Abstract
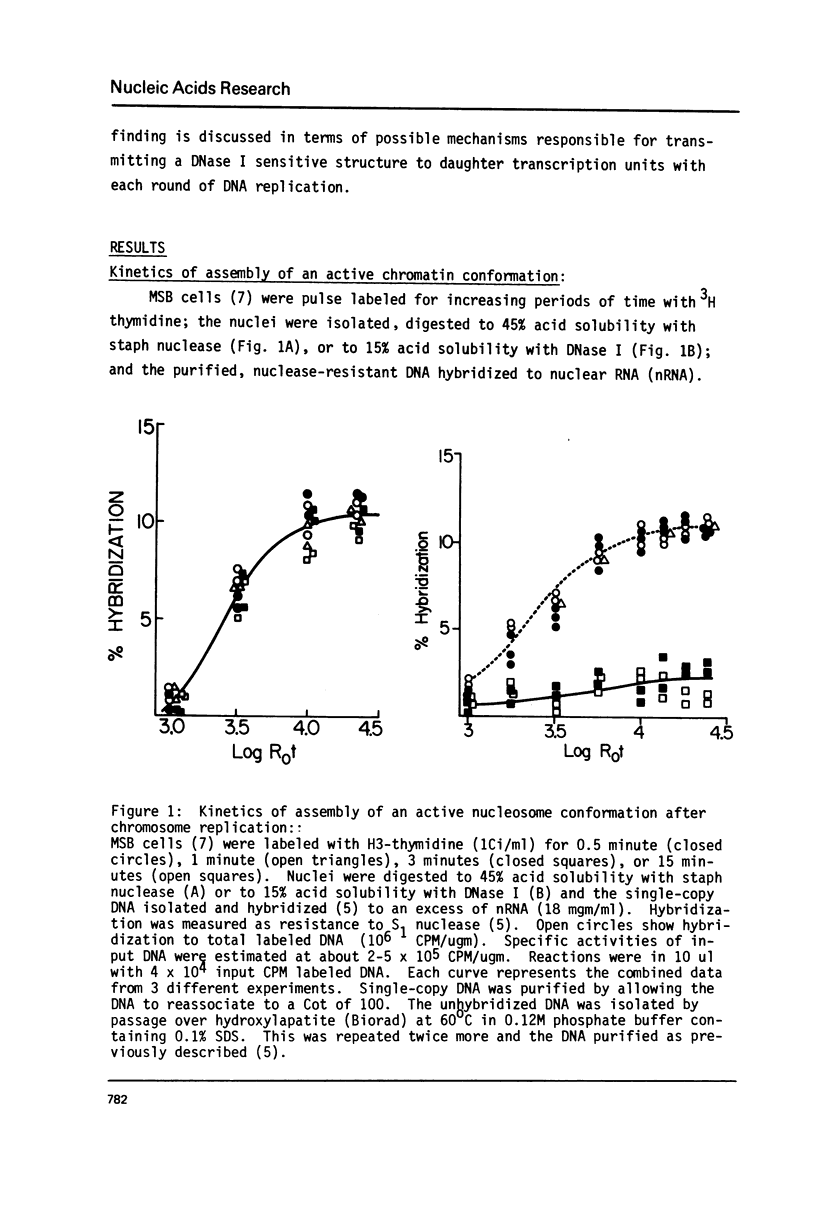
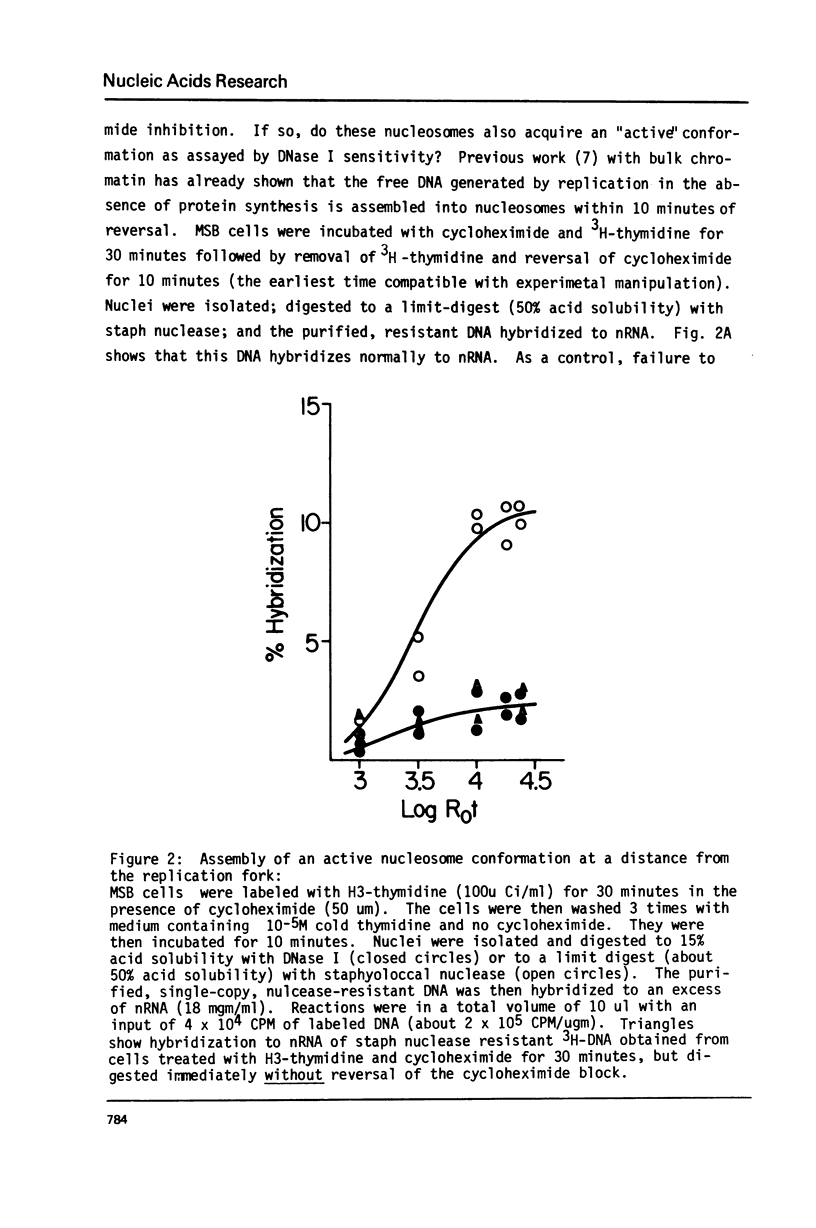
MSB cells were pulse labeled with 3H-thymidine and the isolated nuclei digested with either staphylococcal nuclease (to about 40% acid solubility) or DNase I (to 15% acid solubility). The purified, nuclease resistant single-copy DNA was then hybridized to nuclear RNA (nRNA). The results of these experiments show that actively transcribed genes are assembled into nucleosome-like structures within 5-10 nucleosomes of the replication fork and that they also acquire a conformation characteristic of actively transcribed nucleosomes (ie, a DNase I sensitive structure) within 20 nucleosomes of the fork. Assuming DNA sequence specific interactions are required for establishing a DNase I sensitive conformation on active genes during each round of replication, our results indicate that a specific recognition event can occur very rapidly and very specifically in eukaryotic cells. The results are discussed in terms of the possible mechanisms responsible for propagating active, chromosomal conformations from mother cells to daughter cells.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chao M. V., Gralla J., Martinson H. G. DNA sequence directs placement of histone cores on restriction fragments during nucleosome formation. Biochemistry. 1979 Mar 20;18(6):1068–1074. doi: 10.1021/bi00573a021. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin. Nature. 1978 Jan 12;271(5641):115–122. doi: 10.1038/271115a0. [DOI] [PubMed] [Google Scholar]
- Flint S. J., Weintraub H. M. An altered subunit configuration associated with the actively transcribed DNA of integrated adenovirus genes. Cell. 1977 Nov;12(3):783–794. doi: 10.1016/0092-8674(77)90277-x. [DOI] [PubMed] [Google Scholar]
- Foe V. E., Wilkinson L. E., Laird C. D. Comparative organization of active transcription units in Oncopeltus fasciatus. Cell. 1976 Sep;9(1):131–146. doi: 10.1016/0092-8674(76)90059-3. [DOI] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel A., Zolan M., Axel R. Genes transcribed at diverse rates have a similar conformation in chromatin. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4867–4871. doi: 10.1073/pnas.74.11.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzer H., Rubinstein N., Fellini S., Yeoh G., Chi J., Birnbaum J., Okayama M. Lineages, quantal cell cycles, and the generation of cell diversity. Q Rev Biophys. 1975 Nov;8(4):523–557. doi: 10.1017/s0033583500001980. [DOI] [PubMed] [Google Scholar]
- Lacy E., Axel R. Analysis of DNA of isolated chromatin subunits. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3978–3982. doi: 10.1073/pnas.72.10.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffak I. M., Grainger R., Weintraub H. Conservative assembly and segregation of nucleosomal histones. Cell. 1977 Nov;12(3):837–845. doi: 10.1016/0092-8674(77)90282-3. [DOI] [PubMed] [Google Scholar]
- Lin S., Riggs A. D. The general affinity of lac repressor for E. coli DNA: implications for gene regulation in procaryotes and eucaryotes. Cell. 1975 Feb;4(2):107–111. doi: 10.1016/0092-8674(75)90116-6. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Miller O. L., Jr Electron microscopic analysis of chromatin replication in the cellular blastoderm Drosophila melanogaster embryo. Cell. 1977 Nov;12(3):795–804. doi: 10.1016/0092-8674(77)90278-1. [DOI] [PubMed] [Google Scholar]
- Riley D., Weintraub H. Conservative segregation of parental histones during replication in the presence of cycloheximide. Proc Natl Acad Sci U S A. 1979 Jan;76(1):328–332. doi: 10.1073/pnas.76.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott W. A., Wigmore D. J. Sites in simian virus 40 chromatin which are preferentially cleaved by endonucleases. Cell. 1978 Dec;15(4):1511–1518. doi: 10.1016/0092-8674(78)90073-9. [DOI] [PubMed] [Google Scholar]
- Seale R. L. Studies on the mode of segregation of histone nu bodies during replication in HeLa cells. Cell. 1976 Nov;9(3):423–429. doi: 10.1016/0092-8674(76)90087-8. [DOI] [PubMed] [Google Scholar]
- Tjian R. The binding site on SV40 DNA for a T antigen-related protein. Cell. 1978 Jan;13(1):165–179. doi: 10.1016/0092-8674(78)90147-2. [DOI] [PubMed] [Google Scholar]
- Tsanev R., Sendov B. Possible molecular mechanism for cell differentiation in multicellular organisms. J Theor Biol. 1971 Feb;30(2):337–393. doi: 10.1016/0022-5193(71)90059-2. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Sundin O., Bohn M. A stretch of "late" SV40 viral DNA about 400 bp long which includes the origin of replication is specifically exposed in SV40 minichromosomes. Cell. 1979 Feb;16(2):453–466. doi: 10.1016/0092-8674(79)90021-7. [DOI] [PubMed] [Google Scholar]
- Weintraub H. The assembly of newly replicated DNA into chromatin. Cold Spring Harb Symp Quant Biol. 1974;38:247–256. doi: 10.1101/sqb.1974.038.01.028. [DOI] [PubMed] [Google Scholar]
- Worcel A., Han S., Wong M. L. Assembly of newly replicated chromatin. Cell. 1978 Nov;15(3):969–977. doi: 10.1016/0092-8674(78)90280-5. [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Alberts B. The interaction of estradiol-receptor protein with the genome: an argument for the existence of undetected specific sites. Cell. 1975 Apr;4(4):301–310. doi: 10.1016/0092-8674(75)90150-6. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Revzin A., Gross C. A., Wang A. C. Non-specific DNA binding of genome regulating proteins as a biological control mechanism: I. The lac operon: equilibrium aspects. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4808–4812. doi: 10.1073/pnas.71.12.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]



