Abstract
IGIPT is a web-based integrated platform for the identification of genomic islands (GIs). It incorporates thirteen parametric measures based on anomalous nucleotide composition on a single platform, thus improving the predictive power of a horizontally acquired region, since it is known that no single measure can absolutely predict a horizontally transferred region. The tool filters putative GIs based on standard deviation from genomic average and also provides raw output in MS excel format for further analysis. To facilitate the identification of various structural features, viz., tRNA integration sites, repeats, etc. in the vicinity of GIs, the tool provides option to extract the predicted regions and its flanking regions.
Availability
The database is available for free at http://bioinf.iiit.ac.in/IGIPT/
Keywords: genomic islands, horizontal gene transfer
Background:
A horizontally transferred event is defined as movement of genetic material between phylogenetically unrelated organisms by mechanisms other than vertical descent. These regions from diverse organisms, called Genomic Islands (GIs), are typically 10-200Kb in size (containing clusters of genes). Any biological advantage provided to the recipient organism by transferred DNA creates selective pressure for its retention in the host genome and several pathways of horizontal transfer have been established influencing traits such as antibiotic resistance, symbiosis and fitness, virulence and adaptation [1]. For example, horizontal gene transfer has been demonstrated in many pathogenic strains of bacteria and shown to be responsible for its virulence. The identification of genomic islands also forms the first step in the annotation of newly sequenced genomes. Various bioinformatics approaches have been proposed in their identification [2]. In the genomic era, with availability of large number of bacterial genomes, the preferred methods are based on nucleotide base compositions and comparative genomics. In IGIPT, we have implemented thirteen measures that capture anomaly in nucleotide composition, providing both genome-based and gene-based search on a single platform.
Methodology:
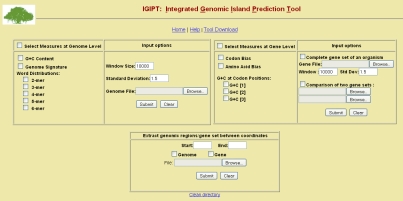
In any genome, vertically transmitted genes experience a particular set of directional mutation pressures mediated by the specific features of the replication machinery of the cell, such as balance of dNTP pools, mutational biases of the DNA polymerases, efficiency of mismatch repair systems and so on [3]. As a result each genome exhibits its own unique signatures, viz., distinct variations in the GC content, dinucleotide relative abundance, variations in usage of k-mer words, codons and amino acids. These measures, called parametric methods, are the most widely used approaches as the putative transferred genes can be identified without relying on comparisons with other organisms, thus providing an independent means of assessing the impact of gene transfer across lineages. The parametric measures implemented in IGIPT are broadly classified as genome-based or gene-based, depending on the analysis (shown as left- and right panel in Figure. 1). These measures are computed in a sliding window and regions deviant from the genomic average by user defined standard deviation (default 1.5σ) are identified as probable GIs.
Figure 1.
Snapshot of the web-server IGIPT.
Measures at Genome Level:
The major advantage of these measures is that they do not require pre-existing annotation or comparison of homologous sequences, and can, therefore, be applied directly to newly sequenced genomes. The input to these measures is the complete genome/contig in Fasta format.
GC content:
It computes the frequency of G and C nucleotides, called the GC content [4].
Genomic signature:
The set of dinucleotide relative abundance values constitutes a “genomic signature” of an organism. Please see supplementary material.
k-mer Distributions:
It has been proposed by Karlin that most horizontally acquired genomic regions have distinct word (k-mer) compositions [5]. Please see supplementary material.
Measures at the Gene Level:
This module identifies horizontally acquired genes in a fully annotated gene set of the organism (in multi-fasta format). In the absence of this information, IGIPT provides comparison of two gene sets, one a representative gene set of the organism and the other whose horizontal acquisition needs to be confirmed (e.g., genes in predicted GIs from genome-based measures). This feature also allows comparison of predicted gene(s) with highly expressed genes of the organism, e.g., ribosomal genes, chaperon genes, etc. to reduce false predictions.
Codon usage Bias:
The unequal usage of synonymous codons has been extensively studied and virtually every codon has been shown to be preferentially used in some organisms and rarely used in others. Please see supplementary material.
Amino Acid Bias:
This bias refers to the deviation in the frequency of usage of individual amino acids over the average usage of all 20 amino acids.Please see supplementary material.
GC Content at Codon Positions:
This involves comparing the frequency of G or C at the three codon positions, GC1, GC2 and GC3, for a given gene set with the core gene set (or genomic average or highly expressed genes) of the organism [8].
IGIPT provides an option to download the predicted horizontally transferred regions/genes and its flanking regions (lower panel in Figure. 1) to facilitate analysis of conserved structural features in the vicinity of probable GIs, e.g., genes coding for integrases or transposases required for chromosomal integration and excision are flanked by direct repeats and are inserted in the vicinity of tRNA and tmRNA genes [9]. This feature is also useful for further analysis such as comparative genomics or phylogenetic analysis of putative GIs. The output of IGIPT is windows/genes filtered based on standard deviation and also provides option to download unfiltered output in MS excel format.
Conclusion:
Evolution of species by horizontal gene transfer is very common not only in prokaryotes but also in eukaryotes. It gives unique functionality to the organism to adapt to different environmental conditions and their identification is particularly useful in pathogens for identifying virulent genes. Since no single measure truly identifies a horizontally acquired region, by integrating numerous parametric measures on a single platform, IGIPT allows the users to analyze the predicted horizontally transferred regions/genes by thirteen different measures simultaneously, thus greatly increasing the confidence of prediction. A drawback of these parametric methods is that regions acquired from donors with similar compositional bias as the host genome will not be identified.
Supplementary material
Footnotes
Citation:Jain et al, Bioinformation 7(6): 307-310 (2011)
References
- 1.EV Koonin, et al. Ann Rev Microbiol. 2001;55:709. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MG Langille, et al. Nat Rev Microbiol. 2010;8:373. doi: 10.1038/nrmicro2350. [DOI] [PubMed] [Google Scholar]
- 3.J Lawrence, et al. Curr Opin Genet Dev. 1999;9:642. doi: 10.1016/s0959-437x(99)00025-8. [DOI] [PubMed] [Google Scholar]
- 4.F Gao, CT Zhang. Nucleic Acids Res. 2006;34:W686. doi: 10.1093/nar/gkl040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.S Karlin, J Mrazek. Proc Natl Acad Sci USA. 1997;94:m10227. [Google Scholar]
- 6.GM Pavlovic-Lazetic, et al. Comput Methods Programs Biomed. 2009;93:241. doi: 10.1016/j.cmpb.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.S Karlin, et al. Trends Microbiol. 2001;9:335. doi: 10.1016/s0966-842x(01)02079-0. [DOI] [PubMed] [Google Scholar]
- 8.SH Yoon, et al. BMC Bioinformatics. 2005;6:184. doi: 10.1186/1471-2105-6-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.U Dobrindt, et al. Nat Rev Microbiol. 2004;2:414. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.