Abstract
The key haematopoietic regulator Myb is essential for coordinating proliferation and differentiation. ChIP-Sequencing and Chromosome Conformation Capture (3C)-Sequencing were used to characterize the structural and protein-binding dynamics of the Myb locus during erythroid differentiation. In proliferating cells expressing Myb, enhancers within the Myb-Hbs1l intergenic region were shown to form an active chromatin hub (ACH) containing the Myb promoter and first intron. This first intron was found to harbour the transition site from transcription initiation to elongation, which takes place around a conserved CTCF site. Upon erythroid differentiation, Myb expression is downregulated and the ACH destabilized. We propose a model for Myb activation by distal enhancers dynamically bound by KLF1 and the GATA1/TAL1/LDB1 complex, which primarily function as a transcription elongation element through chromatin looping.
Keywords: ChIP-sequencing (ChIP-Seq), chromosome conformation capture-sequencing (3C-Seq), erythroid development, long-range interactions, Myb
Introduction
The differentiation of stem and progenitor cells into mature differentiated cells requires a tight control of progenitor cell expansion, proliferation arrest and terminal differentiation. The Myb proto-oncogene encoding the c-Myb transcription factor (TF) is expressed in stem and progenitor cells of all haematopoietic lineages and plays a central role in the control of their proliferation (Mucenski et al, 1991; Sandberg et al, 2005; Vegiopoulos et al, 2006; Ramsay and Gonda, 2008; Lieu and Reddy, 2009). Lack of Myb is lethal (E15) due to the complete absence of definitive erythroid cells (Mucenski et al, 1991). Conditional knockout models revealed additional essential non-erythroid roles of Myb, mainly in the lymphoid system (Bender et al, 2004; Thomas et al, 2005), and the self-renewal and multi-lineage differentiation potential of adult haematopoietic stem cells (Lieu and Reddy, 2009). Myb is highly expressed in immature proliferating haematopoietic cells and is strongly downregulated in terminally differentiating cells (Gonda and Metcalf, 1984; Emambokus et al, 2003), suggesting that Myb is linked to the transition between proliferation and differentiation. Aberrant Myb expression in leukaemic cells is consistent with this idea (Ramsay and Gonda, 2008), correlating with increased proliferation and a loss of differentiation. Despite its importance, the control of Myb expression during haematopoiesis is poorly understood. Early work suggested a regulatory role for sequences in the first intron, primarily in blocking transcription elongation (Bender et al, 1987; Reddy and Reddy, 1989; Hugo et al, 2006). Recently, microRNAs were shown to be involved in regulating c-Myb protein levels (Xiao et al, 2007; Lu et al, 2008). However, the transcriptional regulatory elements and associated trans-acting factors controlling Myb expression during development remain mostly uncharacterized.
The mouse Myb gene on chromosome 10 is flanked by the Ahi1 and Hbs1l genes, which have no known function during haematopoiesis. Several studies pointed out a potential role for the 135 kb Myb-Hbs1l intergenic region in the regulation of Myb: (i) transgene integration within the intergenic region led to severe downregulation of Myb expression (Mukai et al, 2006); (ii) ChIP-on-chip data showed an open chromatin structure (i.e., H3Ac and H4Ac) of the region in human erythroid cells expressing MYB (Wahlberg et al, 2009); and (iii) several studies showed that SNPs in the human MYB-HBS1L intergenic region (possibly affecting MYB expression) were strongly associated with variation in several clinically relevant erythrocyte traits (Thein et al, 2007; Lettre et al, 2008; Ganesh et al, 2009). For example, specific SNPs associate with elevated fetal haemoglobin (HbF), which ameliorates β-haemoglobinopathy severity and has therapeutic potential. Thus, important regulatory elements appear to reside in the Myb-Hbs1l intergenic region, but they have not been localized precisely or characterized in any way.
Erythroid development is controlled by an array of TFs, including GATA1, its associated partners LDB1, TAL1, KLF1 and c-Myb (Cantor and Orkin, 2001). A complex of the haematopoietic TFs GATA1/TAL1/LDB1 together with the ETO2/MTGR1 cofactors (the ‘LDB1 complex’) binds regulatory regions of developmentally regulated genes (Fujiwara et al, 2009; Yu et al, 2009; Kassouf et al, 2010; Soler et al, 2010; Tallack et al, 2010) and controls their activation upon terminal erythroid differentiation (Soler et al, 2010). The LDB1 complex preferentially binds at large distances from promoters (up to 300 kb) in intergenic regions, providing long-range candidate regulatory elements. An example is the long-range control of the β-globin genes by cis-regulatory elements spread over 100 kb, forming the locus control region (LCR). When β-globin is expressed, the LCR folds into a three-dimensional (3D) active chromatin hub (ACH) (Tolhuis et al, 2002; Palstra et al, 2003), where distal enhancers reside in close proximity to the expressed genes. Structural proteins such as CTCF and Cohesin are known to participate in such 3D interactions (Ong and Corces, 2011). TFs also have a role in long-range gene regulation, for example, LDB1, GATA1, FOG1 and KLF1 are required to maintain such interactions within the β-globin locus and other loci (Drissen et al, 2004; Vakoc et al, 2005; Song et al, 2007; Jing et al, 2008).
We report here that the activating LDB1 complex, KLF1 and CTCF occupy multiple regulatory elements within the Myb-Hbs1l intergenic region, which have the chromatin hallmarks of active enhancers. Chromosome Conformation Capture (3C) and high-throughput sequencing (3C-Seq) show that these elements and the actively transcribed Myb gene cluster together in the nuclear space to form an ACH in vivo, bringing the enhancers in close proximity to the Myb gene promoter and first intron. The latter contains a highly conserved CTCF binding site around which productive transcription elongation starts. The ACH is lost when cells terminally differentiate, concomitant with the downregulation of Myb and a decreased binding of TF complexes at the distal enhancers.
Results
The LDB1 complex binds distal enhancers in the Myb-Hbs1l intergenic region
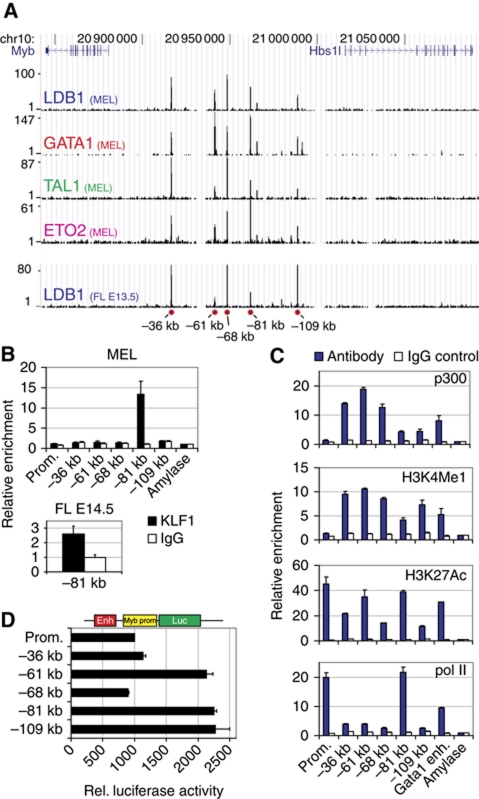
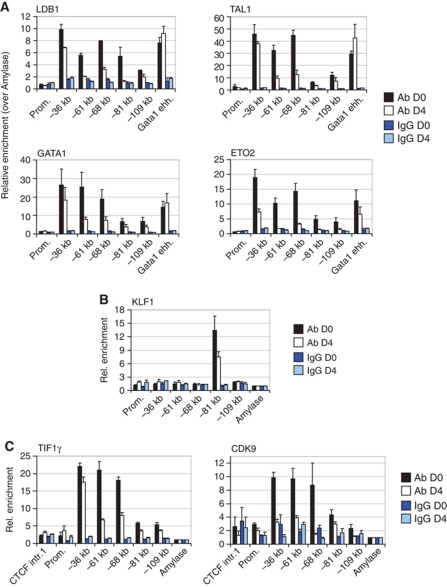
ChIP-Sequencing (ChIP-Seq) was used to identify the genome-wide binding sites of key erythroid TFs in mouse erythroleukaemia (MEL) cells and in primary mouse fetal liver (FL) cells (Soler et al, 2010). This showed preferential intragenic and intergenic binding of the LDB1 complex away from promoter sequences, suggesting it is involved in long-range gene regulation, a hypothesis supported by other studies (Song et al, 2007). Five LDB1 complex binding sites were detected in the Myb-Hbs1l intergenic region, −36, −61, −68, −81 and −109 kb upstream of the Myb transcription start site, in MEL cells and primary mouse erythroid progenitors from E13.5 FL (Figure 1A; Supplementary Figure S1A and B). These intergenic binding sites harboured all components of the activating LDB1 complex (GATA1/LDB1/TAL1/ETO2) in erythroid progenitors, consistent with active transcription of both Myb and Hbs1l genes (Supplementary Figure S1C). Additionally, in MEL and primary FL cells, the −81 kb binding site was found co-occupied by KLF1 (Figure 1B), a key erythroid TF primarily associated with gene activation, in agreement with a recent KLF1 ChIP-Seq experiment performed using primary mouse erythroid progenitors (Tallack et al, 2010; Supplementary Figure S2B and C). None of these TFs were found to bind the Myb or Hbs1l promoters. Next, all intergenic sites were shown to possess characteristic features supporting enhancer activity, that is, the presence of the histone acetyl transferase p300 (Visel et al, 2009), RNA polymerase II (polII), monomethylated histone 3 Lysine 4 (H3K4me1), and acetylated H3K27 (Heintzman et al, 2009; Figure 1C). PolII occupancy was especially abundant on the LDB1/KLF1 bound −81 kb sequence, showing similar enrichments to the highly active Myb promoter. In order to show that these LDB1 binding sites can indeed act as enhancers, they were cloned upstream of a minimal Myb promoter controlling a firefly luciferase reporter gene. Transfection into MEL cells showed that the −61, −81 and −109 kb elements are able to enhance luciferase activity (Figure 1D). In summary, these results suggest that the intergenic LDB1 complex binding sites represent active regulatory elements in erythroid progenitors, some possessing enhancer activity in vitro.
Figure 1.
The Myb-Hbs1l intergenic region contains transcriptional enhancers. (A) ChIP-Seq of the LDB1 complex components LDB1, GATA1, TAL1 and ETO2 in the Myb-Hbs1l locus in MEL cells (MEL). LDB1 binding in primary E13.5 fetal liver erythroid progenitors is also shown (FL E13.5). The position of the intergenic binding sites relative to the Myb TSS is indicated at the bottom (red circles). (B) ChIP analysis showing intergenic KLF1 occupancy in MEL cells and in E14.5 fetal liver cells. (C) ChIP analysis showing the binding of p300, polII and the presence of the enhancer-associated histone modifications H3K4me1 and H3K27ac at the intergenic region in MEL cells. (D) Luciferase reporter assays in MEL cells showing the enhancer activity of the different intergenic elements. ChIP enrichments were calculated versus a negative control region (amylase). Results are presented as the mean±s.e.m. of at least two independent experiments.
In-vivo conformation of the Myb-Hbs1l locus
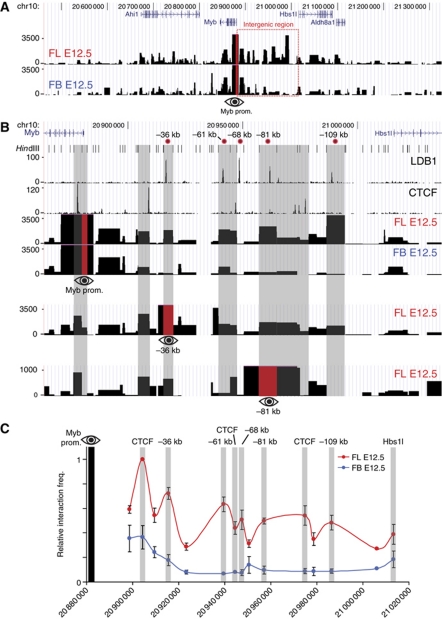
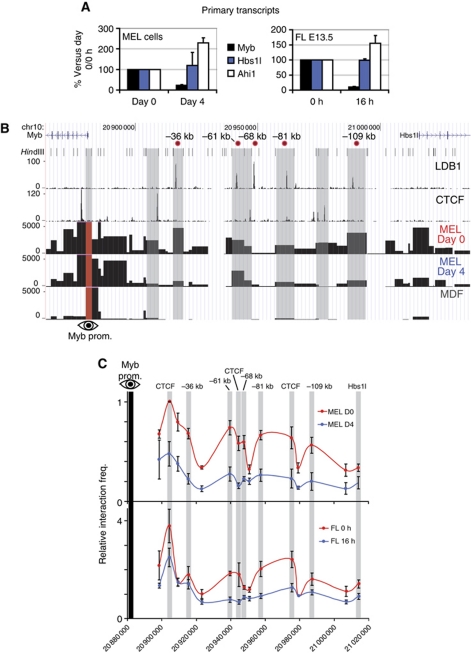
We next performed 3C-Seq (Soler et al, 2010) experiments (Supplementary Figure S3) to investigate whether the Myb promoter was interacting with the intergenic regulatory elements. 3C-Seq was first performed on fresh mouse E12.5 FL tissue (primarily containing erythroid progenitors) using the Myb promoter as the viewpoint. Fetal brain (FB) samples were processed in parallel as a control, since Myb expression is much lower in brain tissue and it lacks the erythroid-specific LDB1 complex. Furthermore, a previous p300 ChIP-Seq performed in FB tissue showed no enrichments within the Myb-Hbs1l intergenic region (Supplementary Figure S2A). Multiple promoter-interacting elements located in the intergenic region were detected in FL, of which most were either absent or showed a low signal in FB (Figure 2A and B), thus revealing erythroid-specific long-range communication between the Myb promoter and intergenic elements. In addition, 3C-Seq signals were shown to correlate with binding of the LDB1 complex, KLF1 and CTCF, which have all been implicated in mediating long-range chromatin interactions (Drissen et al, 2004; Vakoc et al, 2005; Splinter et al, 2006; Song et al, 2007; Figure 2B). Statistical analysis (Poisson distribution/running-mean comparison, P⩽0.001) of the FL and FB 3C-Seq data sets confirmed the erythroid specificity of the majority of intergenic interactions (Supplementary Figure S4). Quantitative 3C-qPCR experiments were carried out to confirm these results. This shows a very similar long-range interaction pattern (Figure 2C), with the exception of the −68-kb LDB1 complex binding site that was not detected by 3C-Seq but was found interacting by 3C-qPCR. These data show in-vivo nuclear proximity between LDB1 complex, KLF1 and CTCF-bound intergenic sequences and the Myb promoter, further implying they represent regulatory elements involved in Myb transcriptional regulation.
Figure 2.
Long-range genomic interactions within the Myb-Hbs1l locus. (A) 3C-Seq analysis of the Myb promoter-associated regions in vivo, using E12.5 mouse fetal liver (FL E12.5) and fetal brain (FB E12.5). Signals are presented as reads per millions per HindIII restriction fragment (vertical axis). The viewpoint (Myb promoter) is indicated by a red bar with an eye symbol. (B) Zoom-in view of the Myb-Hbs1l intergenic region. The ChIP-Seq profiles for LDB1 and CTCF (MEL) are shown together with the 3C-Seq signals obtained using the Myb promoter (top), the −36 kb (middle) and the −81 kb elements (bottom) as viewpoints (indicated by a red bar and eye symbol). Grey shading of HindIII fragments indicates sites where long-range interactions and transcription factor binding colocalize. The position of the HindIII restriction sites and the intergenic enhancers (relative to the Myb TSS) is indicated at the top. (C) Locus-wide crosslinking frequencies analysed by 3C-qPCR using the Myb promoter as viewpoint. Relative crosslinking frequencies observed in E12.5 FL (red) and FB (blue) are shown. Highest crosslinking frequencies per FL/FB pair tested were set to 1. The x axis shows the genomic coordinates of the interacting fragments in the locus. Data are plotted as mean±s.e.m. of at least three independent experiments.
To further confirm the Myb promoter 3C(-Seq) data, the 3C-Seq was repeated using the −36 and −81 kb LDB1 complex binding sites as viewpoints. This showed that both sites interact with the Myb promoter and the adjacent CTCF-bound intron 1 fragment (Figure 2B). Additionally, there were multiple interactions detected between the −36 kb/−81 kb LDB1 complex binding sites and other TF and CTCF binding sites (Figure 2B). Collectively, the 3C data show that the active erythroid Myb promoter and intron 1 cluster with intergenic TF-bound elements to form a complex higher order chromatin structure. Of note, these data indicate that whereas the Myb gene promoter is found in close proximity to the distal enhancers, both the −36 and −81 kb regions also show a strong interaction with the intron 1 CTCF site as well.
The intron 1 CTCF element marks the start of productive transcription elongation and interacts with elongation factor-bound distal enhancers
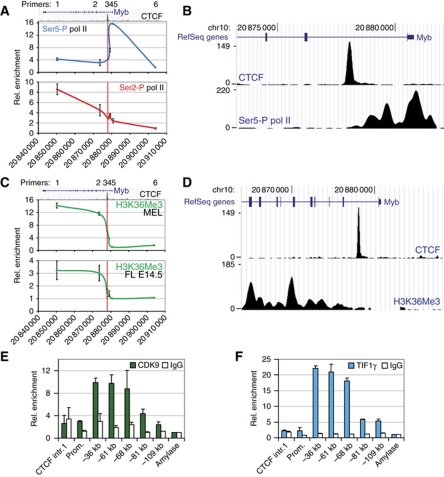
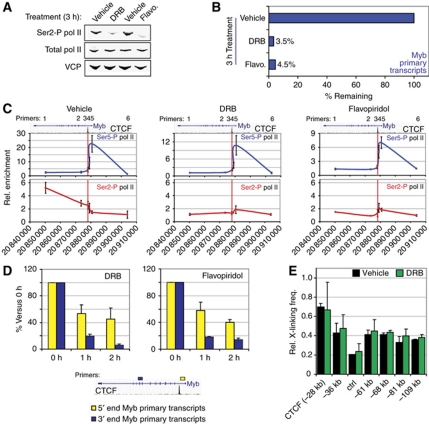
Several studies have shown that Myb expression is regulated at the level of transcription elongation through an attenuation site in the first intron (Bender et al, 1987; Watson, 1988; Reddy and Reddy, 1989; Hugo et al, 2006) ∼2 kb downstream of Myb exon 1, in the vicinity of the CTCF binding site identified in our study. Since this region interacts with the distal −36 and −81 kb elements, the intronic CTCF-bound element may actually mark the site of productive transcription elongation. Hence, ChIP experiments were carried out in erythroid progenitors to map the appearance of Serine 2 (Ser2)-phosphorylated polII (polII Ser2-P) and the H3K36 trimethylation (H3K36me3) mark, which are specifically associated with transcription elongation and peak within the transcribed region of genes (Brookes and Pombo, 2009; Buratowski, 2009; Figure 3A and C). As expected, no polII Ser2-P or H3K36me3 enrichments were detected at the promoter and upstream regions, whereas a sharp increase was seen starting around the CTCF binding site and increasing into the gene body (Figure 3A and C). Ser5-P polII on the other hand, representing the initiating polII state, specifically accumulated upstream of the CTCF site. In order to more precisely localize the transition from transcription initiation to productive elongation, ChIP-Seq for polII Ser5-P and H3K36me3 was performed in MEL cells. As shown in Figure 3B and D, the initiating polII signal covers the 5′ end of the gene and extends up to the intronic CTCF site. In contrast, H3K36me3 starts to appear after the CTCF binding site in MEL cells. In addition, a recently published H3K36me3 data set from mouse primary erythroid progenitors (Wong et al, 2011) and data obtained from the human erythroid cell line K562 show a similar pattern (Supplementary Figure S5). These data suggest that the transition to productive transcription elongation occurs around the intronic CTCF site. ChIP experiments were used next to analyse the presence of the elongation factors CDK9 and TIF1γ at the Myb locus. CDK9 is a kinase that phosphorylates the Ser2 residue of the polII C-terminal domain (CTD), and is known to bind the LDB1 complex (Meier et al, 2006). TIF1γ was recently identified as a component of the LDB1 complex, regulating transcription elongation in haematopoietic cells, at least in part by allowing CDK9 recruitment to its target sites (Bai et al, 2010). CDK9 and TIF1γ showed only minor enrichments at the promoter and first intron (where polII Ser2-P appears), but surprisingly showed a much stronger occupancy at the upstream regulatory elements (Figure 3E and F). These experiments suggest that productive elongation is stimulated around the intronic CTCF site by positive elongation factors bound at the distal enhancer elements. These factors are likely to be brought in physical proximity to the elongation site by dynamic chromatin looping, where they can transiently carry out their enzymatic function. In support of this notion, depletion of TIF1γ in primary human erythroid cells resulted in a severe reduction of Myb mRNA levels (Bai et al, 2010). In addition, in order to prove that the Ser5-P polII enrichments observed in the Myb first intron were specific and independent from productively elongating polII (Ser2-P), MEL cells were treated with the Cdk9 inhibitors DRB or flavopiridol. Under these conditions, a global loss of phosphorylated Ser2 RNA polII was observed (Figure 4A) and Myb transcription was almost completely abolished (primary transcripts are decreased by >95%, Figure 4B). Importantly, ChIP experiments showed that the Ser5-P polII pattern on the Myb promoter and first intron was similar in vehicle-treated cells and cells treated with CDK9 inhibitors, while Ser2-P polII enrichments were lost (Figure 4C). Thus, the Ser5-P polII occupancy of the Myb promoter and first intron up to the CTCF site is independent of ongoing transcriptional elongation.
Figure 3.
Transcription elongation starts in the vicinity of the Myb first intron CTCF element. (A, C) ChIP analysis showing the distribution of (A) polII phosphorylated at Ser5 (Ser5-P) and Ser2 (Ser2-P) in MEL cells, and (C) H3K36me3 in MEL and E14.5 fetal liver cells. UCSC Genome browser pictures depicting the Myb gene and CTCF binding in the first intron (ChIP-Seq) are shown above the graph. Primer pairs (1–6) used for PCR are indicated above the gene. The x axis shows the genomic coordinates and the position of the CTCF binding site is indicated at the top (red vertical line). (B, D) ChIP-Seq profiles of CTCF, (B) Ser5-P pol II and (D) H3K36me3 obtained from MEL cells. (E, F) Occupancy of the Myb-Hbs1l intergenic region by the elongation factors (E) CDK9 and (F) TIF1γ in MEL cells as shown by ChIP. Enrichments were calculated versus a negative control region (amylase) and presented as the mean±s.e.m. of at least two independent experiments.
Figure 4.
Effect of CDK9 inhibition on phosphorylated pol II occupancy, transcription and chromatin looping. (A) Western blot analysis of Ser2-P pol II and total pol II levels in vehicle-, DRB- or Flavopiridol (Flavo.)-treated MEL cells. Valosin Containing Protein (VCP) served as a loading control. (B) Myb primary transcripts measured by RT–qPCR after treatment with the indicated compounds. Signals were normalized to 18S rRNA expression and transcript levels in vehicle-treated cells were set to 100%. (C) ChIP analysis of Ser5-P and Ser2-P pol II binding at the Myb transcriptional unit in vehicle-, DRB- and Flavopiridol-treated MEL cells. Genomic coordinates, gene location, CTCF occupancy and PCR primers are indicated above each graph (as in Figure 3). (D) Time-course CDK9 inhibition in MEL cells using DRB and Flavopiridol. Myb 5′ end and 3′ end transcripts were measured by RT–qPCR. Primer locations within the gene are depicted by coloured rectangles in a schematic below the graphs. (E) 3C-qPCR analysis on vehicle- or DRB-treated MEL cells. The Myb promoter HindIII fragment was used as a viewpoint. Data are plotted as mean±s.e.m. of at least two independent experiments.
In support of this, while Cdk9 inhibition results in a loss of full-length transcripts, transcription of the 5′ end of the gene is maintained and much less sensitive to Cdk9 inhibition (Figure 4D). The 40–50% decrease in 5′ transcripts compared with vehicle-treated cells can be accounted for by the general ∼50% decrease of Ser5-P polII at the promoter under these conditions (Figure 4C). Importantly, these data show that in the absence of Ser2 phosphorylation, RNA polII is still able to engage at the Myb gene and is still transcribing the first ∼2 kb (i.e., up to the CTCF site) but is unable to bypass this site and progress throughout the gene efficiently. Interestingly, the long-range interactions are maintained upon DRB treatment (Figure 4E).
Erythroid differentiation is accompanied by decreased Myb expression and a loss of chromatin looping
In order to correlate the long-range interactions observed in the Myb-Hbs1l locus with Myb transcriptional activity, Myb expression and locus structure were analysed during erythroid differentiation. Differentiation of MEL cells or mouse E13.5 FL primary erythroid progenitors resulted in a strong decrease in Myb, but not Hbs1l or Ahi1 primary transcription (Figure 5A). Erythroid maturation of MEL and FL cells was monitored by analysing the activation of the two terminal differentiation markers Glycophorin A (Gypa) and Beta-Major (Hbb-b1) (Supplementary Figure S6A), as well as the characteristic decrease in cell size of the primary progenitors (Supplementary Figure 6B). Thus, significant downregulation of Myb transcription occurs upon terminal erythroid differentiation, while the flanking genes show stable or modestly increasing expression levels. 3C-Seq was subsequently carried out using the Myb promoter as viewpoint in MEL cells before and after differentiation, representing stages of high and low Myb expression, respectively (Figure 5B). In non-differentiated MEL cells, the Myb promoter showed a long-range interaction pattern very similar to that seen in primary erythroid progenitors (Figure 2B). However, opposite to what was observed for the β-globin locus (Palstra et al, 2003), the frequency of most intergenic contacts was strikingly diminished upon differentiation. This loss of interaction was observed essentially for all LDB1 complex, KLF1- and CTCF-bound fragments of the locus (Figure 5B). 3C-Seq experiments using mouse dermal fibroblasts (MDF, which do not express Myb) confirmed the erythroid-specific nature of the interactions (Figure 5B). The loss of chromatin looping in both MEL cells and primary erythroid progenitors upon erythroid differentiation was confirmed by the more quantitative 3C-qPCR method (Figure 5C). These data show that Myb downregulation upon erythroid differentiation is accompanied by a loss of communication between the Myb promoter and the intergenic TF-bound enhancers.
Figure 5.
Erythroid differentiation is accompanied by a loss of Myb transcription and long-range genomic interactions. (A) Primary transcript levels of Myb, Hbs1l and Ahi1 during terminal differentiation of MEL (left panel) and E13.5 fetal liver (FL) erythroid progenitors (right panel). MEL cells were induced for 4 days in the presence of 2% DMSO. Fetal liver cells were cultured ex vivo for 16 h in differentiation medium. Data are expressed as percentages of expression versus day 0 (MEL) or 0 h (FL) of differentiation. Signals were normalized to Rnh1 or Calr expression, and day 0 (MEL) or 0 h (primary cells) values were set to 100 (i.e., undifferentiated cells). Results are plotted as mean±s.e.m. of three independent experiments. (B) 3C-Seq analysis of the Myb promoter-associated regions in undifferentiated MEL cells (MEL day 0) and differentiated MEL cells (MEL day 4). Mouse dermal fibroblasts (MDFs) were used as a negative control (no Myb expression). Results are represented as in Figure 2. (C) Analysis of the Myb-Hbs1l locus conformation by 3C-qPCR in differentiating MEL and FL cells. See Figure 2C for details.
Decreased transcription and elongation factor occupancy at the intergenic enhancers upon erythroid differentiation
The long-range interactions between Myb and the intergenic enhancers are lost upon differentiation, clearly paralleling Myb downregulation. However, it is unclear what underlies the loss of looping and expression. To address this question, quantitative ChIP experiments were performed in MEL cells before and after differentiation to analyse intergenic TF occupancy during erythroid maturation. An overall decrease in LDB1 complex (Figure 6A), KLF1 (Figure 6B) and elongation factor (Figure 6C) occupancy was seen at the intergenic binding sites upon differentiation. In agreement with this, the levels of enhancer-associated histone modifications and proteins often decrease as well (Supplementary Figure S7A). Furthermore, ChIP-Seq showed changing polII occupancy of the Myb transcription unit during differentiation (Supplementary Figure S7B). In both undifferentiated and differentiated cells, polII accumulates at the promoter and first intron (high signals), up to the CTCF site. In undifferentiated cells, polII actively bypasses this site and progresses into the gene, whereas in differentiated cells a strong reduction of polII beyond the CTCF site is seen (Supplementary Figure S7B). Thus, a loss of activating proteins at the intergenic regulatory elements upon differentiation coincides with losses of long-range interactions, polII progression into the gene body, and expression. Interestingly, initiation still appears to take place, as previously suggested (Bender et al, 1987).
Figure 6.
Erythroid differentiation induces a dramatic decrease of transcription factor occupancy within the Myb-Hbs1l intergenic region. (A–C) MEL cells were treated with 2% DMSO for 4 days to induce erythroid differentiation. Chromatin occupancy of (A) LDB1, TAL1, GATA1, ETO2, (B) KLF1, (C) TIF1γ and CDK9 was examined by ChIP in undifferentiated (D0) and differentiated (D4) MEL cells. Amylase served as a negative control region. Data are presented as mean±s.e.m. of 2–4 independent experiments.
LDB1 and KLF1 are essential for high Myb expression in erythroid progenitors
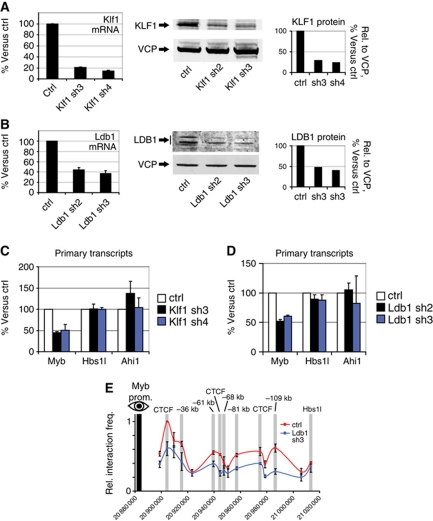
LDB1 and KLF1 have recently been implicated in long-range gene regulation (Drissen et al, 2004; Song et al, 2007; Tallack et al, 2010). KLF1 selectively occupies the −81-kb site in the Myb-Hbs1l intergenic region, while LDB1 binds all five regulatory elements (Figure 1). Since intergenic binding of both proteins decreased as Myb transcription is downregulated, we hypothesized that loss of KLF1 or LDB1 in erythroid progenitors would result in decreased Myb expression. To verify this, short hairpin RNAs (shRNAs) against Klf1 or Ldb1 mRNA were used to reduce their respective protein levels in MEL cells. A 50–80% decrease in mRNA and protein was observed when compared with cells transduced with a control lentivirus (Figure 7A and B). Both knockdowns resulted in a 50% decrease of Myb transcription, while the flanking Hbs1l and Ahi1 genes were not significantly affected (Figure 7C and D). Similarly, knocking down the expression of CTCF also results in a significant reduction of Myb transcription, without affecting Hbs1l or Ahi1 (Supplementary Figure S8C). The decrease in Myb primary transcripts was not caused by cellular differentiation or a change in TF levels due to LDB1 or KLF1 depletion, as we observed no changes compatible with erythroid maturation in the expression of late erythroid markers (Gypa and Hbb-b1) or key erythroid TFs (Supplementary Figure S8A and B). These results are in agreement with reports showing reduced Myb expression in vivo in Klf1−/− FL (Pilon et al, 2008), and a 50% decrease of Myb expression in bone marrow haematopoietic progenitors conditionally depleted for LDB1 (Li et al, 2011). Since LDB1 is a scaffold-like protein important for TF complex assembly and chromatin looping, locus conformation was analysed by 3C-qPCR after LDB1 knockdown (Figure 7E). This showed that LDB1 depletion indeed results in reduced long-range promoter–enhancer contacts (Figure 7E), further emphasizing its key role in chromatin loop formation.
Figure 7.
LDB1 and KLF1 positively regulate Myb expression. (A, B) Two independent shRNAs were used to decrease (A) Klf1 and (B) Ldb1 expression in MEL cells. Knockdown efficiency was measured at the mRNA and protein levels. Results are compared with a non-targeting scrambled shRNA. Valosin Containing Protein (VCP) served as a loading control for protein analysis. (C, D) Effect of (C) Klf1 and (D) Ldb1 knockdowns on Myb, Hbs1l and Ahi1 primary transcript levels. (E) The effect of LDB1 depletion on chromatin looping was measured by 3C-qPCR using the Myb promoter as viewpoint. The interaction frequencies in control and LDB1-depleted samples are shown in red and blue, respectively. Data are plotted as mean±s.e.m. of at least three independent experiments.
In summary, these data suggest that LDB1 and KLF1 form a regulatory module to maintain high levels of Myb transcription, consistent with the gene-activating role described for these factors in erythropoiesis (Soler et al, 2010; Tallack et al, 2010).
Discussion
The expression of the Myb proto-oncogene in haematopoietic cells is subjected to very tight control to properly coordinate cellular proliferation and differentiation. Given that enforced Myb expression impairs haematopoietic differentiation and that aberrant Myb expression associates with haematopoietic malignancies, deciphering Myb transcriptional control is crucial for a better understanding of both normal haematopoietic development and associated disorders.
TF binding and long-range interactions at the Myb-Hbs1l locus
A combination of ChIP-Seq and 3C-Seq was used to map the genome-wide binding sites of critical transcription and structural factors, and to characterize the spatial interactions within the Myb locus. 3C-Seq offers an advantage over array-based 4C technology to map long-range genomic interactions at the level of a single locus (in addition to a genome-wide level), since it does not suffer from saturating signals surrounding the viewpoints (Simonis et al, 2006; Soler et al, 2010). It is therefore well suited to analyse locus-wide chromatin looping within tens of kilobases up to megabases without prior knowledge of the interaction sites. Combining ChIP-Seq and 3C-Seq shows that the Myb-Hbs1l intergenic region harbours important regulatory elements controlling Myb expression, that bind either the structural protein CTCF or the essential erythroid TFs GATA1, LDB1, TAL1 and KLF1. The sites that bind KLF1 and the GATA1/TAL1/LDB1 complex are transcriptional enhancers, confirming the positive role of these factors on erythroid gene expression (Figure 1; Soler et al, 2010; Tallack et al, 2010).The 3C-Seq genomic interaction profiles show an erythroid-specific pattern of interactions between the Myb promoter, first intron and intergenic enhancers (Figure 2), which is highly similar for primary erythroid progenitors and MEL cells. CTCF, KLF1 and GATA1/TAL1/LDB1 binding sites were shown to mark the sites of long-range genomic interactions. The reproducibility between different biological materials, 3C-qPCR validations and the clear overlap between long-range interactions and TF binding further validate the specificity of the 3C-Seq profiles.
Both KLF1 and LDB1 activate Myb expression, and the LDB1 complex is required to establish spatial proximity between Myb and the distal intergenic enhancers
We show here a requirement for KLF1 and LDB1 in maintaining high levels of Myb expression in erythroid progenitors (Figure 7). Reducing the level of either of these factors results in a 50% decrease of Myb transcription without inducing erythroid differentiation (Supplementary Figure S8). This suggests that Myb downregulation coincides with, but is not a driver of differentiation. The DNA-binding erythroid Kruppel-like factor KLF1 is the founding member of the mammalian Kruppel-like family of zinc-finger TFs. It recognizes CACCC-box motifs often found in erythroid-specific gene promoters and is required for their activation. KLF1 binds a single location in the Myb-Hbs1l locus at the −81-kb enhancer, which contains a conserved CACCC-box motif. The positive role of KLF1 on erythroid gene expression is confirmed by our finding that KLF1 activates Myb transcription. Interestingly, Klf1−/− mouse embryos die around E15 from a lack of definitive erythropoiesis, resulting in severe anaemia (Nuez et al, 1995; Perkins et al, 1995). This phenotype shares similarities with the lethal anaemia of Myb−/− embryos which die around E15 (Mucenski et al, 1991). It has been shown that E13.5 Klf1−/− FL-derived erythroid cells fail to progress through the last cell cycles of terminal erythroid differentiation, in part due to misregulation of the G1-to-S phase transition TFs E2F2 and E2F4 (Pilon et al, 2008; Tallack et al, 2009). The phenotypic similarities between Klf1−/− and Myb−/− mouse models, the strong downregulation of Myb in Klf1−/− FL cells (Pilon et al, 2008) and the implication of Myb in the G1-to-S transition (Oh and Reddy, 1999) suggest that Myb misregulation in Klf1−/− cells also contributes significantly to the observed proliferative defect.
The widely expressed nuclear adaptor LDB1 functions as a core component of multiprotein complexes, regulating the development of many tissues. The LDB1 protein itself has no known DNA-binding or enzymatic activities. In erythroid cells, LDB1 forms a complex with the DNA-binding TFs GATA1, TAL1 (SCL), E2A and the cofactors LMO2/LMO4 and ETO2/MTGR1. In addition, the LDB1 complex interacts with transcription elongation factors, like TIF1γ and CDK9, a kinase known to regulate transcription elongation through phosphorylation of the polII CTD at Ser2. Consistent with its essential functions, Ldb1−/− mouse embryos do not develop beyond the E10 stage and show dramatic developmental defects including a lack of haematopoiesis (Mukhopadhyay et al, 2003; Li et al, 2010). Due to the early lethal phenotype, the role played by LDB1 during haematopoiesis in vivo remained largely unexplored. Recent data, however, showed a continuous requirement for LDB1 in the maintenance and differentiation of haematopoietic stem cells, and in the development of the lymphoid, erythroid and megakaryocytic lineages (Li et al, 2010, 2011). LDB1 is required to activate the late erythroid gene expression program (Li et al, 2010; Soler et al, 2010) and it exerts this function at least in part by facilitating long-range interactions between remote enhancers and their target genes (Song et al, 2007). Our analysis of the Myb-Hbs1l locus conformation shows that in erythroid progenitors expressing Myb at high levels, the enhancers are clustered in the nuclear space to form an ACH structure resembling the one observed within the active β-globin locus. We show here that LDB1 is required for the maintenance of the long-range interactions between the Myb gene and the upstream enhancers. Reducing the level of LDB1 in erythroid progenitors results in a decrease of Myb promoter–enhancer interactions and transcription (Figure 7). Interestingly, transcription of the neighbouring genes Hbs1l and Ahi1 remains unaffected under these conditions, even though Ahi1 harbours a binding site for the LDB1 complex in its first intron (Soler et al, 2010). During the course of erythroid differentiation, when Myb transcription is downregulated dramatically, the long-range interactions are reduced, resulting in a loss of the ACH (Figure 5). This loss of long-range communication is explained by the decreased occupancy of the LDB1 complex at the intergenic enhancers (Figure 6). Interestingly, decreasing the level of LDB1 results in a loss of all interactions, not just those bound by LDB1. This suggests that in order to be maintained and stabilized, the chromatin hub requires several if not all the interactions (i.e., the enhancer sites and the CTCF sites). Accordingly, affecting the binding of LDB1 on some sites would induces a destabilization of the whole structure, and thus have an impact on sites not bound by the protein but normally present in the hub.
Strikingly, this observation contrasts with the general increase of binding of the LDB1 complex on induced erythroid genes during terminal differentiation (Soler et al, 2010). A mechanistic explanation for this selective loss of the LDB1 complex from the Myb-Hbs1l locus could be that, in the late stages of differentiation, additional TFs start competing for binding or induce a local destabilization or degradation of the complex.
Heterogeneity between the distal enhancer elements
It is not clear whether Myb requires the entire intergenic region for full activation, because the individual contributions of the different intergenic enhancers are unknown. They could play an additive role to ensure high local concentrations of positive transcriptional regulators and therefore high levels of transcription. Alternatively, they might be required to stabilize the chromatin hub at the Myb gene and first intron. Such a multi-component complex structure has already been observed for developmentally regulated genes like globins. In that case the activity of the elements appears additive, although they are individually clearly different in structure and activity. For the Myb locus, the elements also appear to be different in function. They show different enhancer activity in vitro, and differ in protein occupancy. Indeed, whereas all elements are enriched for the core components of the LDB1 complex and enhancer-associated histone modifications/proteins, the −81-kb enhancer shows a 5- to 7.5-fold higher enrichment for polII and is the only one bound by KLF1 (Figure 1), a factor essential for Myb transcription (Figure 7). The −81-kb element also shows a high degree of sequence conservation between mouse and human. This regulatory element is therefore likely to play a key role in the transcriptional activation of the locus. Conditional deletion of the individual enhancers will provide crucial information about their role(s) in vivo, in particular whether the −81-kb element represents an enhancer with a specialized function.
Transcription and elongation factors at distal regulatory elements: a model for Myb transcriptional activation during development
Our data are in agreement with previous reports highlighting the regulatory potential and the importance of the Myb-Hbs1l intergenic region for Myb transcriptional regulation (Mukai et al, 2006; Wahlberg et al, 2009). In addition, the presence of regulatory elements within the Myb first intron affecting transcription elongation has been reported >20 years ago, although their role is still not fully understood (Bender et al, 1987; Hugo et al, 2006). An attenuation element was mapped in the first intron, where a poly-T tract was predicted to yield a stem-loop structured nascent RNA. Based on this finding, it was speculated that the stable intronic stem-loop transcript might provide a docking site for RNA-binding proteins to overcome the transcription elongation block in a way similar to the HIV TAR stem-loop RNA (Ramsay and Gonda, 2008). Although we cannot exclude this hypothesis, our data indicate that the intronic transcription elongation region corresponds to a domain containing a highly conserved CTCF binding site (coinciding with the start of the Ser2-P polII and H3K36me3 elongation signature, Figure 3), which appears to function in combination with the upstream elements. For example, the −36 and −81 kb enhancers loaded with erythroid TFs, polII and the elongation factors CDK9 and TIF1γ loop towards the Myb intron 1 CTCF site (Figure 2B). As the intergenic elements actively cluster together (Figure 2B) and are also bound by transcription and elongations factors (Figures 1 and 3), they are likely to contribute to the stimulation of transcription elongation. To further support this idea, we carried out CDK9 inhibition experiments (Figure 4). As stated above, CDK9 is primarily bound to the upstream regulatory elements. Its inhibition resulted in a loss of elongating (Ser2-P) polymerase and 3′ Myb transcription, while the initiating (Ser5-P) polymerase and 5′ transcription were retained, without affecting looping. A plausible explanation would therefore be that CDK9 is brought to the intronic transition site by looping, as represented in our model (Figure 8). As the chromatin loops were still able to form under these conditions (Figure 4E), they may have become ‘non-functional’ due to an inability to provide kinase activity.
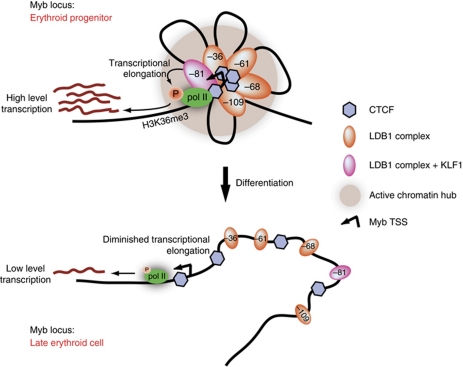
Figure 8.
Model of the dynamic transcriptional regulation of Myb in differentiating erythroid cells. The Myb Active Chromatin Hub (ACH, grey sphere) is a structured nuclear compartment containing clustered cis-regulatory elements enriched for activating transcription factor complexes containing transcription elongation factors (orange and pink ovals) and CTCF (blue diamonds). The ACH provides a local high concentration of polII, transcription and elongation factors around the Myb gene, allowing for high-level expression in erythroid progenitors. During differentiation, intergenic transcription factor occupancy decreases (small ovals) at the cis-regulatory elements, leading to a destabilization of the ACH and a dramatic decrease of Myb transcription, allowing cells to terminally differentiate.
Interestingly, a role for the β-globin LCR in the transition from transcriptional initiation to elongation has been proposed (Sawado et al, 2003). Indeed, both CDK9 and TIF1γ bind the LCR (unpublished observation). It remains to be tested whether the Myb and globin ACHs fulfil similar tasks in the transition to productive elongation. In the Myb locus, the presence of CTCF is likely to play a key role in orchestrating the long-range interactions (Splinter et al, 2006) and its presence is required for high level Myb expression (Supplementary Figure S8C). The intronic CTCF site may mark a transcriptional barrier element preventing polII from progressing further into the gene body (Supplementary Figure S7B). However, when the distal enhancers are loaded with TFs, polII and elongation factors, it would serve as an anchoring site for the enhancers to form an ACH. Clustering all the factors around the Myb promoter and intronic productive elongation site would then override the transcriptional block in erythroid progenitors to allow Myb transcription at a high rate (Figure 8, upper half). The presence of a previously suggested structured nascent RNA (Thompson et al, 1997; Hugo et al, 2006; Ramsay and Gonda, 2008) could locally cause polII to slow down, thereby increasing the chance of phosphorylation by the elongation factors bound at the distal elements. Both mechanisms could thus participate in the elongation checkpoint operating at the Myb intronic attenuation region. During terminal differentiation, the ACH is destabilized due to a loss of intergenic TF occupancy, resulting in decreased Myb transcription to allow the cells to fully mature (Figure 8, lower half).
Implications for development and disease
Since fluctuations in Myb expression are a common feature of differentiating haematopoietic cells, it is expected that similar mechanisms will take place in different lineages, probably using (part of) the intergenic regulatory elements described here, but bound by other lineage-specific TF complexes. Recent genome-wide studies in early haematopoietic stem/progenitor cells revealed the binding of several haematopoietic TFs on some of the Myb intergenic enhancers (Wilson et al, 2010; Li et al, 2011). It will be interesting to track enhancer usage and ACH formation during the course of haematopoietic stem/progenitor cell differentiation to the different lineages (e.g., myeloid versus lymphoid), and to investigate how the locus structure is affected in haematopoietic diseases like leukaemia. Importantly, our data provide a framework for further comparative analysis in human erythroid cells, where MYB-HBS1L allelic variants strongly associate with clinically relevant red blood cell traits and high fetal globin gene expression (Thein et al, 2007; Lettre et al, 2008; Ganesh et al, 2009; Galarneau et al, 2010), a crucial feature decreasing the severity of β-thalassaemia and sickle-cell anaemia. Several intergenic enhancers have high sequence conservation between mouse and human. Considering that the intronic CTCF and transcription elongation transition sites also seem to be conserved in human erythroid cells (Supplementary Figure S5A), a careful examination of the impact of intergenic SNPs on TF binding, chromatin looping and MYB expression in individuals bearing these SNPs will be of primary interest. A preliminary analysis of highly associated SNPs showed that some fall close to or within the conserved intergenic sequences, suggesting that they may affect regulation of MYB expression. However, to date we did not find clear examples where the variants either create or destroy a GATA1/LDB1 binding sequence motif. A more systematic analysis needs to be performed in order to better understand the functional impact of SNPs in the MYB-HBS1L intergenic region. It is likely that the impact of the variants may only have a mild effect on MYB expression, which may complicate the analyses. However, with recent reports implicating c-MYB in the regulation of human fetal haemoglobin expression (Jiang et al, 2006; Sankaran et al, 2011) and the maintenance of leukaemia in mice (Zuber et al, 2011), modulation of c-MYB levels could become an attractive therapeutic approach in the treatment of β-haemoglobinopathies and leukaemia.
Materials and methods
ChIP and ChIP-Seq procedures
ChIP and ChIP-Seq procedures were performed as described (Soler et al, 2010, 2011). ChIP-Seq samples were sequenced (36 bp reads) on the Illumina GAII platform and analysed by NARWHAL (Brouwer et al, 2011). Data were visualized using a local mirror of the UCSC genome browser.
3C and 3C-Seq procedures
The 3C and 3C-Seq libraries were prepared as described previously (Simonis et al, 2006; Soler et al, 2010; Supplementary Figure S3). HindIII was used as the primary restriction endonuclease. The 3C PCR signals were normalized as described (Palstra et al, 2003), with the highest crosslinking frequency set to 1. For 3C-Seq, either NlaIII (Myb prom and −36 kb viewpoints) or DpnII (−81 kb viewpoint) were used as secondary restriction enzymes. The 3C-Seq library was sequenced (76 bp reads) on the Illumina GAII platform.
For more detailed Materials and methods, see the Supplementary data.
Accession codes
The ChIP-Seq and 3C-Seq data sets were deposited to the Sequence Read Archive (the accession numbers for the ChIP-Seq were previously published (Soler et al, 2010). 3C-Seq data can be obtained using accession number SRA048225).
Supplementary Material
Acknowledgments
We are grateful to Sjaak Philipsen for critical reading of the manuscript and to Jean-Christophe Andrau for helpful discussions. We thank Zeliha Ozgür for technical assistance and Rutger Brouwer for bioinformatics support. This work was supported by the EU-FP7 Eutracc consortium, the Netherlands Cancer Genomics Center (CGC) and the DFG SFB/Transregio5.
Author contributions: ES and FG conceived the study; RS, RJP, FG and ES designed the experiments; RS, CAS, EdB, AvdH, MS and ES performed the experiments; DE provided critical reagents and helpful comments; BL supervised informatics analyses; BL and ST designed the 3C-Seq analysis pipeline; ST, JCB and BL performed ChIP-Seq analysis; CK, AvdS and WvIJ performed ChIP-Seq and 3C-Seq DNA library preparation and Illumina sequencing; MvdH performed Illumina sequences alignments and data export; ES, FG and RS wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bai X, Kim J, Yang Z, Jurynec MJ, Akie TE, Lee J, LeBlanc J, Sessa A, Jiang H, DiBiase A, Zhou Y, Grunwald DJ, Lin S, Cantor AB, Orkin SH, Zon LI (2010) TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell 142: 133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender TP, Kremer CS, Kraus M, Buch T, Rajewsky K (2004) Critical functions for c-Myb at three checkpoints during thymocyte development. Nat Immunol 5: 721–729 [DOI] [PubMed] [Google Scholar]
- Bender TP, Thompson CB, Kuehl WM (1987) Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation. Science 237: 1473–1476 [DOI] [PubMed] [Google Scholar]
- Brookes E, Pombo A (2009) Modifications of RNA polymerase II are pivotal in regulating gene expression states. EMBO Rep 10: 1213–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer RWW, van den Hout MCGN, Grosveld FG, van Ijcken WFJ (2011) NARWAHL, a primary analysis pipeline for NGS data. Bioinformatics (advance online publication, 8 November 2011; doi:; DOI: 10.1093/bioinformatics/btr613) [DOI] [PubMed] [Google Scholar]
- Buratowski S (2009) Progression through the RNA polymerase II CTD cycle. Mol Cell 36: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor AB, Orkin SH (2001) Hematopoietic development: a balancing act. Curr Opin Genet Dev 11: 513–519 [DOI] [PubMed] [Google Scholar]
- Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W (2004) The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev 18: 2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J (2003) Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J 22: 4478–4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, O’Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH (2009) Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell 36: 667–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau G, Palmer CD, Sankaran VG, Orkin SH, Hirschhorn JN, Lettre G (2010) Fine-mapping at three loci known to affect fetal hemoglobin levels explains additional genetic variation. Nat Genet 42: 1049–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesh SK, Zakai NA, van Rooij FJ, Soranzo N, Smith AV, Nalls MA, Chen MH, Kottgen A, Glazer NL, Dehghan A, Kuhnel B, Aspelund T, Yang Q, Tanaka T, Jaffe A, Bis JC, Verwoert GC, Teumer A, Fox CS, Guralnik JM et al. (2009) Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat Genet 41: 1191–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda TJ, Metcalf D (1984) Expression of myb, myc and fos proto-oncogenes during the differentiation of a murine myeloid leukaemia. Nature 310: 249–251 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M et al. (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo H, Cures A, Suraweera N, Drabsch Y, Purcell D, Mantamadiotis T, Phillips W, Dobrovic A, Zupi G, Gonda TJ, Iacopetta B, Ramsay RG (2006) Mutations in the MYB intron I regulatory sequence increase transcription in colon cancers. Genes Chromosomes Cancer 45: 1143–1154 [DOI] [PubMed] [Google Scholar]
- Jiang J, Best S, Menzel S, Silver N, Lai MI, Surdulescu GL, Spector TD, Thein SL (2006) cMYB is involved in the regulation of fetal hemoglobin production in adults. Blood 108: 1077–1083 [DOI] [PubMed] [Google Scholar]
- Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA (2008) Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell 29: 232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassouf MT, Hughes JR, Taylor S, McGowan SJ, Soneji S, Green AL, Vyas P, Porcher C (2010) Genome-wide identification of TAL1's functional targets: insights into its mechanisms of action in primary erythroid cells. Genome Res 20: 1064–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre G, Sankaran VG, Bezerra MA, Araujo AS, Uda M, Sanna S, Cao A, Schlessinger D, Costa FF, Hirschhorn JN, Orkin SH (2008) DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc Natl Acad Sci USA 105: 11869–11874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Jothi R, Cui K, Lee JY, Cohen T, Gorivodsky M, Tzchori I, Zhao Y, Hayes SM, Bresnick EH, Zhao K, Westphal H, Love PE (2011) Nuclear adaptor Ldb1 regulates a transcriptional program essential for the maintenance of hematopoietic stem cells. Nat Immunol 12: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lee JY, Gross J, Song SH, Dean A, Love PE (2010) A requirement for Lim domain binding protein 1 in erythropoiesis. J Exp Med 207: 2543–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieu YK, Reddy EP (2009) Conditional c-myb knockout in adult hematopoietic stem cells leads to loss of self-renewal due to impaired proliferation and accelerated differentiation. Proc Natl Acad Sci USA 106: 21689–21694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Guo S, Ebert BL, Zhang H, Peng X, Bosco J, Pretz J, Schlanger R, Wang JY, Mak RH, Dombkowski DM, Preffer FI, Scadden DT, Golub TR (2008) MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell 14: 843–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier N, Krpic S, Rodriguez P, Strouboulis J, Monti M, Krijgsveld J, Gering M, Patient R, Hostert A, Grosveld F (2006) Novel binding partners of Ldb1 are required for haematopoietic development. Development 133: 4913–4923 [DOI] [PubMed] [Google Scholar]
- Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott WJ Jr, Potter SS (1991) A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 65: 677–689 [DOI] [PubMed] [Google Scholar]
- Mukai HY, Motohashi H, Ohneda O, Suzuki N, Nagano M, Yamamoto M (2006) Transgene insertion in proximity to the c-myb gene disrupts erythroid-megakaryocytic lineage bifurcation. Mol Cell Biol 26: 7953–7965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay M, Teufel A, Yamashita T, Agulnick AD, Chen L, Downs KM, Schindler A, Grinberg A, Huang SP, Dorward D, Westphal H (2003) Functional ablation of the mouse Ldb1 gene results in severe patterning defects during gastrulation. Development 130: 495–505 [DOI] [PubMed] [Google Scholar]
- Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F (1995) Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature 375: 316–318 [DOI] [PubMed] [Google Scholar]
- Oh IH, Reddy EP (1999) The myb gene family in cell growth, differentiation and apoptosis. Oncogene 18: 3017–3033 [DOI] [PubMed] [Google Scholar]
- Ong CT, Corces VG (2011) Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet 12: 283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W (2003) The beta-globin nuclear compartment in development and erythroid differentiation. Nat Genet 35: 190–194 [DOI] [PubMed] [Google Scholar]
- Perkins AC, Sharpe AH, Orkin SH (1995) Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature 375: 318–322 [DOI] [PubMed] [Google Scholar]
- Pilon AM, Arcasoy MO, Dressman HK, Vayda SE, Maksimova YD, Sangerman JI, Gallagher PG, Bodine DM (2008) Failure of terminal erythroid differentiation in EKLF-deficient mice is associated with cell cycle perturbation and reduced expression of E2F2. Mol Cell Biol 28: 7394–7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay RG, Gonda TJ (2008) MYB function in normal and cancer cells. Nat Rev Cancer 8: 523–534 [DOI] [PubMed] [Google Scholar]
- Reddy CD, Reddy EP (1989) Differential binding of nuclear factors to the intron 1 sequences containing the transcriptional pause site correlates with c-myb expression. Proc Natl Acad Sci USA 86: 7326–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg ML, Sutton SE, Pletcher MT, Wiltshire T, Tarantino LM, Hogenesch JB, Cooke MP (2005) c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Dev Cell 8: 153–166 [DOI] [PubMed] [Google Scholar]
- Sankaran VG, Menne TF, Scepanovic D, Vergilio JA, Ji P, Kim J, Thiru P, Orkin SH, Lander ES, Lodish HF (2011) MicroRNA-15a and -16-1 act via MYB to elevate fetal hemoglobin expression in human trisomy 13. Proc Natl Acad Sci USA 108: 1519–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawado T, Halow J, Bender MA, Groudine M (2003) The beta -globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes Dev 17: 1009–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W (2006) Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 38: 1348–1354 [DOI] [PubMed] [Google Scholar]
- Soler E, Andrieu-Soler C, Boer E, Bryne JC, Thongjuea S, Rijkers E, Demmers J, Ijcken W, Grosveld F (2011) A systems approach to analyze transcription factors in mammalian cells. Methods 53: 151–162 [DOI] [PubMed] [Google Scholar]
- Soler E, Andrieu-Soler C, de Boer E, Bryne JC, Thongjuea S, Stadhouders R, Palstra RJ, Stevens M, Kockx C, van Ijcken W, Hou J, Steinhoff C, Rijkers E, Lenhard B, Grosveld F (2010) The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev 24: 277–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SH, Hou C, Dean A (2007) A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol Cell 28: 810–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W (2006) CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev 20: 2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallack MR, Keys JR, Humbert PO, Perkins AC (2009) EKLF/KLF1 controls cell cycle entry via direct regulation of E2f2. J Biol Chem 284: 20966–20974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallack MR, Whitington T, Yuen WS, Wainwright EN, Keys JR, Gardiner BB, Nourbakhsh E, Cloonan N, Grimmond SM, Bailey TL, Perkins AC (2010) A global role for KLF1 in erythropoiesis revealed by ChIP-seq in primary erythroid cells. Genome Res 20: 1052–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein SL, Menzel S, Peng X, Best S, Jiang J, Close J, Silver N, Gerovasilli A, Ping C, Yamaguchi M, Wahlberg K, Ulug P, Spector TD, Garner C, Matsuda F, Farrall M, Lathrop M (2007) Intergenic variants of HBS1L-MYB are responsible for a major quantitative trait locus on chromosome 6q23 influencing fetal hemoglobin levels in adults. Proc Natl Acad Sci USA 104: 11346–11351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MD, Kremer CS, Ravichandran KS, Rajewsky K, Bender TP (2005) c-Myb is critical for B cell development and maintenance of follicular B cells. Immunity 23: 275–286 [DOI] [PubMed] [Google Scholar]
- Thompson MA, Flegg R, Westin EH, Ramsay RG (1997) Microsatellite deletions in the c-myb transcriptional attenuator region associated with over-expression in colon tumour cell lines. Oncogene 14: 1715–1723 [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W (2002) Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell 10: 1453–1465 [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA (2005) Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell 17: 453–462 [DOI] [PubMed] [Google Scholar]
- Vegiopoulos A, Garcia P, Emambokus N, Frampton J (2006) Coordination of erythropoiesis by the transcription factor c-Myb. Blood 107: 4703–4710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA (2009) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlberg K, Jiang J, Rooks H, Jawaid K, Matsuda F, Yamaguchi M, Lathrop M, Thein SL, Best S (2009) The HBS1L-MYB intergenic interval associated with elevated HbF levels shows characteristics of a distal regulatory region in erythroid cells. Blood 114: 1254–1262 [DOI] [PubMed] [Google Scholar]
- Watson RJ (1988) A transcriptional arrest mechanism involved in controlling constitutive levels of mouse c-myb mRNA. Oncogene 2: 267–272 [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, Pimanda JE, de Bruijn MF, Gottgens B (2010) Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell 7: 532–544 [DOI] [PubMed] [Google Scholar]
- Wong P, Hattangadi SM, Cheng AW, Frampton GM, Young RA, Lodish HF (2011) Gene induction and repression during terminal erythropoiesis are mediated by distinct epigenetic changes. Blood 118: e128–e138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K (2007) MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131: 146–159 [DOI] [PubMed] [Google Scholar]
- Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, Bernstein BE, Fraenkel E, Cantor AB (2009) Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell 36: 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Rappaport AR, Luo W, Wang E, Chen C, Vaseva AV, Shi J, Weissmueller S, Fellmann C, Taylor MJ, Weissenboeck M, Graeber TG, Kogan SC, Vakoc CR, Lowe SW (2011) An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev 25: 1628–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.