Abstract
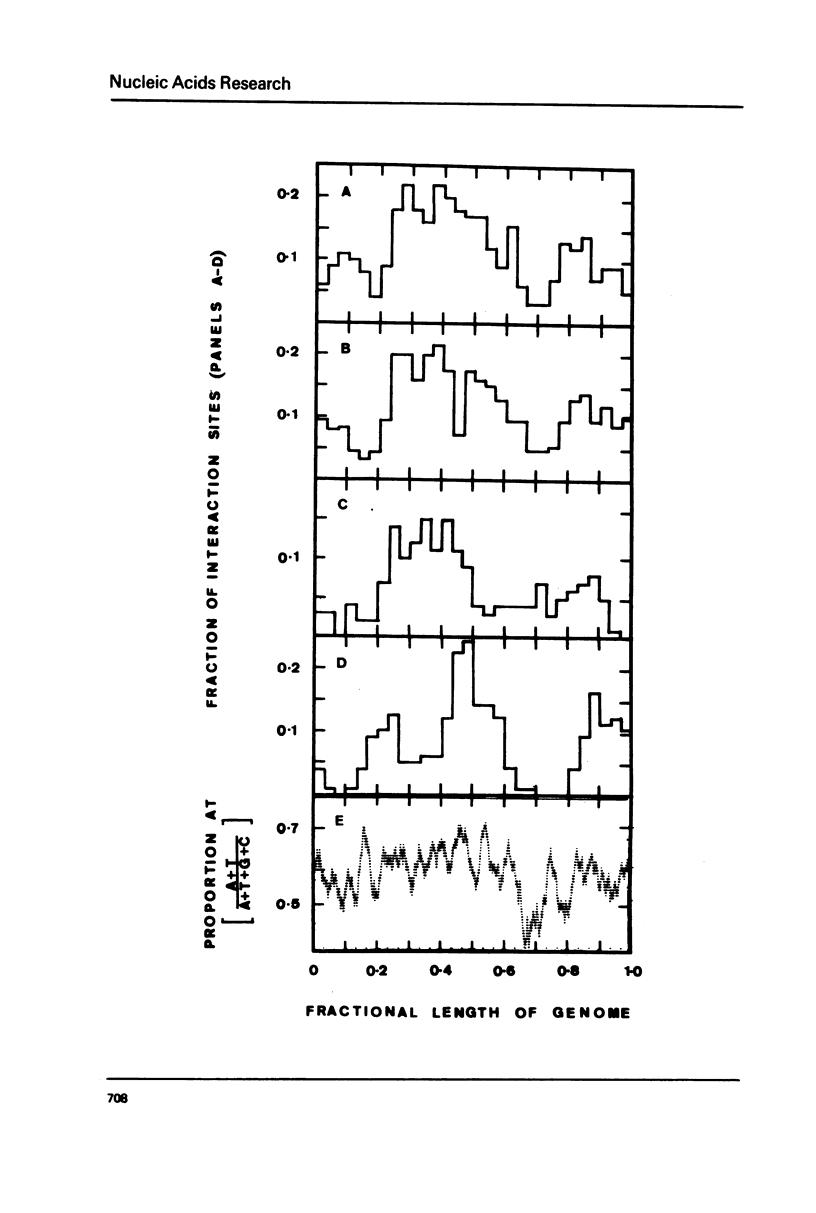
We have found that nucleosomes reconstituted from histone octamers and SV40 DNA Form I by progressively decreasing the salt concentration from 2 M NaCl are formed preferentially around 0.27, 0.37, 0.50 and 0.85 on SV40 DNA (relative to the EcoRI site). When SV40 DNA Form III is used, the nucleosomes form mainly at 0.28, 0.38, 0.61 and 0.83. These sites are very close to both the sites of RNA chain initiation by calf thymus RNA polymerase B on SV40 DNA Form I (0.25, 0.35, 0.42 and 0.88) and the regions of the supercoiled DNA which are readily denaturable by T4 gene 32 protein (0.25, 0.47 and 0.88), and correspond to AT-rich regions as deduced from the nucleotide sequence of SV40 DNA. The physiologically important region around 0.67 is an unfavourable site for all three types of proteins, and corresponds to a GC-rich region surrounding a 17 base pair AT cluster.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beard P., Morrow J. F., Berg P. Cleavage of circular, superhelical simian virus 40 DNA to a linear duplex by S1 nuclease. J Virol. 1973 Dec;12(6):1303–1313. doi: 10.1128/jvi.12.6.1303-1313.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao M. V., Gralla J., Martinson H. G. DNA sequence directs placement of histone cores on restriction fragments during nucleosome formation. Biochemistry. 1979 Mar 20;18(6):1068–1074. doi: 10.1021/bi00573a021. [DOI] [PubMed] [Google Scholar]
- Delius H., Mantell N. J., Alberts B. Characterization by electron microscopy of the complex formed between T4 bacteriophage gene 32-protein and DNA. J Mol Biol. 1972 Jun 28;67(3):341–350. doi: 10.1016/0022-2836(72)90454-8. [DOI] [PubMed] [Google Scholar]
- Dubochet J., Ducommun M., Zollinger M., Kellenberger E. A new preparation method for dark-field electron microscopy of biomacromolecules. J Ultrastruct Res. 1971 Apr;35(1):147–167. doi: 10.1016/s0022-5320(71)80148-x. [DOI] [PubMed] [Google Scholar]
- Fareed G. C., Davoli D. Molecular biology of papovaviruses. Annu Rev Biochem. 1977;46:471–522. doi: 10.1146/annurev.bi.46.070177.002351. [DOI] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Haegemann G., Rogiers R., Van de Voorde A., Van Heuverswyn H., Van Herreweghe J., Volckaert G., Ysebaert M. Complete nucleotide sequence of SV40 DNA. Nature. 1978 May 11;273(5658):113–120. doi: 10.1038/273113a0. [DOI] [PubMed] [Google Scholar]
- Hossenlopp P., Oudet P., Chambon P. Animal DNA-dependent RNA polymerases. Studies on the binding of mammalian RNA polymerases AI and B to Simian virus 40 DNA. Eur J Biochem. 1974 Jan 16;41(2):397–411. doi: 10.1111/j.1432-1033.1974.tb03281.x. [DOI] [PubMed] [Google Scholar]
- Laub O., Bratosin S., Horowitz M., Aloni Y. The initiation of transcription of SV40 DNA at late time after infection. Virology. 1979 Jan 30;92(2):310–323. doi: 10.1016/0042-6822(79)90136-3. [DOI] [PubMed] [Google Scholar]
- Morrow J. F., Berg P. Location of the T4 gene 32 protein binding site on simian virus 40 DNA. J Virol. 1973 Dec;12(6):1631–1632. doi: 10.1128/jvi.12.6.1631-1632.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy V. B., Thimmappaya B., Dhar R., Subramanian K. N., Zain B. S., Pan J., Ghosh P. K., Celma M. L., Weissman S. M. The genome of simian virus 40. Science. 1978 May 5;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Scott W. A., Wigmore D. J. Sites in simian virus 40 chromatin which are preferentially cleaved by endonucleases. Cell. 1978 Dec;15(4):1511–1518. doi: 10.1016/0092-8674(78)90073-9. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Streeck R. E., Zachau H. G. Nucleosome formation abolishes base-specific binding of histones. Nature. 1975 Dec 4;258(5534):447–450. doi: 10.1038/258447a0. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Streeck R. E., Zachau H. G. Reconstituted histone--DNA complexes. Philos Trans R Soc Lond B Biol Sci. 1978 May 11;283(997):259–268. doi: 10.1098/rstb.1978.0022. [DOI] [PubMed] [Google Scholar]
- Sümegi J., Breedveld D., Hossenlopp P., Chambon P. A rapid procedure for purification of EcoRI endonuclease. Biochem Biophys Res Commun. 1977 May 9;76(1):78–85. doi: 10.1016/0006-291x(77)91670-9. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Sundin O., Bohn M. A stretch of "late" SV40 viral DNA about 400 bp long which includes the origin of replication is specifically exposed in SV40 minichromosomes. Cell. 1979 Feb;16(2):453–466. doi: 10.1016/0092-8674(79)90021-7. [DOI] [PubMed] [Google Scholar]
- Waldeck W., Föhring B., Chowdhury K., Gruss P., Sauer G. Origin of DNA replication in papovavirus chromatin is recognized by endogenous endonuclease. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5964–5968. doi: 10.1073/pnas.75.12.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B., Thevenin G., Oudet P., Chambon P. Transcription of in vitro assembled chromatin by Escherichia coli RNA polymerase. J Mol Biol. 1979 Mar 5;128(3):411–440. doi: 10.1016/0022-2836(79)90095-0. [DOI] [PubMed] [Google Scholar]
- Wilson G. A., Young F. E. Isolation of a sequence-specific endonuclease (BamI) from Bacillus amyloliquefaciens H. J Mol Biol. 1975 Sep 5;97(1):123–125. doi: 10.1016/s0022-2836(75)80028-3. [DOI] [PubMed] [Google Scholar]



