ABSTRACT
Cell wall proteins (CWPs) mediate important cellular processes in fungi, including adhesion, invasion, biofilm formation, and flocculation. The current model of fungal cell wall organization includes a major class of CWPs covalently bound to β-1,6-glucan via a remnant of a glycosylphosphatidylinositol (GPI) anchor. This model was established by studies of ascomycetes more than a decade ago, and relatively little work has been done with other fungi, although the presumption has been that proteins identified in the cell wall which contain a predicted GPI anchor are covalently linked to cell wall glucans. The pathogenic basidiomycete Cryptococcus neoformans encodes >50 putatively GPI-anchored proteins, some of which have been identified in the cell wall. One of these proteins is chitin deacetylase 2 (Cda2), an enzyme responsible for converting chitin to chitosan, a cell wall polymer recently established as a virulence factor for C. neoformans infection of mammalian hosts. Using a combination of biochemistry, molecular biology, and genetics, we show that Cda2 is GPI anchored to membranes but noncovalently associated with the cell wall by means independent of both its GPI anchor and β-1,6-glucan. We also show that Cda2 produces chitosan when localized to the plasma membrane, but association with the cell wall is not essential for this process, thereby providing insight into the mechanism of chitosan biosynthesis. These results increase our understanding of the surface of C. neoformans and provide models of cell walls likely applicable to other undercharacterized basidiomycete pathogenic fungi.
IMPORTANCE
The surface of a pathogenic microbe is a major interface with its host. In fungi, the outer surface consists of a complex matrix known as the cell wall, which includes polysaccharides, proteins, and other molecules. The mammalian host recognizes many of these surface molecules and mounts appropriate responses to combat the microbial infection. Cryptococcus neoformans is a serious fungal pathogen that kills over 600,000 people annually. It converts most of its chitin, a cell wall polysaccharide, to chitosan, which is necessary for virulence. Chitin deacetylase enzymes have been identified in the cell wall, and our studies were undertaken to understand how the deacetylase is linked to the wall and where it has activity. Our results have implications for the current model of chitosan biosynthesis and further challenge the paradigm of covalent linkages between cell wall proteins and polysaccharides through a lipid modification of the protein.
Introduction
The fungal cell wall is a complex organelle, essential for maintaining cell morphology and viability under stress and for mediating interactions with the environment and, in the case of pathogenic fungi, the host. Current understanding of cell wall organization is based primarily upon studies of ascomycetes, including the model yeast Saccharomyces cerevisiae and the fungal pathogens Candida albicans and Aspergillus spp. (1–3). There is much variety in cell wall composition between and even within species, but the common features include an interconnected matrix of polysaccharides with associated glycoproteins. Cell wall glycoproteins are abundant and mediate a variety of cellular processes, such as maintenance of osmotic stability, carbohydrate/glycan modification, protection, adhesion, and iron uptake (4). Covalently bound cell wall proteins (CWPs) are transported to the wall via a classical secretory mechanism and are heavily glycosylated by N-linked and O-linked glycosylation in Ser/Thr-rich regions (4). Members of the major class are called glycosylphosphatidylinositol (GPI) CWPs because they are linked to β-1,6-glucan via a remnant of a GPI anchor (4). Mechanisms of protein attachment other than that of GPI CWP class members, including disulfide bonding (5) and noncovalent interactions (6), have been described. Some GPI CWPs are also secreted, likely released by endogenous enzymes, and are thought to exist in the cell wall solely in transit to the extracellular space (6).
Cryptococcus neoformans is an environmental basidiomycete causing opportunistic pulmonary infection and potentially fatal meningoencephalitis in immunocompromised individuals. C. neoformans encodes more than 50 putative GPI-anchored mannoproteins (7). Of 30 proteins identified by mass spectrometry (MS) in the C. neoformans extracellular proteome, 17 were found to contain a putative GPI anchor attachment signal (8), known as an ω site, and thus far the presumption has been that these proteins are covalently bound to β-1,6-glucan as in yeast. Although not identified in this MS study, the most well characterized GPI-anchored protein in C. neoformans is the virulence factor phospholipase B1 (Plb1). Plb1 has been detected in the cell wall, membrane, and secreted fractions by Western blot analysis, and the GPI anchor is an established determinant of this localization (9). There is mounting evidence that Plb1 follows the GPI CWP model established for yeast. Levels of Plb1 are decreased in the cell wall fraction and increased in secretions upon loss of its GPI anchor and also in β-1,6-glucan-deficient strains (9, 10). Plb1 released from cell walls by β-1,3-glucanase digestion contained a β-1,6-glucan moiety, as evidenced by pulldown assays (11). These results are consistent with Plb1 being covalently bound to β-1,6-glucan via its GPI anchor and suggest that other GPI CWPs also exist in C. neoformans.
Three additional putatively GPI-anchored proteins that were identified by mass spectrometry in the C. neoformans extracellular proteome (8) are the chitin deacetylases (Cdas). These enzymes are responsible for converting the GlcNAc homopolymer chitin to its more soluble derivative, chitosan (12). In contrast to the model yeast S. cerevisiae, which contains chitosan only during sporulation (13), C. neoformans contains significant levels of chitosan during vegetative growth (14). Loss of chitosan, accomplished by deletion of all three CDA genes, has detrimental consequences, causing cells to be slow growing and to have incomplete bud separation and increased sensitivity to chemical agents that challenge cell wall integrity (12). Cda2, also known as MP98, was shown previously to be an immunogenic extracellular mannoprotein (7). More recently, we demonstrated that chitosan is necessary for virulence in a mouse model of C. neoformans infection (15). Chitosan synthesis represents a promising target for anticryptococcal therapeutics, as chitosan is required for virulence and absent from the human host. Over 600,000 people die every year from cryptococcosis, and current therapies are insufficient. Studies delineating the mechanisms of chitosan production are necessary to better exploit chitosan synthesis as a novel therapeutic target for treatment of these patients.
To gain insight into the mechanism of chitosan synthesis in C. neoformans, we generated a monoclonal antibody to Cda2. We found that Cda2 is secreted, consistent with previous findings, and also is distributed in the plasma membrane and cell wall. Given that Cda2 is predicted to be GPI anchored, the current cell wall protein model predicts that Cda2 is covalently bound to β-1,6-glucan. However, our data demonstrate that this enzyme is noncovalently associated with the wall and that neither the GPI anchor nor the β-1,6-glucan is required for this association. We also provide evidence that the membrane-bound, but not the cell wall-associated, Cda2 is necessary and sufficient for chitosan production during vegetative growth.
RESULTS
Chitin deacetylase 2 is secreted and localized to the membrane and cell wall.
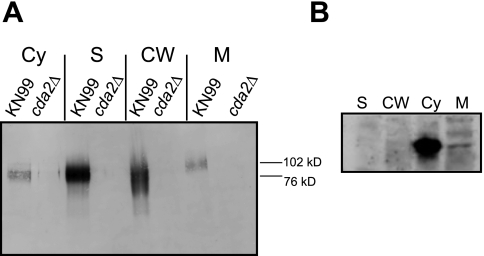
The presence on Cda2 of a putative N-terminal signal sequence and C-terminal ω site suggests that it is localized to the plasma membrane or cell wall or secreted, or all three, as was the case for the well-characterized cryptococcal GPI-anchored protein Plb1 (9). Cda2 (MP98) was detected in the proteomic analysis of the cell wall (8) and was detected as an immunogenic component of C. neoformans culture filtrates, indicating that it is secreted (7). To determine the subcellular localization of Cda2 (CNAG_10230.2, Broad Institute) we generated a monoclonal antibody to a Cda2 peptide (TDDWAAGTNGVTEQDVTN). This peptide was chosen to maximize specificity because it is unique to Cda2 and not found in Cda1 or Cda3, which share ~40% identity and ~50% similarity with Cda2. BLASTP analysis also demonstrated that the peptide was not highly homologous to any other C. neoformans protein; no hits were obtained using an E value of <10 as a cutoff. The peptide is also from a region predicted to be free of glycosylation, which may interfere with antibody binding. This antibody was used in Western blot analysis of subcellular fractions, including the cytosolic, secreted, crude membrane, and cell wall fractions. Its specificity was confirmed using a cda2Δ deletion strain (Fig. 1A). Cda2 was detected in all subcellular fractions (Fig. 1A). The protein migrated at a higher molecular mass than that predicted by the polypeptide (~48 kDa), consistent with the presence of glycosylation as previously demonstrated by periodic acid-Schiff (PAS) staining (7). Cda2 detected from membranes migrated slightly more slowly (~102 kDa) than that from the secreted and cell wall fractions (~76 kDa), suggesting the possible presence of a modification(s) on the enzyme (explored further below). Although Cda2 was detected in the soluble cytosolic fraction, it is improbable that the cytosol is a biological source of the enzyme due to the presence of a signal sequence and putative GPI anchor. The appearance of Cda2 in this fraction is likely a consequence of shearing from the wall during sample preparation. Although the subcellular fractionation method utilized was the same accepted in the literature for establishing localization patterns of Plb1, a primary concern with this type of analysis is the potential for abundant cytosolic proteins ending up in the secreted fractions due to cell lysis or in the cell wall fractions due to association during the lysing procedure. To assess this, we analyzed all fractions for the known cytosolic proteins actin and Mpk1 and found them present in the soluble cytosolic fraction but not in cell walls, membranes, or secretions (Fig. 1B and data not shown). Also, the slower migration of Cda2 from the membrane than from the remaining fractions suggests that the membrane fraction is in fact distinct.
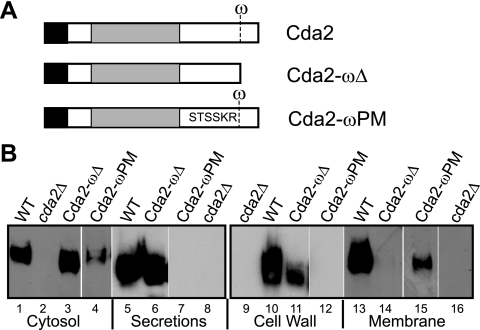
FIG 1 .
Subcellular localization of Cda2 in C. neoformans. (A) Western blot analysis of fractions collected from wild-type serotype A strain KN99 and a cda2Δ strain in the same background probed with monoclonal antibodies directed against a Cda2-specific peptide. Cy, cytosol; S, secretions; CW, cell wall; M, membrane. (B) Control blots containing fractions from KN99 demonstrating the presence of actin in the cytosolic fraction and it absence from the remaining fractions.
Cda2 localization is not disrupted in β-1,6-glucan-deficient mutants.
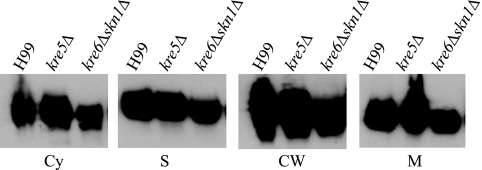
In S. cerevisiae, β-1,6-glucan serves to covalently attach GPI-anchored proteins to the cell wall matrix (4). Since Cda2 has been predicted to be modified by the addition of a GPI anchor, we hypothesized that Cda2 localization would be disrupted upon loss of β-1,6-glucan. To address this hypothesis, we analyzed Cda2 in subcellular fractions isolated from kre5Δ and kre6Δ skn1Δ strains, two mutants in which β-1,6-glucan is undetectable and the localization of the known GPI-anchored cryptococcal protein, Plb1, is disrupted; specifically, Plb1 displays decreased cell wall localization (10). Surprisingly, in contrast to what we previously observed for Plb1, we detected Cda2 in all fractions from both mutants with no observable differences from what was seen in fractions derived from wild-type cells (Fig. 2).
FIG 2 .
Subcellular localization of Cda2 in β-1,6-glucan-deficient mutants. Western blot analysis of Cda2 was performed as described in the legend of Fig. 1, and fractions collected from the kre5Δ mutant, the kre6Δ skn1Δ mutant, and wild-type serotype A background strain H99 were compared. Cda2 in each fraction migrated at the same apparent molecular mass as shown in Fig. 1. Cy, cytosol; S, secretions; CW, cell wall; M, membrane.
Cda2 is released from cell walls by SDS.
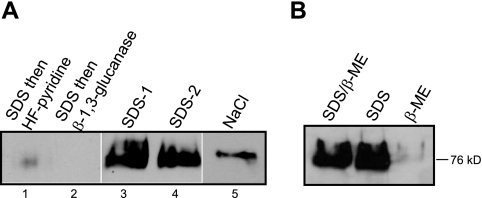
Since localization to the cell walls of β-1,6-glucan-deficient mutants does not eliminate the possibility that this polymer anchors Cda2, we further assessed the attachment of Cda2 to the cell wall in wild-type KN99. In the Western blot shown in Fig. 1, Cda2 was solubilized from the cell wall matrix by β-1,3-glucanase digestion, which is considered to release primarily covalently bound proteins but may also simply serve to loosen the polysaccharide network and facilitate the release of noncovalent interactions. To directly determine whether Cda2 is GPI anchored to β-1,6-glucan, we treated isolated cell walls with hydrogen fluoride (HF)-pyridine, which specifically cleaves the GPI anchor remnant and is a primary means by which GPI CWPs in ascomycetes have been classified (16). Cell walls were first boiled in SDS–β-mercaptoethanol (β-ME) to remove proteins tightly associated through noncovalent interactions prior to HF-pyridine treatment according to a published protocol (17). HF-pyridine extraction did not release Cda2 from isolated cell walls (Fig. 3A, lane 1); however, Cda2 was detected in the SDS–β-ME washes (Fig. 3A, lanes 3 and 4), consistent with noncovalent association. No additional Cda2 was released from SD–β-ME-treated cell walls by the β-1,3-glucanase treatment utilized in the experiment detailed in Fig. 1 (Fig. 3A, lane 2). Treatment with SDS–β-ME also solubilized Cda2 from the kre5Δ mutant and the kre6Δ skn1Δ β-1,6-glucan-deficient mutant (data not shown). To distinguish between noncovalent and disulfide bond-mediated associations of Cda2, cell walls were extracted with SDS or β-ME alone and in combination (Fig. 3B). Cda2 was released from walls only by treatments containing SDS, not by β-ME alone (Fig. 3B).
FIG 3 .
Extraction of Cda2 from isolated cell walls. (A) Cell walls collected from KN99 were subjected to various treatments (400 µg [dry weight] equivalents per sample) as described in Materials and Methods; two samples were boiled twice in SDS–β-ME (solubilized proteins from these extractions were loaded in lanes 3 and 4), followed by either HF-pyridine extraction (lane 1) or β-1,3-glucanase digestion (lane 2). (B) Isolated cell walls were boiled in SDS and β-ME either alone or in combination, as indicated.
Cda2 is GPI anchored to membranes.
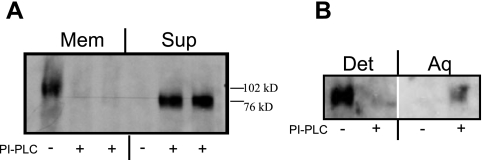
The results presented above are in contrast to what is has been established regarding covalently bound GPI-anchored cell wall proteins in other fungi and for Plb1 in C. neoformans. These results caused us to question whether Cda2 actually contains a GPI anchor, since the presence of this modification has been predicted in silico but never demonstrated biochemically. We assessed the anchoring of Cda2 to membranes using classical methods that establish the presence of a GPI anchor. Membrane fractions were collected as before and then either mock treated or treated with the GPI anchor-cleaving enzyme phosphatidylinositol-specific phospholipase C (PI-PLC). Digestion with PI-PLC released Cda2, which ran at ~76 kDa by SDS-PAGE, from membranes into the supernatant (Fig. 4A, lanes 5 and 6), consistent with cleavage of the GPI anchor. For confirmation of this result, we used the detergent Triton X-114, which separates membrane-bound proteins (detergent phase) from soluble proteins (aqueous phase). GPI-anchored proteins remain in the detergent phase unless released by PI-PLC digestion to partition in the aqueous phase. Cda2 released by PI-PLC partitioned to the aqueous phase upon Triton X-114 treatment (Fig. 4B, lane 4), while it remained in the detergent phase in mock-treated samples (Fig. 4B, lane 1). Together, these results for the first time establish the presence of a GPI anchor facilitating membrane attachment of Cda2 in C. neoformans.
FIG 4 .
Analysis of membrane anchoring of Cda2. (A) Total membranes were isolated and either mock treated (1st and 4th lanes) or digested with 1 U of purified PI-PLC (2nd, 3rd, 5th and 6th lanes). Samples were subsequently subjected to ultracentrifugation to separate the membrane (Mem)-bound proteins (1st to 3rd lanes) from soluble proteins in the supernatant (Sup) (4th to 6th lanes). (B) Total membranes were isolated and either mock treated (1st and 3rd lanes) or digested with PI-PLC (2nd and 4th lanes). Samples were subjected to phase partitioning with Triton X-114 into detergent (Det) (1st and 2nd lanes) and aqueous (Aq) (3rd and 4th lanes) phases.
Loss of the GPI anchor disrupts the membrane, but not the cell wall, attachment of Cda2.
We also took a genetic approach to further understand the requirement of a GPI anchor for Cda2 localization. We generated strains expressing a truncated version of CDA2 lacking the putative ω site, which we have termed cda2-ωΔ, as illustrated in Fig. 5A. The cda2-ωΔ construct was transformed into a cda1Δ cda2Δ cda3Δ strain to permit assessment of the implication of potential Cda2 mislocalization on chitosan production, since the presence of any CDA gene individually is sufficient for this process (12). We isolated subcellular fractions from two cda1Δ cda2-ωΔ cda3Δ isolates and determined the Cda2-ωΔ protein localization pattern by Western blotting (Fig. 5B and data not shown). Cda2-ωΔ was present in the secreted (lane 6) and cell wall (extracted with β-1,3-glucanase [lane 11] or SDS [data not shown]) fractions but absent from membranes (lanes 14). These results are consistent with the data presented above and provide further evidence that Cda2 is GPI anchored to membranes but that this anchor is not required for association with the cell wall.
FIG 5 .
Localization of Cda2 ω site mutants. (A) Schematic illustrating the domain organization of wild-type CDA2 and indicating the presence of the ω site. The cda2-ωΔ construct encodes a truncated version of Cda2 lacking the ω site and subsequent hydrophobic tail. In cda2-ωPM, the ω-1 through ω-5 sequence has been replaced with the corresponding sequence from S. cerevisiae Yps1. (B) Western blot analysis detecting Cda2 in fractions from KN99 (wild type [WT]) and the cda2Δ, cda1Δ cda2-ωΔ cda3Δ, and cda1 Δcda2-ωPM cda3Δ mutants. Cda2 in each fraction migrated at the same apparent molecular mass as shown in Fig. 1.
Loss of the GPI anchor prevents chitosan production by Cda2.
We demonstrated previously that the cryptococcal cell wall contains significant levels of chitosan during vegetative growth (14), in contrast to the model yeast S. cerevisiae, which contains chitosan only during sporulation (13). Furthermore, chitosan is important for maintaining cryptococcal cell wall integrity and virulence (12, 15). Consequently, C. neoformans is an ideal organism for investigating the mechanism of chitosan biosynthesis, and such studies have the potential to uncover novel avenues for therapeutic intervention. To this end, we investigated the impact of Cda2 localization on its ability to generate chitosan. We considered three potential models for chitosan production, illustrated in Fig. 6A, which we have named “membrane,” “cell wall,” and “hybrid,” based upon the localization of the Cda2 that functions to enzymatically convert chitin to chitosan. We tested these models utilizing strains expressing Cda2-ωΔ to determine whether cell wall localization alone is sufficient for Cda2 to generate chitosan. If a strain expressing Cda2-ωΔ is capable of chitosan production in the absence of other chitin deacetylases, this result would support the cell wall model, whereas chitosan deficiency in this strain would be consistent with both the membrane and hybrid models. The cda1Δ cda2-ωΔ cda3Δ strain phenotypically mimicked a cda1Δ cda2Δ cda3Δ strain in vitro, being highly sensitive to the cell wall inhibitors SDS and NaCl (Fig. 6B); it also lacked chitosan, as determined by eosin Y staining (chitosan-specific dye) (12) and biochemical analysis (Fig. 6C and D). A cda2-ωΔ cda3Δ strain that still encodes CDA1 and displays the same localization pattern for Cda2-ωΔ protein observed previously (data not shown) behaved like the wild type (Fig. 6B to D), serving as a control confirming that expression of the cda2-ωΔ mutant does not result in an untoward disruption of the intrinsic ability of the cell to produce chitosan.
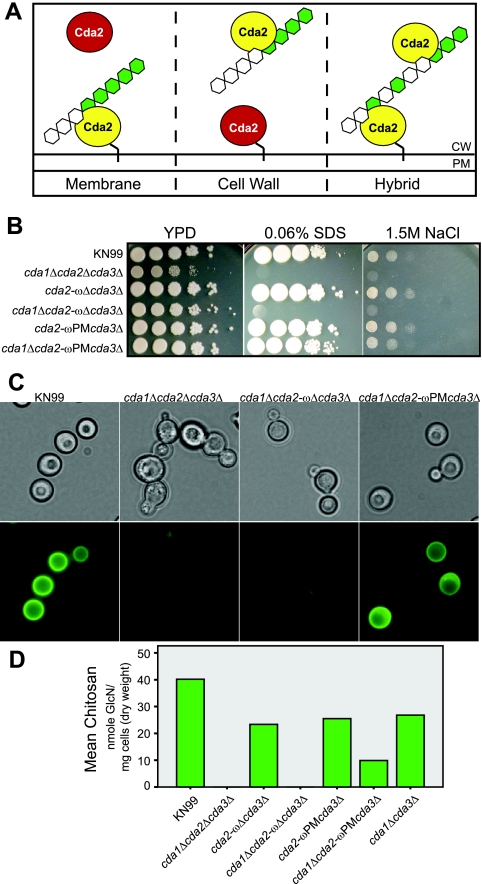
FIG 6 .
Implications of Cda2 localization for chitosan synthesis. (A) Proposed models of chitosan production with respect to Cda2 localization in C. neoformans. Ovals represent Cda2, with yellow indicating the enzyme that functions to produce cell wall chitosan. The black line linking Cda2 to the membrane represents the GPI anchor. CW, cell wall; PM, plasma membrane. (B) Plating experiment illustrating the sensitivity of the cda1Δ cda2-ωΔ cda3Δ mutant, which is analogous to that of the chitosan-deficient cda1Δ cda2Δ cda3Δ strain, and the wild-type growth of the cda1Δ cda2-ωPM cda3Δ mutant. (C) Eosin Y staining of Cda2 ω site mutants. (D) Chitosan levels in cell walls from Cda2 ω site mutants. The cda1Δ cda3Δ strain was included as a control for the chitosan produced by a strain expressing wild-type CDA2 in the absence of the other two chitin deacetylases. Values are averages of results from two independent biological replicates for each strain.
Membrane restriction of Cda2 allows chitosan production.
The results described above suggest that the presence of Cda2 in the membrane is necessary for chitosan production but fail to rule out a role for cell wall-localized Cda2, which may first require the action of the membrane-bound enzyme. We next sought to generate a strain that would retain Cda2 in the membrane and prevent its translocation to the wall. The absence of chitosan in a strain expressing membrane-restricted Cda2 would support the hybrid model, while the presence of chitosan would be consistent with the membrane model. To achieve membrane restriction of Cda2, we used what was previously established for ascomycetes with regard to GPI-anchored proteins. In several ascomycete species studied, plasma membrane GPI-anchored proteins contain a dibasic motif immediately N terminal of the ω site that is absent in GPI proteins directed to the wall (17, 18) and is absent in Cda2. In S. cerevisiae, replacement of the sequence preceding the ω site of a known cell wall protein with that of the plasma membrane protein Yps1 resulted in its loss from the wall and subsequent accumulation in the membrane (19). Based on these results, we replaced the ω-1 through ω-5 sequence of Cda2 (GSNNA) with that of Yps1 (TSSKR) and termed the resulting construct cda2-ωPM (PM for plasma membrane) (Fig. 5A). To enable analysis of potential effects on chitosan production as for the cda2-ωΔ mutant, we generated the cda2-ωPM cda3Δ (contains CDA1) and cda1Δ cda2-ωPM cda3Δ strains. Western blot results confirmed that these strains displayed the same Cda2-ωPM protein localization pattern. Cda2-ωPM protein was detected in the membrane fraction (Fig. 5B, lane 15) but was absent from the cell wall fraction (Fig. 5B, lane 12). Unlike the cda1Δ cda2-ωΔ cda3Δ strain, a strain expressing cda2-ωPM in the absence of CDA1 and CDA3 displayed growth akin to that of the wild type in the presence of cell wall inhibitors (Fig. 6B). Furthermore, the cda1Δ cda2-ωPM cda3Δ mutant was able to produce chitosan, as demonstrated by eosin Y staining and biochemical analysis (Fig. 6C and D). These data establish that membrane localization of Cda2 is not only necessary but also sufficient for Cda2 to generate chitosan in C. neoformans and are consistent with the membrane model of chitosan synthesis (Fig. 6A).
DISCUSSION
In this study, we establish that Cda2 is a GPI-anchored protein active in the plasma membrane and also noncovalently associated with the cell wall in a manner independent of both the GPI anchor and β-1,6-glucan in C. neoformans. It has been established in the literature that a covalent linkage exists between GPI CWPs and β-1,6-glucan in the fungal cell walls of several ascomycetes, including S. cerevisiae, C. albicans, Aspergillus niger, and Schizosaccharomyces pombe. This conclusion is based upon two main lines of evidence: the identification of a residue in the S. cerevisiae cell wall that contains β-1,6-glucan covalently bound to a moiety with the composition of a GPI remnant (20) and identification of GPI-anchored proteins in proteomic analyses of cell walls. Although convincing biochemical and immunological evidence of this linkage exists for a few specific proteins, other studies have presumed that GPI proteins are covalently bound to β-1,6-glucan but have provided no direct evidence of this linkage, and even the existence of the GPI anchor is often based upon in silico predictions. Some existing evidence in the literature suggests that that the GPI CWP model may not be absolute, even in ascomycetes. For example, the majority of the GPI-anchored proteins of A. fumigatus, a filamentous ascomycete, reside in the plasma membrane (21). For the few apparent GPI CWPs in this organism it has been suggested that there may be alternative modes of attachment, such as to β-1,3-glucan or another cell wall polymer, since A. fumigatus contains no detectable β-1,6-glucan in the alkali-insoluble portion of its cell wall (22). The GPI-anchored acid phosphatase PhoAp was released from the A. fumigatus cell wall by SDS–β-ME, suggesting that it may be noncovalently associated with the cell wall (21), as we have observed here for Cda2. Directly determining whether a given protein is covalently or noncovalently associated with the cell wall is not a trivial detail but has important implications when one considers potential mechanisms for their secretion. Covalent binding of GPI CWPs to the polysaccharide matrix requires that their release into the “secretome” involve enzymatic release not only from the membrane but also from the glucans.
The foundation of our current understanding of fungal cell wall proteins is based primarily upon the investigations of the ascomycetous fungi described above, and data from basidiomycetous fungi are limited. Recent reviews have cautioned against extrapolating the model of cell wall structure and organization from ascomycetes to basidiomycetes (23, 24). The observations in this study regarding the pattern of Cda2 localization in C. neoformans may reveal an important distinction regarding the characteristics of CWPs between these fungal phyla. Future investigations of CWPs in other basidiomycetes will reveal whether noncovalent association of GPI CWPs is a common feature. Of particular interest, the genome of the basidiomycete Ustilago maydis contains 55 genes predicted to encode GPI-anchored proteins, but investigations have failed to detect covalently bound GPI proteins in the U. maydis cell wall (23). The majority of U. maydis GPI proteins are most closely related to proteins from other basidiomycetes, including C. neoformans, suggesting that U. maydis GPI CWPs may be noncovalently associated, like Cda2.
Along with having broad implications, the data from this study provide important insight into cell wall organization in C. neoformans specifically. Previous chemical analysis of isolated cryptococcal cell walls revealed the presence of only glucose and hexosamine (25), indicating the near absence of covalently bound proteins. This observation may suggest that the majority of cryptococcal CWPs are associated with the cell wall in the manner observed for Cda2. Several proteins predicted to contain a GPI anchor have been identified in C. neoformans (8). Thus far, the assumption has been that these proteins are covalently cross-linked to β-1,6-glucan. This conclusion has been further justified by evidence that the known GPI-anchored virulence factor, Plb1, appears to follow the yeast paradigm of a GPI CWP. The results presented here for Cda2 reveal important distinctions from Plb1. While both Cda2 and Plb1 are GPI anchored in the membrane, this anchor is required for the cell wall attachment of Plb1 but dispensable for the cell wall association of Cda2. The cell wall localization of Plb1, but not Cda2, was decreased in β-1,6-glucan-deficient mutants (10). These observations suggest that two classes of GPI CWPs may exist in C. neoformans, with some linked to β-1,6-glucan, as in ascomycetes, and with others tightly associated via noncovalent interactions. Alternatively, it is possible that the apparent distinctions between Cda2 and Plb1 could be an artifact of the different types of analyses performed for each protein. For example, although current evidence is consistent with covalent attachment of Plb1 to the cell wall, ultimate proof of this linkage requires chemical analysis of its C-terminal peptide, as was performed for Tip1 in S. cerevisiae (26). The possibility of a fraction of cell wall Plb1 displaying noncovalent association has not been excluded. Some Plb1 remained associated with the cell walls of β-1,6-glucan-deficient mutants (10) and was released from wild-type cell walls by NaCl (11), consistent with a fraction of Plb1 being noncovalently associated with the cell wall independently of β-1,6-glucan. It remains to be determined whether other putatively GPI-anchored proteins in the cryptococcal cell wall share the features of Cda2 described here. Our findings caution against the current assumption that all cryptococcal GPI CWPs are covalently bound to β-1,6-glucan.
Prior to this study, it was unknown whether basidiomycete fungi utilize a dibasic motif N terminal to the ω site to sort GPI-anchored proteins. Cda2 does not contain a dibasic motif upstream of its ω site and is therefore predicted to localize primarily to the cell wall, according to what has been established for ascomycetes. Although present in the cell wall, the active form of Cda2 appears to be localized to the membrane. Conversion of the amino acids upstream of the Cda2 ω site to that of an S. cerevisiae GPI-anchored membrane protein resulted in its retention in the plasma membrane. These results indicate that C. neoformans is capable of utilizing the dibasic motif as a signal to sort proteins modified by GPI anchor addition and suggest that other basidiomycetes may also employ this mechanism.
The results of this investigation provide significant insight into the mechanism of chitosan production. Strains expressing cell wall-localized Cda2-ωΔ failed to generate chitosan, while those expressing membrane-restricted Cda2-ωPM contained chitosan. These data establish that membrane localization of Cda2 is not only necessary but also sufficient for Cda2 to generate chitosan in C. neoformans. These results are inconsistent with the cell wall model of chitosan synthesis and support the membrane model (Fig. 6A). Chitosan production begins with the synthesis of a chitin substrate from cytoplasmic pools of UDP-GlcNAc, facilitated by a chitin synthase (Chs) enzyme present in the membrane, in conjunction with a cognate regulator (chitin synthase regulator, Csr). Once formed, chitin is then deacetylated via the action of Cda enzymes. C. neoformans encodes eight Chs and three Csr enzymes; however, a single pair, Chs3 and Csr2, produce the majority of the chitin that is converted to chitosan (14). This preferential deacetylation has led to the hypothesis that a complex may form between Chs3, Csr2, and the Cdas, thus facilitating the observed specificity. The observation that Cda2 appears to require membrane localization to produce chitosan is consistent with this hypothesis, as Cda2 localized to the membrane has the potential to sustain an interaction(s) with Chs3. The observation that Cda2 is active in the membrane is consistent with it being a conventional GPI-anchored membrane protein. The lack of apparent activity of cell wall-associated Cda2 may be due to a variety of factors, not all of which are mutually exclusive. The enzyme may be associated in the cell wall in a configuration that renders it catalytically inactive. Alternatively, Cda2 in the cell wall may maintain enzymatic activity but fail to obtain access to its substrate. Finally, Cda2 may be active in the cell walls producing a particular type of chitosan, which could not be distinguished by the methods utilized here. Based on the data in this study, the physiological role, if any, of the cell wall-associated enzyme remains obscure. It is often cited in the literature that cell wall proteins play an important structural role, a notion refuted with several compelling lines of evidence in a recent review (27). The fact that strains lacking Cda2 in the wall displayed no observable defects in cell integrity argues against it serving this function. One possibility is that Cda2 is transiently present in the cell wall on its way to being secreted. It is important to note, however, that although the loss of Cda2 from the wall did not prevent chitosan production, the levels of chitosan present in strains expressing membrane-restricted Cda2-ωPM were lower than in a strain expressing wild-type Cda2. There are multiple potential explanations for this observation. One possibility could be altered expression of Cda2, since the cda2-ωPM construct inserted ectopically in the genome, as opposed to at the endogenous CDA2 locus. Western blot analyses suggest that the level of Cda2 is decreased in the cda2-ωPM mutants, although this method is not definitively quantitative. Also, it is unknown whether mutation of the sequence N terminal of the ω site affects the overall enzymatic activity of Cda2. Alternatively, the failure of Cda2-ωPM to acquire full chitosan levels may suggest a role for wall-associated Cda2 in chitosan production, such as that illustrated in the hybrid model (Fig. 6A). In this model, chitosan production would be processive, such that Cda2 in the membrane would initially deacetylate chitin, thereby generating a partially deacetylated substrate that could be further deacetylated via the action of Cda2 in the wall. Such a scenario could explain why the cda2-ωΔ mutants contain no chitosan while the cda2-ωPM strains contain the polymer. Chitosan derived from different fungal or insect species is known to vary in degree of acetylation (DA), and this property of cryptococcal chitosan is yet to be determined. Future analysis of the DA of the chitosan present in wild-type versus cda2-ωPM strains may support or refute the hybrid model. Ultimately, the observation that cda2-ωPM strains display no observable growth defects indicates that the chitosan produced in the absence of cell wall-associated Cda2 is sufficient to maintain cell integrity.
In summary, we have determined the localization of Cda2 in C. neoformans. Our data demonstrate for the first time that Cda2 is GPI anchored to the plasma membrane and that this anchor, as well as β-1,6-glucan, is dispensable for cell wall association. We probed the mechanism of chitosan production with respect to Cda2 localization, revealing that the membrane association of this Cda is required for it to generate chitosan. These findings have important implications for our understanding of the cell surfaces of pathogenic fungi and set the groundwork for future investigations into fungal chitosan synthesis.
MATERIALS AND METHODS
Strains and media.
All strains used are in the C. neoformans serotype A H99 and KN99 backgrounds. C. neoformans was grown on YPD (1% yeast, 2% Bacto peptone, 2% dextrose), with solid media containing 2% Bacto agar and those used in sensitivity assays (Fig. 6B) containing NaCl or SDS at the appropriate concentrations.
Subcellular fractionation.
Cryptococcal secretions and subcellular fractions were prepared essentially as described previously (10, 11), with minor modifications. Cryptococcal cells, grown as YPD agar lawns for 72 h, were harvested, washed with saline, and resuspended in secretion buffer (10 mM imidazole, pH 5.0, 2% glucose) as a concentrated suspension (approximately 1 to 2 ml buffer and a 5-ml packed cell volume). Incubation was carried out overnight at 30°C. Cells were pelleted by centrifugation and the secretions collected. Cell pellets were washed once with saline and once with MES (morpholineethanesulfonic acid)-buffered saline (MBS; 25 mM MES, 150 mM NaCl, 2 mM EDTA [pH 6.5]), flash frozen in liquid nitrogen, and stored at −80°C. Following thawing on ice, pellets were resuspended in 5 ml of prechilled MBS containing 0.1% (vol/vol) Triton X-100 and Roche complete protease inhibitors, and 1-ml aliquots were disrupted in prechilled tubes and lysed by bead beating for two cycles of 12 min at 4°C. Lysates were transferred to a fresh prechilled Eppendorf tube. The beads were washed with 0.5 ml MBS containing 0.1% Triton X-100 and protease inhibitors, and the wash was combined with the lysate to maximize protein recovery. Cell lysates were centrifuged at 3,500 × g for 10 min at 4°C and the supernatants collected and set aside on ice. Pellets were resuspended in 1 ml of the same buffer and further disrupted by probe sonication for 5 cycles of 10 s on, 10 s off, with at an output of 3 to 5. Samples were centrifuged again at 3,500 × g for 10 min at 4°C, and the supernatants were combined with that from the previous spin. The cell wall-containing pellets after centrifugation at 3,500 × g were washed three times with 30 ml saline and resuspended in 1 ml of β-1,3-glucanase (lysing enzyme; 20 mg/ml was made up in water and protease inhibitors; Sigma catalog no. L1412). Incubation was carried out for 1 h with agitation at 37°C, and the supernatant, containing released proteins and defined as the cell wall fraction, was collected by centrifugation (14,000 × g for 15 min at 4°C). The supernatants after centrifugation at 3,500 × g were ultracentrifuged at 135,000 × g for 1 h at 4°C to separate the pellet (crude membranes) from the supernatant (cytosol). The protein concentration of each fraction was analyzed using Quick Start Bradford reagent (Sigma) with bovine serum albumin (BSA) as a standard.
Solubilization of CWPs.
KN99 was grown overnight in YPD broth at 30°C, and cells were collected and disrupted as described above to isolate cell walls. For this analysis, aliquots of cell walls were washed with NaCl and lyophilized to be consistent with published protocols using HF-pyridine (28). Cell walls (equivalent of 400 mg [dry cell weight] per sample) were treated twice with 50 mM Tris-HCl, pH 7.8, 2% SDS, 100 mM Na-EDTA, 40 mM β-mercaptoethanol for 5 min at 100°C and washed three times with sterile water. The insoluble pellet was either extracted with 10 ml HF-pyridine per mg (dry weight) for 3 h at 0°C (lane 1) or digested with β-1,3-glucanase as described above (lane 2). Solubilized proteins were collected and analyzed by Western blotting. Following each treatment, samples were centrifuged at 13,000 rpm in a microcentrifuge to separate the cell wall debris from the solubilized proteins. The supernatant was removed, and samples were flash frozen and stored at −80°C until analyzed by Western blotting (see below).
PI-PLC and Triton X-114 phase partitioning of GPI-anchored proteins.
KN99 was grown overnight in YPD broth at 30°C, and cells were collected and disrupted as described above to collect crude membranes. GPI-anchored proteins were released from membranes as described previously (11). Briefly, crude membranes were resuspended in 200 µl of ice-cold Tris-HCl, pH 7.5, 0.5 mM EDTA with protease inhibitors and GPI-anchored proteins were released by incubation with 1 U (G)PIPLC from Bacillus cereus at 37°C for 1 to 2 h with agitation. The released GPI-anchored proteins were collected by ultracentrifugation, and supernatants and pellets (membranes) were frozen and stored at −80°C prior to analysis by Western blotting (Fig. 4A). For phase partitioning, membrane samples were treated with (G)PI-PLC and then diluted with 100 µl ice-cold buffer with protease inhibitors, followed by the addition of 60 µl Triton X-114 (from which contaminating detergents had been removed) and chilled on ice for 1 h. Samples were centrifuged for 10 min at 10,000 × g at 4°C to remove insoluble debris. The supernatant was heated to 37°C for 30 min to achieve phase separation. Samples were spun for 5 min at 14,000 rpm in a microcentrifuge, and the upper (aqueous) phase was removed from the lower (detergent) phase. The detergent phase was washed three times to remove residual aqueous material. Samples were frozen and stored at −80°C until analyzed by Western blotting (see below). Proteins in each fraction were trichloroacetic acid (TCA) precipitated, separated by SDS-PAGE, and transferred to nitrocellulose.
SDS-PAGE and Western blot analysis.
For the analyses of subcellular fractions shown in Fig. 1, 2, and 5, 25 µg total protein of each fraction was precipitated with TCA, resuspended in 1× sample buffer (Bio-Rad) with β-ME, and separated on 4 to 20% TGX gels (Bio-Rad), and proteins were transferred to nitrocellulose membranes. For the cell wall protein extractions and GPI-anchored membrane analyses whose results are shown in Fig. 3 and 4, entire samples of each were TCA precipitated and analyzed to ensure that any Cda2 released would be detected. Blots were probed overnight with a monoclonal antibody (generated by the Saint Louis University hybridoma facility) to a Cda2-specific peptide (TDDWAAGTNGVTEQDVTN; synthesized by GenScript, Piscataway, NJ) in Tris-buffered saline with Tween (TBST) with 5% milk and then for 1 h with a goat anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibody in TBST. Signal was detected with ECL-plus reagent for autoradiography. Due to the absence of specific proteins useful as loading controls for each fraction (such as actin for use in analyses of cytosolic proteins), equal protein loadings and transfers were confirmed by staining membranes with Ponceau S or Swift reagent (AB Biosciences) total protein stains.
Generation of CDA2 ω site mutants.
Constructs were generated by overlap PCR using primers. The cda2-ωΔ construct consisted of a 5′ fragment (primers CDA2truncA and CDA2-GPIrev) and a 3′ fragment (primers CDA2truncB and CDA2-GPIfor). The cda2-ωΔ construct was transformed into LBCN458 to generate NGCN 1057 and into LBCN632 to achieve NGCN 1058 and NGCN 1059, and insertion at the endogenous CDA2 locus was confirmed using CDA2truncB and CDA2truncD. Loss of the ω site and of the subsequent sequence encoding the C-terminal hydrophobic domain was confirmed by sequencing (primer CDA2seq1 through CDA2seq8). The cda2-ωPM construct consisted of a 5′ fragment (primers CDA2truncA and CDA2PlMemGPIrev) and a 3′ fragment (primers CDA2truncB and CDA2PlMemGPIfor). The cda2-ωPM construct was transformed into LBCN458 and integrated ectopically into the genome, not at the CDA2 locus, to generate JLCN 814. The presence of the mutation upstream of the CDA2 ω site was confirmed by sequencing (primer CDA2seq1-8). JLCN 814 and was crossed with LBCN369 to achieve JLCN 849 and JLCN 855. Progeny were PCR screened extensively to confirm the absence of wild-type CDA1, CDA2, or CDA3 using primers within each open reading frame and the presence of cda2-ωPM using CDA2truncB and CDA2truncD.
Chitosan analysis.
Strains were stained with eosin Y as described previously (9). Biochemical measurements of chitosan levels were performed using Trichoderma viride chitinase, essentially as previously described (10, 12, 14).
ACKNOWLEDGMENTS
We thank Maureen Donlin for assistance with figure preparation and Woei Lam and Roger Herr for technical advice and assistance.
This work was supported by Public Health Service grant AI072195 from the National Institute of Allergy and Infectious Disease to J.K.L. and an American Heart Association predoctoral fellowship to N.M.G.
Footnotes
Citation Gilbert NM, Baker LG, Specht CA, Lodge JK. 2012. A glycosylphosphatidylinositol anchor is required for membrane localization but dispensable for cell wall association of chitin deacetylase 2 in Cryptococcus neoformans. mBio 3(1):e00007-12. doi:10.1128/mBio.00007-12.
REFERENCES
- 1. Klis FM, Mol P, Hellingwerf K, Brul S. 2002. Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 26:239–256 [DOI] [PubMed] [Google Scholar]
- 2. Klis FM, de Groot P, Hellingwerf K. 2001. Molecular organization of the cell wall of Candida albicans. Med. Mycol. 39(Suppl.):11–18 [PubMed] [Google Scholar]
- 3. Mouyna I, Fontaine T. 2009. Cell wall of Aspergillus fumigatus: a dynamic structure, p. 169–183 In Latge JP, Steinback WJ, Aspergillus fumigatus and aspergillosis. ASM Press, Washington, DC [Google Scholar]
- 4. De Groot PWJ, Ram AF, Klis FM. 2005. Features and functions of covalently linked proteins in fungal cell walls. Fungal Genet. Biol. 42:657–675 [DOI] [PubMed] [Google Scholar]
- 5. Jaafar L, Moukadiri I, Zueco J. 2003. Characterization of a disulphide-bound Pir-cell wall protein (Pir-CWP) of Yarrowia lipolytica. Yeast 20:417–426 [DOI] [PubMed] [Google Scholar]
- 6. Chaffin WL. 2008. Candida albicans cell wall proteins. Microbiol. Mol. Biol. Rev. 72(3):495–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Levitz SM, Specht CA. 2006. The molecular basis for the immunogenicity of Cryptococcus neoformans mannoproteins. FEMS Yeast Res. 6:513–524 [DOI] [PubMed] [Google Scholar]
- 8. Eigenheer RA, Jin Lee Y, Blumwald E, Phinney BS, Gelli A. 2007. Extracellular glycosylphosphatidylinositol-anchored mannoproteins and proteases of Cryptococcus neoformans. FEMS Yeast Res. 7:499–510 [DOI] [PubMed] [Google Scholar]
- 9. Djordjevic JT, Del Poeta M, Sorrell TC, Turner KM, Wright LC. 2005. Secretion of cryptococcal phospholipase B1 (PLB1) is regulated by a glycosylphosphatidylinositol (GPI) anchor. Biochem. J. 389:803–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gilbert NM, et al. 2010. KRE genes are required for beta-1,6-glucan synthesis, maintenance of capsule architecture and cell wall protein anchoring in Cryptococcus neoformans. Mol. Microbiol. 76:517–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Siafakas AR, et al. 2007. Cell wall-linked cryptococcal phospholipase B1 is a source of secreted enzyme and a determinant of cell wall integrity. J. Biol. Chem. 282:37508–37514 [DOI] [PubMed] [Google Scholar]
- 12. Baker LG, Specht CA, Donlin MJ, Lodge JK. 2007. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot. Cell 6:855–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Briza P, Ellinger A, Winkler G, Breitenbach M. 1988. Chemical composition of the yeast ascospore wall. The second outer layer consists of chitosan. J. Biol. Chem. 263:11569–11574 [PubMed] [Google Scholar]
- 14. Banks IR, et al. 2005. A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot. Cell 4:1902–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baker LG, Specht CA, Lodge JK. 2011. Cell wall chitosan is necessary for virulence in the opportunistic pathogen Cryptococcus neoformans. Eukaryot. Cell 10:1264–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kapteyn JC, et al. 1996. Retention of Saccharomyces cerevisiae cell wall proteins through a phosphodiester-linked beta-1,3-/beta-1,6-glucan heteropolymer. Glycobiology 6:337–345 [DOI] [PubMed] [Google Scholar]
- 17. Hamada K, Fukuchi S, Arisawa M, Baba M, Kitada K. 1998. Screening for glycosylphosphatidylinositol (GPI)-dependent cell wall proteins in Saccharomyces cerevisiae. Mol. Gen. Genet. 258:53–59 [DOI] [PubMed] [Google Scholar]
- 18. Caro LHP, et al. 1997. In silico identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 13:1477–1489 [DOI] [PubMed] [Google Scholar]
- 19. Frieman MB, Cormack BP. 2003. The omega-site sequence of glycosylphosphatidylinositol-anchored proteins in Saccharomyces cerevisiae can determine distribution between the membrane and the cell wall. Mol. Microbiol. 50:883–896 [DOI] [PubMed] [Google Scholar]
- 20. Kollár R, et al. 1997. Architecture of the yeast cell wall. Beta(1→6)-glucan interconnects mannoprotein, beta(1 →)3-glucan, and chitin. J. Biol. Chem. 272:17762–17775 [DOI] [PubMed] [Google Scholar]
- 21. Bernard M, et al. 2002. Characterization of a cell-wall acid phosphatase (PhoAp) in Aspergillus fumigatus. Microbiology 148:2819–2829 [DOI] [PubMed] [Google Scholar]
- 22. Fontaine T, et al. 2000. Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. J. Biol. Chem. 275:27594–27607 [DOI] [PubMed] [Google Scholar]
- 23. Ruiz-Herrera J, Ortiz-Castellanos L. 2010. Analysis of the phylogenetic relationships and evolution of the cell walls from yeasts and fungi. FEMS Yeast Res. 10:225–243 [DOI] [PubMed] [Google Scholar]
- 24. Xie X, Lipke PN. 2010. On the evolution of fungal and yeast cell walls. Yeast 27:479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. James PG, Cherniak R, Jones RG, Stortz CA, Reiss E. 1990. Cell-wall glucans of Cryptococcus neoformans Cap 67. Carbohydr. Res. 198:23–38 [DOI] [PubMed] [Google Scholar]
- 26. Fujii T, Shimoi H, Iimura Y. 1999. Structure of the glucan-binding sugar chain of Tip1p, a cell wall protein of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1427:133–144 [DOI] [PubMed] [Google Scholar]
- 27. Latgé JP. 2007. The cell wall: a carbohydrate armour for the fungal cell. Mol. Microbiol. 66:279–290 [DOI] [PubMed] [Google Scholar]
- 28. Yin QY, et al. 2005. Comprehensive proteomic analysis of Saccharomyces cerevisiae cell walls: identification of proteins covalently attached via glycosylphosphatidylinositol remnants or mild alkali-sensitive linkages. J. Biol. Chem. 280:20894–20901 [DOI] [PubMed] [Google Scholar]