Abstract
We present the development of a web server, a protein short motif search tool that allows users to simultaneously search for a protein sequence motif and its secondary structure assignments. The web server is able to query very short motifs searches against PDB structural data from the RCSB Protein Databank, with the users defining the type of secondary structures of the amino acids in the sequence motif. The output utilises 3D visualisation ability that highlights the position of the motif in the structure and on the corresponding sequence. Researchers can easily observe the locations and conformation of multiple motifs among the results. Protein short motif search also has an application programming interface (API) for interfacing with other bioinformatics tools.
Availability
The database is available for free at http://birg3.fbb.utm.my/proteinsms
Keywords: Protein short motif search, protein secondary structure, visualization, application programming interface (API)
Background:
Motifs are frequently observed in biological sequences, such as transcription factor binding sites in DNA sequences and catalytic sites in protein sequences. The purpose of our tool is to allow users to simultaneously search for a sequence motif and its secondary structure assignments. Because a protein sequence motif is identified on the basis of sequence similarity and without the knowledge of the function that is conferred by the structural conformation represented by its assignment, it is important to determine where the conserved amino acids lie in the three-dimensional (3D) structure and to what extent these conserved amino acids represent known functional regions. There are many sequence motif search engines available online, but they have varied limitations. Most search functions in motif databases are limited to previously identified motifs such as InterPro [1], BLOCKS [2] and PRINTS [3]. The majority of the motif search tools and databases do not have 3D visualisation and present their results as sequences. The position of the motif in the spatial arrangement is either visualised using third party tools, such as Jmol or using a mash-up that combines sequence searching and 3D visualisation, such as ScanProsite for PROSITE [4] and Motif3D for PRINTS [5]. However, ScanProsite only displays a static GIF image of the motif, whereas Motif3D does not have the ability to query the ultrashort linear motifs typically found in SLiM [6] and Mini Motif Miner [7]. Recent developments of 3D motif visualisation tools allow interactive 3D visualisation within the conformational structure; these tool include seeMotif [8], 3MATRIX and 3MOTIF [9], and PDBeMotif [10]. However, PDBeMotif only allow users to add secondary structure patterns and not to specifically assign secondary structures to the amino acids in their motif queries.
Database development:
Data downloaded from Protein Data Bank (PDB) was stored on a single server that serves as the web server and perform the search function. Powered by Ubuntu 9.04 Server Edition, the server runs on top of Lighttpd 1.4.19 with FastCGI in addition to a backup web server of FastCGI-enabled Apache 2.2.4. Development uses Perl (version 5.10) with MySQL for database management, JavaScript, Yahoo User Interface (YUI), AJAX, JSON and Perl DBIx::Class. The website was built modularly, following a Model-View-Controller (MVC) framework as a FastCGI application. The server also acts as a PDB file server catering to Jmol requests and PDB files are streamed to the Jmol applet for each successful query through MySQL table.
Software input:
A user will enter a sequence motif and its corresponding secondary structure for the amino acids into the submission box. The queries will then be searched against PDB structure files, which are continuously updated. There are several variables that can narrow the search possibilities. An example search for the sequence motif “PEEL” that exists in beta sheets requires the user to enter “PEEL” in the sequence query and EEEE (or H for helices) in the secondary structure assignment to search for all occurrences of the sequence and its corresponding secondary assignment. Users can use the wildcard “O” for any type of secondary structure or the wildcard “X” for unassigned secondary structures, which are usually found in undefined regions of the protein.
Software output:
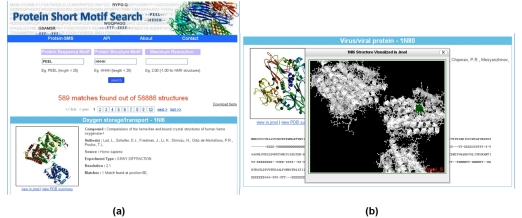
The results page (Figure 1(a)) shows the query on the top half of the page and highlights the number of matches against the PDB structural data from the RCSB. A brief description of the protein is provided and the users can also download all of the matching structures in FASTA format. The results page also shows a sequence alignment of the match and its corresponding secondary structure. We also added visualisation capability using Jmol in which the structure is loaded in a new window, and the position match highlighted in the structure (Figure 1(b)). Users can explore and export the structure using Jmol functionalities. An application programming interface (API) for Protein Short Motif Search was created to allow other developers to parse their data.
Figure 1.
(a) Protein SMS Search Results and (b) Jmol Visualisation of the Results.
Caveat:
The search is conducted against PDB files downloaded weekly from the PDB.
Conclusion:
Protein short motif search unique functionality is the ability to search short motifs where the secondary structure of each amino acid in the motif can be specifically assigned. It is aimed to complement other search tools with the API allowing users to automate parsing high throughput data.
Future Development:
We intend to improve by adding functionalities and annotations such as solvent accessibility value cluster, the results according to SCOP or CATH classification and link to other protein database.
Acknowledgments
The authors acknowledge the Malaysian Ministry of Science, Technology and Innovation (MOSTI) and its E-Science Fund grant no. 02-01-06-SF0302 for supporting of the development of Protein Short Motif Search.
Footnotes
Citation:Venkataraman et al, Bioinformation 7(6): 304-306 (2011)
References
- 1.S Hunter, et al. Nucleic Acids Res. 2009;37:D211. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.JG Henikoff, et al. Nucleic Acids Res. 2000;28:228. doi: 10.1093/nar/28.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.TK Attwood, et al. Nucleic Acids Res. 2003;31:400. doi: 10.1093/nar/gkg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.E De Castro, et al. Nucleic Acids Res. 2006;34:W632. [Google Scholar]
- 5.A Gaulton, TK Attwood. Nucleic Acids Res. 2003;31:3333. doi: 10.1093/nar/gkg534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.NE Davey, et al. Nucleic Acids Res. 2006;34:3546. doi: 10.1093/nar/gkl486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.S Rajasekaran, et al. Nucleic Acids Res. 2009;37:D185. doi: 10.1093/nar/gkn865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DT Chang, et al. Nucleic Acids Res. 2009;37:W552. [Google Scholar]
- 9.SP Bennett, et al. Nucleic Acids Res. 2003;31:3328. doi: 10.1093/nar/gkg564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.A Golovin, K Henrick. BMC Bioinformatics. 2008;9:312. doi: 10.1186/1471-2105-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]