pNG162-ccdB (p204 promoter [IPTG-inducible], pSC101 ori, spectinomycin resistance) (16, 44).
To generate TEM-1 fusions for the translocation assay, the Shigella and Sodalis effectors were introduced into pDSW206-TEM-1-ccdB (ColE1 ori, ampicillin resistance) (a gift from John Leong and Loranne Magoun, University of Massachusetts Medical School)
Finally, for chaperone stability, chaperones were tagged with Myc at their N termini and introduced onto the pNG162-ccdB plasmid.
All oligonucleotide primers used in the constructs described here are described in Table S2 in the supplemental material.
Conventional (manual) PIP assays.
Yeast assays were preformed as previously described (16). Yeast cells were visualized 4 to 6 h after galactose induction using a Nikon TE300 microscope with Chroma Technology filters and a 100× objective. Images were captured digitally using a black-and-white Sensys charge-coupled-device (CCD) camera and IP LAB software (Scanalytics).
High-content image analysis (automated PIP assays).
The yeast cells were grown in 96-well microtiter plates (Nunc) as previously described (16) and transferred into 96-well microtiter plates (BD). Each experimental condition was performed in duplicate on a plate. Three sets of identical plates were examined, bringing the total number of replicates to six per condition. The cells were imaged at a magnification of ×40 in each well using the IN Cell Analyzer 1000 (GE Healthcare, Piscataway, NJ). Briefly, the IN Cell Analyzer 1000 is an epifluorescence microscope with fully automated image acquisition capabilities, including a motorized stage, motorized filter wheel, and computer-controlled CCD camera. Three fields were preselected to avoid sampling bias throughout the plate. Exposure parameters were empirically optimized for GFP fluorescence to ensure that images fall within the linear range of exposure. Following exposure optimization, images were collected in selected wells and stored for analysis. Images were analyzed using IN Cell Workstation Software (GE Healthcare). Cells were segmented using the nucleus “Top Hat” algorithm and a 1-µm cell collar. Cell detection was determined by endogenous GFP expression within the yeast. Foci or pip were identified as an organelle between 0.2 µm and 2.0 µm with a mean organelle intensity/cell intensity ratio above 1.5. Output parameters included the following: cell count, nucleus area (whole yeast cell), nucleus intensity (whole yeast cell intensity), organelle count, organelle intensity, organelle intensity/cell intensity, percent pip formation, and total number of pip formation. Cells were classified as pip positive if cells expressed ≥1 organelle; otherwise, cells were classified as pip negative. Cells not expressing GFP were not analyzed.
Secretion and chaperone stability assays.
The pDSW206-based plasmids encoding each of the IPTG-inducible FLAG-tagged effectors were transformed into the designated Shigella strains. Complementation experiments were performed with a Shigella Δspa15 mutant strain, where homologous chaperones were expressed under an IPTG-inducible pNG162 vector. Congo red type 3 secretion assays were conducted as previously described. Basically, the total cell and supernatant fractions were separated by two centrifugations (each centrifugation at 20,000 × g for 2 min). The cell pellet of the first centrifugation was taken as the total cell fraction. Proteins in the supernatant of the second centrifugation were precipitated with trichloroacetic acid (TCA) (10% [vol/vol]). Protein content of the pellet and supernatant fraction were assessed by Western blotting with anti-FLAG antibody (Sigma). For loading control, membranes were probed with anti-IcsA (an autotransporter that is cleaved and released into the media once it reaches the surface of Shigella). We have previously established that lysis is not an issue using this protocol, as we rarely, if ever, detect evidence of cytoplasmic proteins (16) or unprocessed IcsA.
High-throughput TEM-1-based translocation assay.
Translocation of effectors into HeLa cells was performed as previously described (36) with minor modifications. Basically, bacteria were grown overnight in tryptic soy broth medium (BD Scientific) in a 96-well plate. In the morning, the cultures were back diluted (1/100), and 1.0 mM IPTG was added when the bacterial cultures reached an optical density at 600 nm (OD600) of 0.6. Thirty minutes after induction, bacteria were centrifuged, and the pellet was resuspended in phosphate-buffered saline (PBS). HeLa cells (1.5 × 104 cells/well, in 96-well black plates with clear bottom [Costar]) preloaded with CCF4/AM according to the manufacturer’s instructions (Invitrogen) were infected at an MOI of 100. The plates were centrifuged for 10 min to promote contact of the bacteria with HeLa cells. IPTG (0.1 mM) was added to the medium, and the plates were incubated at 37°C for 2 h. Subsequently, the infected cells were fixed with paraformaldehyde, and the percentage of effector translocated was assessed via fluorescence microscopy (Nikon TE300 microscope with Chroma Technology filters and a 40× objective) by determining the percentage of cells that fluoresce blue.
SUPPLEMENTAL MATERIAL
Shigella and Salmonella class IB chaperones specifically bind each other’s class IB-dependent effectors. Potential interactions between Salmonella InvB and Shigella effectors known to interact with Spa15 (A), Shigella Spa15 and Salmonella InvB with Salmonella effectors known to interact with InvB (B), or Salmonella InvB and Shigella effectors that do not interact with Spa15 (C) were investigated using the PIP assay. The images shown are representative of yeast expressing the designated effector fused to GFP and designated class IB chaperone fused to μNS. The yeast cells were visualized 4 to 5 h after induction of each protein pair. IpgB2 is extremely toxic when expressed in yeast and is not visible when expressed as a GFP fusion protein (16). Thus, a catalytic inactive allele of IpgB2_W62A (IpgB2*) was used in this study. In addition, since full-length SipA colocalizes with yeast actin cables (46), a truncated allele of SipA (amino acids 1 to 254) was used in this assay. Download Figure S1, PDF file, 0.2 MB.
The CCBD sequence uniquely defines interactions between effectors and class IB chaperones. (A) Alignment of N termini of effectors previously proposed to contain a β-strand (11) which is indicated in red. (B) Each of the images is representative of yeast expressing the designated effector fused to GFP and μNS fused to either Salmonella InvB or Shigella Spa15. The yeast cells were visualized 4 to 5 hours after induction of the fusion proteins. (C) Representative images of yeast coexpressing class IA chaperones, E. coli CesT and Salmonella SPI2 SrcA, fused to μNS and Spa15-dependent effectors fused to GFP. Download Figure S2, TIFF file, 0.9 MB.
All class IB homologs except for those from the B. mallei and S. glossinidius SSR1 T3SSs directly bind Shigella CCBD-containing effectors. (A) Summary of automated PIP data. Images of yeast coexpressing the designated CCBD-containing effectors fused to GFP and the class IB chaperones fused to μNS were obtained using an automated microscope. The percentage of yeast with fluorescent foci (pips) was determined. The values shown are means plus SEM (error bars) and are representative of 3 independent experiments conducted on 2 days. At least 6,000 cells were counted for each condition. (B) The class IB chaperones from the B. mallei and S. glossinidius SSR1 T3SSs are capable of localizing to μNS-generated foci when expressed in yeast. Images of yeast expressing class IB chaperones from the B. mallei and S. glossinidius T3SSs each fused to CFP-μNS observed 4 to 5 hours after induction of expression of the fusion proteins are shown. Download Figure S3, TIFF file, 0.9 MB.
All class IB homologs except for those from the B. mallei and S. glossinidius SSR1 T3SSs facilitate translocation of Shigella CCBD-containing effectors from the Shigella Δspa15 mutant. (A) Summary of high-throughput TEM-1 translocation data. HeLa cells preloaded with CCF4/AM were infected with the Shigella Δspa15 mutant coexpressing the designated CCBD-containing effectors fused to TEM-1 and class IB chaperones for 2 hours. The cells were fixed, and the ratio of blue cells to green cells was determined. Values are representative of 3 independent experiments done in triplicate, and at least 600 cells were counted for each sample (means plus SEM shown). (B) Immunoblots of extracts of the Shigella Δspa15 mutant expressing myc-tagged versions of the class IB chaperones from the S. flexneri, B. mallei, and S. glossinidius T3SSs. The blots were probed with anti-OspF antibody as a loading control. Download Figure S4, TIFF file, 1 MB.
Sodalis SSR2_InvB complements secretion of Shigella effectors from the Shigella Δspa15 mutant. S. flexneri strains were grown under conditions that induce T3SS. The S. flexneri strains used were the wild type, Δspa15 mutant, and the Δspa15 mutant complemented with Shigella Spa15 or the class IB chaperone from the Sodalis SSR2 T3SS and expressing FLAG-tagged Shigella effectors. The supernatant proteins were probed with anti-FLAG and anti-IcsA antibodies. The blots shown are representative of at least three experiments. Download Figure S5, TIFF file, 0.2 MB.
Identification of the first effectors from Sodalis. (A) Alignment of N termini of hypothetical Sodalis ORFs identified by a pattern search algorithm for CCBD sequence. The putative CCBD domain is shown in red. (B) The wild-type S. flexneri and S. flexneri ΔmxiM mutant strains expressing the designated FLAG-tagged Sodalis proteins were grown under conditions that induce T3SS. The supernatant proteins were probed with anti-FLAG and anti-IcsA antibodies. The blots shown are representative of at least three experiments. Download Figure S6, TIFF file, 0.3 MB.
Summary of Y2H Spa15-OspD1 interactions.
List of oligonucleotides used to generate fragments via PCR for cloning into Gateway entry vectors.
ACKNOWLEDGMENTS
This work was conducted through support from the NIH (R01 AI064285) and a Charles E. Culpeper Medical Scholarship from the Rockefeller Brothers Fund and Goldman Philanthropic Partnerships awarded to C.F.L.
We thank Stephen Lory, Luisa Stamm, Analise Zaunbrecher, and Seth Maleri for critically reading the manuscript. We are also grateful to multiple investigators who provided genomic DNA for these studies: Colin Dale, Mauricio Pontes, Serap Aksoy, and Brian Weiss (S. glossinidius), Stephen Lory (Burkholderia dolosa), Joan Mecsas (Y. enterocolitica), Harold Mobley (P. mirabilis), and Patrick Degnan (H. defensa). We also thank Marcia Goldberg for providing anti-IcsA antibody and Dan Zurawski and Anthony Maurelli for supplying anti-OspF antibody.
Footnotes
Citation Costa SCP, et al. 2012. A new means to identify type 3 secreted effectors: functionally interchangeable class IB chaperones recognize a conserved sequence. mBio 3(1):e00243-11. doi:10.1128/mBio.00243-11.
REFERENCES
- 1. Troisfontaines P, Cornelis GR. 2005. Type III secretion: more systems than you think. Physiology (Bethesda) 20:326–339 [DOI] [PubMed] [Google Scholar]
- 2. Schesser K, Frithz-Lindsten E, Wolf-Watz H. 1996. Delineation and mutational analysis of the Yersinia pseudotuberculosis YopE domains which mediate translocation across bacterial and eukaryotic cellular membranes. J. Bacteriol. 178:7227–7233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sory MP, Boland A, Lambermont I, Cornelis GR. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc. Natl. Acad. Sci. U. S. A. 92:11998–12002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buchko GW, et al. 2010. A multi-pronged search for a common structural motif in the secretion signal of Salmonella enterica serovar Typhimurium type III effector proteins. Mol. Biosyst. 6:2448–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arnold R, et al. 2009. Sequence-based prediction of type III secreted proteins. PLoS Pathog. 5:e1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Löwer M, Schneider G. 2009. Prediction of type III secretion signals in genomes of gram-negative bacteria. PLoS One 4:e5917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Samudrala R, Heffron F, McDermott JE. 2009. Accurate prediction of secreted substrates and identification of a conserved putative secretion signal for type III secretion systems. PLoS Pathog. 5:e1000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parsot C, Hamiaux C, Page AL. 2003. The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol. 6:7–14 [DOI] [PubMed] [Google Scholar]
- 9. van Eerde A, Hamiaux C, Pérez J, Parsot C, Dijkstra BW. 2004. Structure of Spa15, a type III secretion chaperone from Shigella flexneri with broad specificity. EMBO Rep. 5:477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Birtalan SC, Phillips RM, Ghosh P. 2002. Three-dimensional secretion signals in chaperone-effector complexes of bacterial pathogens. Mol. Cell 9:971–980 [DOI] [PubMed] [Google Scholar]
- 11. Lilic M, Vujanac M, Stebbins CE. 2006. A common structural motif in the binding of virulence factors to bacterial secretion chaperones. Mol. Cell 21:653–664 [DOI] [PubMed] [Google Scholar]
- 12. Schubot FD, et al. 2005. Three-dimensional structure of a macromolecular assembly that regulates type III secretion in Yersinia pestis. J. Mol. Biol. 346:1147–1161 [DOI] [PubMed] [Google Scholar]
- 13. Stebbins CE, Galán JE. 2001. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature 414:77–81 [DOI] [PubMed] [Google Scholar]
- 14. Hachani A, et al. 2008. IpgB1 and IpgB2, two homologous effectors secreted via the Mxi-Spa type III secretion apparatus, cooperate to mediate polarized cell invasion and inflammatory potential of Shigella flexneri. Microbes Infect. 10:260–268 [DOI] [PubMed] [Google Scholar]
- 15. Page AL, Sansonetti P, Parsot C. 2002. Spa15 of Shigella flexneri, a third type of chaperone in the type III secretion pathway. Mol. Microbiol. 43:1533–1542 [DOI] [PubMed] [Google Scholar]
- 16. Schmitz AM, Morrison MF, Agunwamba AO, Nibert ML, Lesser CF. 2009. Protein interaction platforms: visualization of interacting proteins in yeast. Nat. Methods 6:500–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reference deleted. [Google Scholar]
- 18. Ehrbar K, Friebel A, Miller SI, Hardt WD. 2003. Role of the Salmonella pathogenicity island 1 (SPI-1) protein InvB in type III secretion of SopE and SopE2, two Salmonella effector proteins encoded outside of SPI-1. J. Bacteriol. 185:6950–6967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ghosh P. 2004. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68:771–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kenny B, Warawa J. 2001. Enteropathogenic Escherichia coli (EPEC) Tir receptor molecule does not undergo full modification when introduced into host cells by EPEC-independent mechanisms. Infect. Immun. 69:1444–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rosqvist R, Håkansson S, Forsberg A, Wolf-Watz H. 1995. Functional conservation of the secretion and translocation machinery for virulence proteins of yersiniae, salmonellae and shigellae. EMBO J. 14:4187–4195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. de Groot JC, et al. 2011. Structural basis for complex formation between human IRSp53 and the translocated intimin receptor tir of enterohemorrhagic E. coli. Structure 19:1294–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bronstein PA, Miao EA, Miller SI. 2000. InvB is a type III secretion chaperone specific for SspA. J. Bacteriol. 182:6638–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ehrbar K, Hapfelmeier S, Stecher B, Hardt WD. 2004. InvB is required for type III-dependent secretion of SopA in Salmonella enterica serovar Typhimurium. J. Bacteriol. 186:1215–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Estojak J, Brent R, Golemis EA. 1995. Correlation of two-hybrid affinity data with in vitro measurements. Mol. Cell. Biol. 15:5820–5829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Alto NM, et al. 2006. Identification of a bacterial type III effector family with G protein mimicry functions. Cell 124:133–145 [DOI] [PubMed] [Google Scholar]
- 28. Cooper CA, et al. 2010. Structural and biochemical characterization of SrcA, a multi-cargo type III secretion chaperone in Salmonella required for pathogenic association with a host. PLoS Pathog. 6:e1000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thomas NA, et al. 2005. CesT is a multi-effector chaperone and recruitment factor required for the efficient type III secretion of both LEE- and non-LEE-encoded effectors of enteropathogenic Escherichia coli. Mol. Microbiol. 57:1762–1779 [DOI] [PubMed] [Google Scholar]
- 30. Pearson MM, Mobley HL. 2007. The type III secretion system of Proteus mirabilis HI4320 does not contribute to virulence in the mouse model of ascending urinary tract infection. J. Med. Microbiol. 56:1277–1283 [DOI] [PubMed] [Google Scholar]
- 31. Stevens MP, et al. 2002. An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol. Microbiol. 46:649–659 [DOI] [PubMed] [Google Scholar]
- 32. Haller JC, Carlson S, Pederson KJ, Pierson DE. 2000. A chromosomally encoded type III secretion pathway in Yersinia enterocolitica is important in virulence. Mol. Microbiol. 36:1436–1446 [DOI] [PubMed] [Google Scholar]
- 33. Dale C, Jones T, Pontes M. 2005. Degenerative evolution and functional diversification of type-III secretion systems in the insect endosymbiont Sodalis glossinidius. Mol. Biol. Evol. 22:758–766 [DOI] [PubMed] [Google Scholar]
- 34. Toh H, et al. 2006. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 16:149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Degnan PH, Yu Y, Sisneros N, Wing RA, Moran NA. 2009. Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc. Natl. Acad. Sci. U. S. A. 106:9063–9068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Charpentier X, Oswald E. 2004. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J. Bacteriol. 186:5486–5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Biegert A, Mayer C, Remmert M, Söding J, Lupas AN. 2006. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 34:W335–W339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schuch R, Sandlin RC, Maurelli AT. 1999. A system for identifying post-invasion functions of invasion genes: requirements for the Mxi-Spa type III secretion pathway of Shigella flexneri in intercellular dissemination. Mol. Microbiol. 34:675–689 [DOI] [PubMed] [Google Scholar]
- 39. McDermott JE, et al. 2011. Computational prediction of type III and IV secreted effectors in gram-negative bacteria. Infect. Immun. 79:23–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rodgers L, Mukerjea R, Birtalan S, Friedberg D, Ghosh P. 2010. A solvent-exposed patch in chaperone-bound YopE is required for translocation by the type III secretion system. J. Bacteriol. 192:3114–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Akeda Y, Galán JE. 2005. Chaperone release and unfolding of substrates in type III secretion. Nature 437:911–915 [DOI] [PubMed] [Google Scholar]
- 42. Slagowski NL, Kramer RW, Morrison MF, LaBaer J, Lesser CF. 2008. A functional genomic yeast screen to identify pathogenic bacterial proteins. PLoS Pathog. 4:e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Alberti S, Gitler AD, Lindquist S. 2007. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast 24:913–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Goehring NW, Gonzalez MD, Beckwith J. 2006. Premature targeting of cell division proteins to midcell reveals hierarchies of protein interactions involved in divisome assembly. Mol. Microbiol. 61:33–45 [DOI] [PubMed] [Google Scholar]
- 45. Pavlidis P, Noble WS. 2003. Matrix2png: a utility for visualizing matrix data. Bioinformatics 19:295–296 [DOI] [PubMed] [Google Scholar]
- 46. Lesser CF, Miller SI. 2001. Expression of microbial virulence proteins in Saccharomyces cerevisiae models mammalian infection. EMBO J. 20:1840–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Shigella and Salmonella class IB chaperones specifically bind each other’s class IB-dependent effectors. Potential interactions between Salmonella InvB and Shigella effectors known to interact with Spa15 (A), Shigella Spa15 and Salmonella InvB with Salmonella effectors known to interact with InvB (B), or Salmonella InvB and Shigella effectors that do not interact with Spa15 (C) were investigated using the PIP assay. The images shown are representative of yeast expressing the designated effector fused to GFP and designated class IB chaperone fused to μNS. The yeast cells were visualized 4 to 5 h after induction of each protein pair. IpgB2 is extremely toxic when expressed in yeast and is not visible when expressed as a GFP fusion protein (16). Thus, a catalytic inactive allele of IpgB2_W62A (IpgB2*) was used in this study. In addition, since full-length SipA colocalizes with yeast actin cables (46), a truncated allele of SipA (amino acids 1 to 254) was used in this assay. Download Figure S1, PDF file, 0.2 MB.
The CCBD sequence uniquely defines interactions between effectors and class IB chaperones. (A) Alignment of N termini of effectors previously proposed to contain a β-strand (11) which is indicated in red. (B) Each of the images is representative of yeast expressing the designated effector fused to GFP and μNS fused to either Salmonella InvB or Shigella Spa15. The yeast cells were visualized 4 to 5 hours after induction of the fusion proteins. (C) Representative images of yeast coexpressing class IA chaperones, E. coli CesT and Salmonella SPI2 SrcA, fused to μNS and Spa15-dependent effectors fused to GFP. Download Figure S2, TIFF file, 0.9 MB.
All class IB homologs except for those from the B. mallei and S. glossinidius SSR1 T3SSs directly bind Shigella CCBD-containing effectors. (A) Summary of automated PIP data. Images of yeast coexpressing the designated CCBD-containing effectors fused to GFP and the class IB chaperones fused to μNS were obtained using an automated microscope. The percentage of yeast with fluorescent foci (pips) was determined. The values shown are means plus SEM (error bars) and are representative of 3 independent experiments conducted on 2 days. At least 6,000 cells were counted for each condition. (B) The class IB chaperones from the B. mallei and S. glossinidius SSR1 T3SSs are capable of localizing to μNS-generated foci when expressed in yeast. Images of yeast expressing class IB chaperones from the B. mallei and S. glossinidius T3SSs each fused to CFP-μNS observed 4 to 5 hours after induction of expression of the fusion proteins are shown. Download Figure S3, TIFF file, 0.9 MB.
All class IB homologs except for those from the B. mallei and S. glossinidius SSR1 T3SSs facilitate translocation of Shigella CCBD-containing effectors from the Shigella Δspa15 mutant. (A) Summary of high-throughput TEM-1 translocation data. HeLa cells preloaded with CCF4/AM were infected with the Shigella Δspa15 mutant coexpressing the designated CCBD-containing effectors fused to TEM-1 and class IB chaperones for 2 hours. The cells were fixed, and the ratio of blue cells to green cells was determined. Values are representative of 3 independent experiments done in triplicate, and at least 600 cells were counted for each sample (means plus SEM shown). (B) Immunoblots of extracts of the Shigella Δspa15 mutant expressing myc-tagged versions of the class IB chaperones from the S. flexneri, B. mallei, and S. glossinidius T3SSs. The blots were probed with anti-OspF antibody as a loading control. Download Figure S4, TIFF file, 1 MB.
Sodalis SSR2_InvB complements secretion of Shigella effectors from the Shigella Δspa15 mutant. S. flexneri strains were grown under conditions that induce T3SS. The S. flexneri strains used were the wild type, Δspa15 mutant, and the Δspa15 mutant complemented with Shigella Spa15 or the class IB chaperone from the Sodalis SSR2 T3SS and expressing FLAG-tagged Shigella effectors. The supernatant proteins were probed with anti-FLAG and anti-IcsA antibodies. The blots shown are representative of at least three experiments. Download Figure S5, TIFF file, 0.2 MB.
Identification of the first effectors from Sodalis. (A) Alignment of N termini of hypothetical Sodalis ORFs identified by a pattern search algorithm for CCBD sequence. The putative CCBD domain is shown in red. (B) The wild-type S. flexneri and S. flexneri ΔmxiM mutant strains expressing the designated FLAG-tagged Sodalis proteins were grown under conditions that induce T3SS. The supernatant proteins were probed with anti-FLAG and anti-IcsA antibodies. The blots shown are representative of at least three experiments. Download Figure S6, TIFF file, 0.3 MB.
Summary of Y2H Spa15-OspD1 interactions.
List of oligonucleotides used to generate fragments via PCR for cloning into Gateway entry vectors.