Abstract
Non-LTR retrotransposons comprise significant portion of the plants genome. Their complete characterization is thus necessary if the sequenced genome is to be annotated correctly. The long and short interspersed nucleotide repetitive elements (LINE and SINE) may be responsible for alteration in the expression mechanism of neighboring genes, the complete identification of these elements in the rice genome is essential in order studying their putative functional interactions with the plant genes and its role in genome composition. The main emphasis of this work is to assemble a comprehensive dataset of nonLTR (LINEs and SINEs) and the map of completely inserted LINEs and SINE type of retroelement by both intact ends (3' and 5' ends). The assembled information and work may help for further research in this direction.
Keywords: LINEs, SINEs, Retroelement, Oryza sativa
Background:
Transposable elements are found in all eukaryotic genomes. A particular class of these elements, the Non-LTRretrotransposons, is also the component of large plant genomes as LTR retrotransposons [1]. But the LINE and SINE retroelements also contributed some part of plants genome composition. Our observation suggested the rice genome has seven LINE and twelve SINE types of retroelements most frequently dispersed throughout in all twelve chromosomes as characterized in supplementary material File 2 (S2). These elements transpose via RNA intermediate through a copy/paste mechanism, this results in the amplification or genomic expansion [2]. Rice genome (Oryza sativa) is about 430 MB in size having twelve chromosomes and only two percent of total retrotransposons in all chromosomes are LINE and SINE type of elements but they successfully and completely dispersed as characterized in Table 1 and 2 (see supplementary material). Previous studies of eukaryotic genomes suggested that the analysis of these LINE and SINE type of retroelements is very significant for revealing the secrets of genomic organization and evolution of any genome therefore we used the computational approach for the characterization and distribution analysis of these non-LTR elements in the vast rice genome. Transposable elements are separated into two major groups (class I and class II) depending on their mode of transposition [3, 4]. The analysis suggested that rice also has notable amount of these nonLTR retrotransposons.
Retrotransposons are subclass of transposable element which is further classified into LTR and nonLTR. These retrotransposons are particularly abundant in plants where they are principal component of nuclear DNA for example maize, wheat etc. The main objective of our work is to map the LINE and SINE type of retroelements which come under the category of non-LTR retrotransposons.
Methodology:
The genome of rice (Oryza sativa) was collected from FTP server of NCBI and the retroelement sequences were taken from REPBASE database [5]. Standalone Blast was used for searching the copies of all LINE and SINE type of repetitive element in rice genome [6]. The BioPERL program was developed and used to extract the information required for the map generation and extraction of upstream sequences to the insertion site of these repetitive elements [7]. The total output file of developed BioPERL program is provided in supplementary material File 2 (available with authors). M.S. Excel was used for managing and graphical representation of data. WEB LOGO was used to generate the upstream sequence logo for further analysis [8].
Results and Discussion:
The survey of all transposable elements revealed that 60% are retroelements, 22% transposons, 18% are MITES, and only 2% of retroelements observed as LINE and SINE type of elements in the rice genome. The percentage of these LINE and SINE retroelements are much less in comparison to other eukaryotic genome such as human genome where these LINE and SINE elements are highly dispersed and constituted the major part of total genome.
About 15047 copies of total LINE and SINE type retroelements are found uniformly distributed in the rice genome, out of 15047, 13487 copies are of SINE type of elements (Table 1, see supplementary material) and 1560 copies are LINE type of elements (Table 2, see supplementary material). There are twelve SINE type and seven LINE type of retroelements found in the genome (Oryza sativa), 13487 copies scattered throughout the genome of all SINE type elements and the LINE type of elements were observed in a total 1560 copies, in the genome. The F524 type of SINE element showed very significant result. It was found that 119 copies successfully inserted both intact ends which further revealed that the F524 is the most successful dispersed SINE type of retroelement in comparison to other SINE elements as indicated in Table 1 (see supplementary material).
The highest populated SINE type of element was observed as SINE3_OS which have more than seven thousand copies distributed throughout in the genome but it has only ten both intact ends copies as shown in Table 1 (see supplementary material). The following order represents the population density of each and every SINE type of retroelements scattered in the rice genome i.e., SINE03_OS> SINE9_OS> f524> pSINE1_OS1> p-SINE1_OS> SINE1r5_OS> SINE16_OS> CaSINE> SINE6_OS> SINE1OS> ormosia> SINE8_OS, where SINE03_OS element populated maximum and SINE8_OS has minimum population. In case of LINE elements 1560 copies are dispersed in the genome, out of these 2.5% dispersed elements have both intact ends, 79.55% showed both truncated ends, 8.14% have 3' truncated ends and 9.8% of LINE type elements showed 5' truncated ends, which further revealed that the most of the dispersed element are not successfully inserted by both intact ends. Only very few elements such as LINE5A, LINE-5 showed complete insertion in rice genome having both intact ends than other LINEs but LINE-1 and LINE-3 also have maximum copies distributed in the rice genome beside this it has very less number of complete insertion by both ends (intact ends) as shown in Table 1 (see supplementary material).
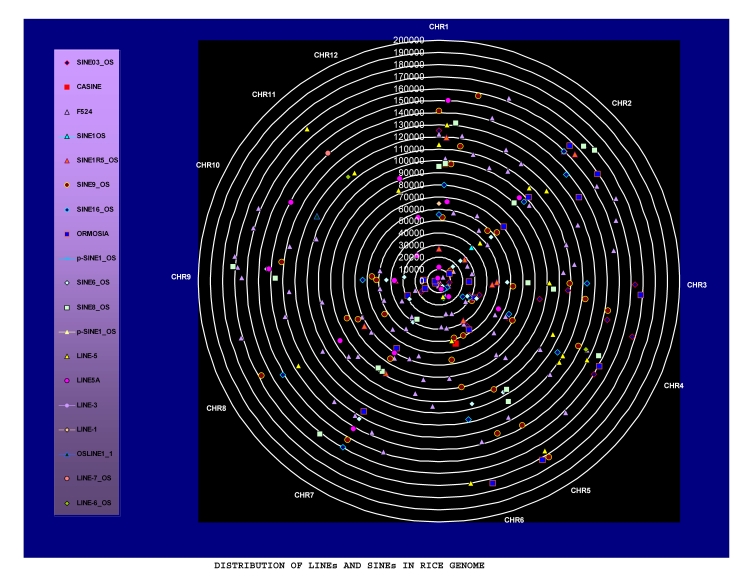
To enhance our understanding about the genome evolution in rice, it is necessary to develop a well organized map of repetitive elements, therefore we have developed the graphical map of whole rice genome further classified by the twelve chromosomes populated with nineteen different LINE and SINE type of retroelements as shown in Figure 2. The map represents only the successful both intact ends copies distributed in the rice genome of all LINE and SINE retroelements. The distribution datasheet of total LINE and SINE retroelements (truncated and intact ends) are provided in supplementary File 2 (available with authors). The datasheet helps the researchers to map its own interest of intact or truncated elements for further studies.
Figure 2.
Map of rice genome populated with all successful SINEs and LINEs
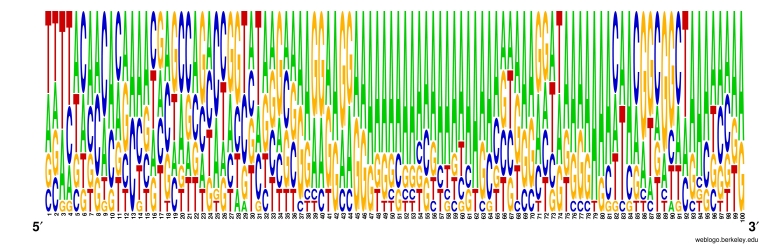
During the analysis, we observed that the copies of non-LTR elements of upstream sequence in the insertion site showed specific and unique pattern in their nucleotide sequence of 40th to 100th base pair position. The AG (Adenine, Guanine) rich region was found in all LINE type of retroelements which further represent the selection site of high AG density in 40th to 100th base pair of upstream region.
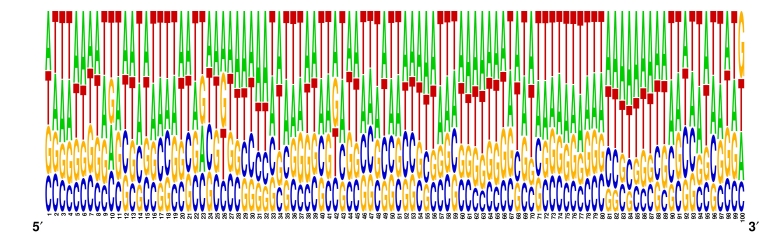
In case of SINEs the similar kind of pattern is found but A, T (Adenine, Thymine) concentration rich region was clearly visible instead of A, G. In other words we can say that A, G rich region can be a signal to select the insertion site by LINE types of retroelement for their insertion and A, T pattern in upstream region can be a insertion site selection signal used by SINE type of retroelements for their insertion in the host genome. Figure 2 Figure 3 and Table 3 (see supplementary material) also supports the above interpretation as its values indicated i.e., total A, G (Adenine, Guanine) concentration is more than 68 percent which is dominant over T, C (Thymine, Cytosine) in LINEs and in case of SINE 63% A, T rich concentration was found in upstream sequences of all SINE type of retroelements.
Figure 3.
Frequency distribution LOGO of upstream sequences to the insertion site of LINEs in rice genome
Conclusion:
The main emphasis of this work is to map the successfully inserted LINEs and SINEs retroelement as shown in Figure 2. Another objective of this work is to check the status of these elements in the rice genome; whether they are completely inserted (Both intact ends) or truncated (break) end. As expected these Non-LTR type of retroelements (LINEs, SINEs) were inserted successfully in hundreds of copies but more than 97% of LINEs and SINEs are truncated in the form of both truncated ends and one side truncated ends (either 3'or 5'end). The observed percentage of LINEs and SINEs having both intact ends is very less in comparison to total truncated ends elements dispersed in the genome. As shown in map (Figure 2), the all LINEs and SINEs laid up to 200000 base pair position in all twelve chromosomes while most of the chromosomes of rice have more than 400000 base pair long which means the all LINEs and SINEs is present inside the half range of all chromosomes and still amplifying its status towards the boundaries of chromosomes in rice genome. It was reported during the analysis of upstream regions that the insertion site of completely inserted (both intact ends) LINE and SINE retroelements are heavily populated with A,T and A,G concentration. The A,T rich upstream region is responsible for the insertion of SINEs elements while the A,G rich region is required for the insertion of LINEs elements as indicated in the Figure 3, Figure 4 and Table 3 (see supplementary material).
Figure 4.
Frequency distribution LOGO of upstream sequences to the insertion site of SINEs in rice genome
Supplementary material
Figure 1.
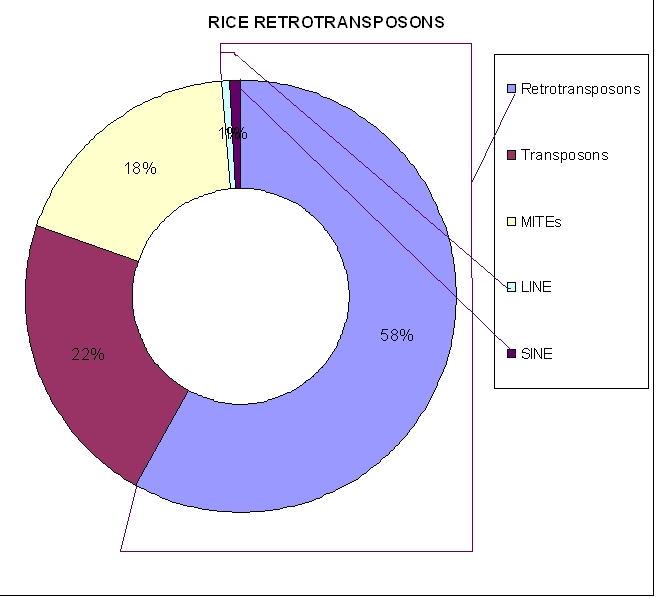
Percentage distribution of transposable element in rice genome
Footnotes
Citation:Faheem Khan et al, Bioinformation 7(6): 276-279 (2011)
References
- 1.A Kumar, JL Bennetzen. Annu Rev Genet. 1999;33:479. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- 2.B Piegu, et al. Genome Res. 2006;16:1262. doi: 10.1101/gr.5290206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DJ Finnegan, et al. Trends Genet. 1989;5:103. [Google Scholar]
- 4.AJ Flavell, et al. Curr Opin Genet Dev. 1994;4:838. [Google Scholar]
- 5.J Jurka, et al. Cytogenet Genome Res. 2005;110:462. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 6.SF Altschul, et al. J Mol Biol. 1990;215:403. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 7.JE Stajich, et al. Methods Mol Biol. 2007;406:535. doi: 10.1007/978-1-59745-535-0_26. [DOI] [PubMed] [Google Scholar]
- 8.GE Crooks, et al. Genome Res. 2004;14:1188. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.