Abstract
The activating E2F-transcription factors are best known for their dependence on the Retinoblastoma protein and their role in cellular proliferation. E2F3 is uniquely amplified in specific human tumours where its expression is inversely correlated with the survival of patients. Here, E2F3B interaction partners were identified by mass spectrometric analysis. We show that the SNF2-like helicase HELLS interacts with E2F3A in vivo and cooperates with its oncogenic functions. Depletion of HELLS severely perturbs the induction of E2F-target genes, hinders cell-cycle re-entry and growth. Using chromatin immmunoprecipitation coupled to sequencing, we identified genome-wide targets of HELLS and E2F3A/B. HELLS binds promoters of active genes, including the trithorax-related MLL1, and co-regulates E2F3-dependent genes. Strikingly, just as E2F3, HELLS is overexpressed in human tumours including prostate cancer, indicating that either factor may contribute to the malignant progression of tumours. Our work reveals that HELLS is important for E2F3 in tumour cell proliferation.
Keywords: cell-cycle re-entry, E2F3, HELLS, H3K27me3, prostate tumour
Introduction
The E2F-transcription factor family is well conserved and widely known for its role in proliferation and cell-cycle progression (van den Heuvel and Dyson, 2008). Among this family, E2F1–E2F3 are most intriguing, since these E2Fs directly associate with and are antagonized by the pRB (retinoblastoma) tumour suppressor protein (Burkhart and Sage, 2008). Genetic mouse models provided first evidence that E2F3 largely contributes to pRb-attributed tumourigenic events, such as the reduction of Rb-deficient pituitary adenomas in E2F3-deficient mice (Ziebold et al, 2003), but also preneoplastic lesions of the lung (Parisi et al, 2007). In addition, the loss of E2F3, in combination with E2F1 and E2F2, reduces hyperplasia in intestinal epithelia (Chong et al, 2009). Also in human patients, aberrant E2F3-expression was linked to cancer. In some of these tumours such as invasive urinary bladder carcinoma, it was suggested that the loss of pRB as well as amplification of E2F3 are obligate events for tumourigenesis (Feber et al, 2004; Oeggerli et al, 2004; Hurst et al, 2008). In advanced prostate cancers, E2F3 is highly overexpressed and the highest E2F3 levels determine the worst clinical outcome for the patient (Foster et al, 2004). Apart from inappropriate E2F3-expression, the whole pRB/E2F-pathway is regarded as being deregulated in nearly every human tumour (Hanahan and Weinberg, 2011).
In the past, the transcriptional programmes of specific E2Fs were established after overexpression of E2Fs in mammalian cells and subsequent microarray (DeGregori and Johnson, 2006). These analyses led to the identification of many DNA synthesis, checkpoint control, DNA repair and apoptosis-associated targets and suggested that there are similarities but also specificity among the individual E2Fs (Kong et al, 2007). Using the more sophisticated chromatin immunoprecipitation (ChIP) assays coupled to microarrays, it was concluded that there are functional overlaps between the E2Fs, but also that isoform-specific functions exist (Takahashi et al, 2000; Ren et al, 2002; Asp et al, 2009). Owing to these studies many new E2F-targets were identified, but the impact of most targets for the oncogenic E2F function is still elusive.
Recently, more attempts were made to deconstruct the mechanistic basis of the pRB/E2F complex (Blais and Dynlacht, 2007). Various molecules were identified that are tethered by pRB/E2F, acting both locally and globally, to modify the chromatin of E2F-dependent promoters. Most of the numerous pRB-interacting molecules, such as the histone methyltransferase SUV39H1 contribute to repression. pRB-tethered SUV39H1 leads to heterochromatinization and silencing of S-phase genes (Nielsen et al, 2001; Narita et al, 2003). In order for a cell to proceed through the cell-cycle E2F-dependent promoters have to be inverted from this repressive chromatin state into a state favouring transcription. E2F1–E2F3 commence this transition by recruiting chromatin regulators such as HCF-1 in complex with the mixed-lineage leukaemia (MLL1) protein that possess methyltransferase activity towards histone H3 lysine 4, H3K4 (Tyagi et al, 2007). To alleviate the repressive state of the chromatin, HCF-1 also enlists the histone lysine demethylase PHF8 (Liu et al, 2010). That a majority of promoters bound by PHF8 overlap with E2F-bound promoters was demonstrated by ChIP coupled with deep sequencing technology (ChIP-Seq). Anticipating that all molecules that assist the E2F-transactivating function or revert the state of the chromatin at E2F-dependent promoters harbour immense potential as drug targets in human tumours, we explored such molecules in the context of oncogenic E2F3.
Here, we identified the SNF2-like, ‘lymphoid-specific’ helicase HELLS (also called SMARCA6, PASG or LSH) as a novel E2F3-interaction partner. Previously, it was shown that HELLS is required for the silencing of constitutive heterochromatic regions such as retrotransposons or the pericentromer (Dennis et al, 2001; Sun et al, 2004) via an interaction with DNA methyltransferases (Myant and Stancheva, 2008). Now, we demonstrate that HELLS binds to E2F3 in vivo, aiding induction of E2F-target genes and cell-cycle re-entry. Furthermore, we provide evidence that HELLS, akin to E2F3, is overexpressed in several human tumours. In prostate carcinomas, HELLS/E2F3 co-expression is marking the most aggressive stages. The vast overlap of the identified co-bound promoters suggests that E2F3 and HELLS co-regulate target genes. Notably, a highly HELLS-enriched promoter is the trithorax-related MLL1. We suggest that in cancer cells MLL1, that is important for histone methylation and activation of key cyclin genes, depends itself on E2F3:HELLS.
Results
HELLS is a novel E2F3-associated factor
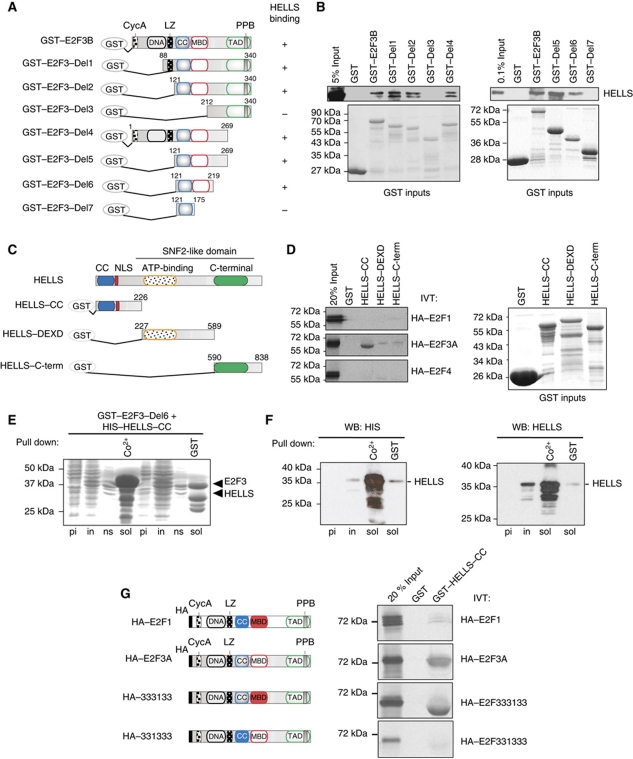
To explore mechanisms that aid in activating E2F-responsive genes, we purified E2F3-containing complexes from highly proliferative HCT116 cells. Bacterially expressed GST-tagged E2F3B was used as bait. The putative interaction partners complexed to E2F3B were separated by SDS–PAGE (Supplementary Figure S1A) and identified by mass spectrometry. We identified >50 candidate interaction partners (Supplementary Table SI) alongside the critical E2F-partner pRB (Burkhart and Sage, 2008), the histone acetyltransferase GCN5 (Lang et al, 2001) and TRRAP (McMahon et al, 1998), validating our approach. As one of the candidates we identified the SNF2-like helicase HELLS. Loss of HELLS function creates several striking phenotypes that resemble those of E2F3-loss. The Hells-deficient as well as the E2f3-deficient mouse embryonic fibroblasts (MEFs) age prematurely with characteristic up-regulation of senescence-associated p19Arf and p21Cip1 (Fan et al, 2003; Aslanian et al, 2004; Sun et al, 2004). Moreover, loss of either protein leads to diminished proliferation rates and enhanced genomic instability (Humbert et al, 2000; Saavedra et al, 2003; Sun et al, 2004). The mass spectrometry results were validated by mapping the domains of HELLS and E2F3 that are required for complex formation using various deletion constructs (Figure 1A–D). As before, GST-pulldowns were performed with HCT116 nuclear extracts incubated with GST–E2F3B full-length protein or its truncated forms (Figure 1A). Interestingly, a region in E2F3–Del6 encompassing both the coiled-coil domain as well as the marked box was essential for the interaction to HELLS (Figure 1B). In contrast, deleting the DNA-binding, leucine-zipper or the transactivation domain did not diminish the binding of E2F3 to HELLS. The E2F3-coiled-coil domain itself is insufficient for HELLS binding. We conclude that a sequence encompassing the marked box and the coiled-coil domain is proficient to interact with HELLS. For the reverse approach, we created constructs (a) with an N-terminal deletion of HELLS containing its coiled-coil domain, (b) enclosing the ATP-binding domain and (c) coding for the C-terminal part of the SNF2-domain (Figure 1C). These three HELLS peptides were created as GST-fusion proteins and incubated with two activator E2Fs, HA–E2F1 and HA–E2F3A, or the repressive HA–E2F4. All E2Fs were 35S-labelled by in-vitro translation, and exposed to the HELLS peptides (Figure 1D). An interaction between the N-terminal part of HELLS and HA–E2F3A was readily observed, but not with the GST controls. We also detected a dramatically reduced association of E2F3A with the other two regions of HELLS. However, the interaction with HA–E2F1 or HA–E2F4 with any of the HELLS constructs was even more reduced, demonstrating that HELLS shows strong preference for E2F3.
Figure 1.
HELLS is a novel E2F3-interacting protein. (A) Schematic representation of GST–E2F3 deletion constructs used for the mapping of the interaction of HELLS with E2F3. A positive interaction was labelled (+), a negative interaction (−). (B) The same constructs shown in (A) were used as baits, incubated with HCT116 extracts and GST-pulldowns performed. HELLS was subsequently detected by western blot (upper panel). The near equal loading of the E2F3 GST-constructs is shown by the Coomassie staining (GST inputs). (C) Scheme of HELLS and the three GST-HELLS constructs used for the mapping. (D) IVT-E2F1, -E2F3A or -E2F4 proteins were probed for their HELLS interaction and separated via SDS–PAGE with subsequent autoradiography (left). The amounts of the fusion proteins were determined by SDS–PAGE following Coomassie staining (right). (E, F) E2F3–Del6 and HELLS-CC co-expressed from a singular vector are capable to interact in vitro. (E) Various bacterial extracts containing GST–E2F3–Del6 and HIS–HELLS–CC were separated via SDS–PAGE and stained with Coomassie before induction (pi), after IPTG induction (in), the non-soluble protein fraction (ns) as well as the soluble fractions (sol). The latter fractions were subjected to pull downs using either metal affinity chromatography resin (Co2+) or GST-beads (GST). (F) The presence of HIS–HELLS in the direct (Co2+) or the indirect (GST)-pulldowns was identified using either a HIS-specific (left) or a HELLS-specific antibody (right). (G) Schematic drawing of E2F1 and E2F3A used for in vitro translation (IVT). In both WT proteins, the major domains are depicted: Haemagglutinin-tag (HA), Cyclin A-binding domain (CycA), DNA-binding domain (DNA), leucine-zipper domain (LZ), coiled-coil domain (CC), marked box domain (MBD), transactivating domain (TAD) and the pocket protein binding domain (PPB). Two swapping mutants HA–333133 and HA–331333 are shown. These E2F3A molecules carry amino-acid exchanges to resemble either the E2F1 CC or MBD. IVT HA–E2F1, HA–E2F3A, HA–333133 or HA–331333 proteins were probed for their HELLS interaction and separated via SDS–PAGE with subsequent autoradiography. Figure source data can be found in Supplementary data.
To confirm the results of the interaction studies, bacterially expressed GST-tagged E2F3–Del6 and the HIS-tagged HELLS CC-domain were used. A single vector co-expression system was applied (Supplementary data) that is capable of co-expressing the peptides simultaneously (Figure 1E). Performing pulldowns with metal affinity beads led to the precipitation of significant amounts of the HIS–HELLS (Figure 1F), but also co-precipitated E2F3–del6. Importantly, using the reverse GST-pulldown, we also detected HELLS-CC by western blotting (Figure 1F), demonstrating that the sole E2F3–Del6 and the N-terminus of HELLS are sufficient to interact. Moreover, using the same co-expression system, a ternary interaction between E2F3:HELLS:DP2 was confirmed (Supplementary Figure S1C–E). Although this is not quantifiable, the simultaneous co-expression of all three molecules seems to stabilize the E2F3:HELLS interaction.
To address which sequences within E2F3–Del6 (E2F3-coiled-coil domain or E2F3-marked box) are essential to provide specificity to the E2F3:HELLS binding interface, specific amino-acid exchanges were introduced. Similar to previous studies (Halstrom and Nevins, 2003), the E2F3-coiled-coil or marked box were swapped with the respective E2F1 domains. Surprisingly, the E2F3-swapping mutant containing the E2F1-coiled-coil domain (331333) was unable to stabilize the complex with HELLS (Figure 1G). The inability of 331333 to interact with HELLS is not due to a general misfolding effect since this peptide interacts with DP2 efficiently (Supplementary Figure S1F and G). This approach clearly demonstrates that the E2F3-marked box may enhance the interaction (Figure 1B), but it is the E2F3-coiled-coil domain that is essential for the specificity in the interaction of E2F3 with HELLS.
HELLS interacts with E2F3 in vivo
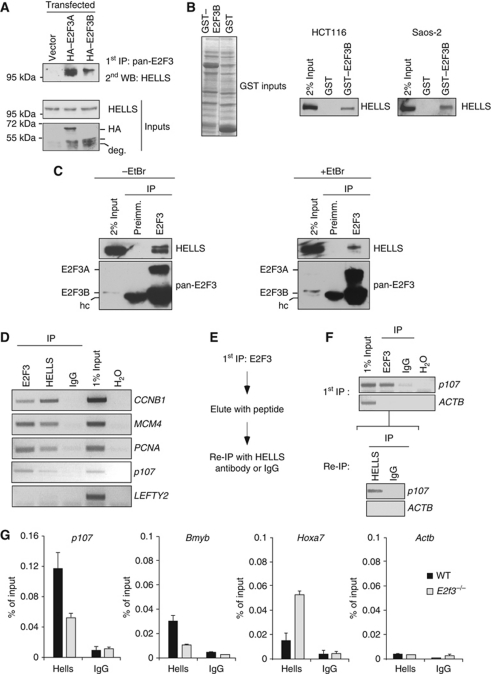
To substantiate that the in vitro interaction between HELLS and E2F3 is meaningful, we analysed a possible complex formation in vivo. To be able to detect both existing E2F3 isoforms, we created an antiserum recognizing E2F3A and E2F3B specifically, but neither E2F1 nor E2F4 (Supplementary Figure S2A). After overexpression of the HA-tagged E2F3 isoforms in HeLa cells (Figure 2A), both were precipitated with the pan-E2F3 antiserum. HELLS was complexed to HA–E2F3A or HA–E2F3B to a similar degree. As the critical coiled-coil domain of E2F3 is identical in both proteins, this likely explains this finding. Since E2F3B is known to interact with pRB just as E2F3A (Leone et al, 2000), we assayed if the E2F3:HELLS complex formation depends on this protein. In independent GST–E2F3B pulldowns, we detected HELLS by western blot using extracts of either the HCT116 or the pRB-deficient SAOS-2 cell line (Figure 2B), but not in GST controls. Hence, the interaction between E2F3 and HELLS is not per se dependent on the presence of pRB. Next, we raised the question if endogenous E2F3:HELLS complexes exist. Using pan-E2F3 antiserum, we consistently co-immunoprecipitated HELLS alongside E2F3A or E2F3B in HCT116 cells, but not using preimmune serum, indicating that the interaction occurs in vivo (Figure 2C). These analyses were also performed in the presence of ethidium bromide, resulting in decreased amounts of co-precipitated HELLS. This decrease is consistent with the idea that endogenous E2F3:HELLS complexes partly depend on or are bridged via chromatin (Figure 2C). This finding is consistent with the ability of E2F3, DP2 and HELLS to form ternary complexes and prompted us to question if HELLS contributes to the regulation of E2F-dependent targets. We prepared chromatin from HCT116 cells growing asynchronously and performed ChIP assays using antibodies specific for pan-E2F3, HELLS or control IgG. Clearly, both E2F3 and HELLS were found on specific genomic regions, consistent with chromatin interactions, but no enrichment was seen if control serum was used. Not only E2F3, but also HELLS was detected at promoters of cell-cycle genes such as CCNB1, MCM4, PCNA or p107 but not at non-E2F-targets (Figure 2D). Since both factors bind E2F-associated promoters, Re-ChIP analyses were performed to address if endogenous E2F3:HELLS complexes co-occupy selected promoters. The Re-ChIP is a modified ChIP procedure (Figure 2E), whereby E2F3-bound chromatin is collected and probed for its enrichment for E2F-dependent promoters such as p107 (Figure 2F). The E2F3-bound chromatin is eluted with a surplus of E2F3 peptide and used for an additional ChIP using the HELLS-specific antibody. Importantly, HELLS was detected at the p107 promoter, verifying that E2F3 and HELLS can co-occupy this promoter. Next, we questioned if the loss of E2F3 could lead to a change of HELLS binding to E2F-target genes. To do so, we deployed a well-established mouse knockout-line that is deficient in both E2f3a and E2f3b, for simplicity termed E2f3−/− (Humbert et al, 2000). Performing ChIP assays in primary murine fibroblasts (MEFs) derived from E2f3−/− embryos we detected a two-fold reduced occupancy of Hells on the E2F-dependent promoters of p107 or Bmyb as compared with wild-type (WT) MEFs (Figure 2G). The decrease is specific for E2F-regulated target genes since the binding of the HELLS protein to the Hoxa7 promoter, a locus that is repressed by HELLS (Xi et al, 2007), was not diminished. Importantly, the Hox locus is hardly bound by E2F3. The observed reduction of HELLS-associated promoters in E2f3−/− MEFs is restrained and is not caused by altered HELLS expression, which remains unchanged (Supplementary Figure S2B). Based on these data, we suggest that E2F3 and HELLS can be found complexed in vivo and are also likely to be found on certain promoters together. Hence, the loss of E2F3 appears to cause a diminished, but not an abolished, chromatin association of HELLS.
Figure 2.
HELLS binds to E2F3 in vivo and it is also found on E2F-promoters. (A) HeLa cells were transfected with either HA–E2F3A, HA–E2F3B or a control (vector). Nuclear extracts from these cells were immunoprecipitated with a pan-E2F3 antibody and co-precipitated endogenous HELLS was detected by western blot. Both E2F3 isoforms were detected with a HA-antibody and show a band running close to HA–E2F3b, which is a known degradation product (deg.). (B) The interaction of HELLS:E2F3 is not perturbed by the absence of pRB. GST–E2F3B was used as bait, incubated with either HCT116 or Saos-2 nuclear extract, GST-pulldowns performed and HELLS was detected by western blot. The near equal loading of the E2F3 GST-constructs is shown by the Coomassie staining (GST inputs). (C) The HELLS:E2F3 interaction per se is not dependent on chromatin. Endogenous E2F3 was immunoprecipitated from HCT116 in the absence of ethidium bromide (EtBr) or presence of EtBr, which dissociates protein:DNA interactions. E2F3 or E2F3-bound HELLS was detected with an E2F3 or HELLS-specific antibody by western blot. As a control, a corresponding preimmune serum was used. The interaction was reduced but not absent in the presence of ethidium bromide (+EtBr). The heavy chain from the antiserum is labelled (hc). (D) ChIP was performed in HCT116 cells with antibodies against E2F3, HELLS or IgG as a control. The precipitated DNA was subsequently used for semiquantitative PCRs in the promoter regions of the E2F-targets CCNB1, MCM4, PCNA and p107. The LEFTY2 promoter served as a negative control. (E) Scheme for the Re-ChIP procedure. (F) ChIP in HCT116 cells using an E2F3-specific antibody or IgG control. The E2F3 precipitate is eluted afterwards with an excess of the corresponding peptide. The eluate was subsequently immunoprecipitated with a HELLS-specific antibody or an IgG control. For both ChIPs, the binding to the p107 promoter was determined by semiquantitative PCR. The β-actin (ACTB) promoter served as negative control. (G) HELLS is found at the E2F-targets p107 or Bmyb as determined by quantitative ChIP analysis in WT and E2f3−/− MEFs. The Hoxa7 promoter served as E2F independent but HELLS-positive control. The β-actin promoter (Actb) served as a negative control. Figure source data can be found in Supplementary data.
HELLS activity is essential for early cell-cycle entry
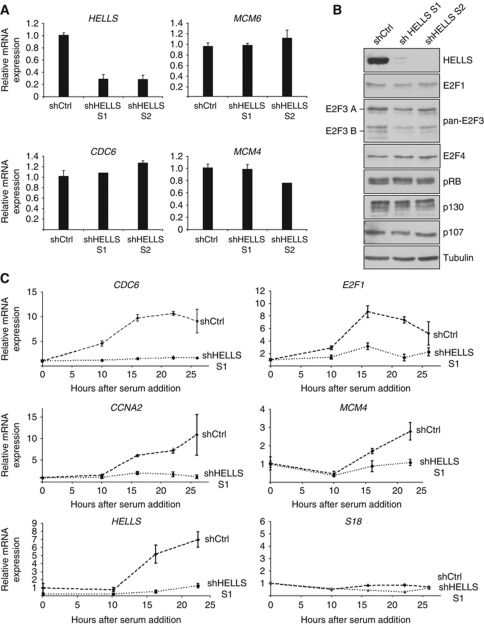
To assess a possible cooperation of HELLS and E2F3 in regulating E2F-target genes, we deployed a lentiviral strategy to deliver small hairpin RNA (shRNA) targeting human HELLS. Two independent shRNAs strongly reduced the HELLS expression in T98G cells also visibly diminishing its protein (Figure 3A and B). Since HELLS-depletion is known to inappropriately sensitize the p53-axis in primary cells (Fan et al, 2003; Sun et al, 2004), it is important to note, that T98G cells have compromised p53 and neither express senescence-associated p14ARF nor p16INK4A (Simon et al, 1999). Hence in T98G cells, HELLS-dependent transcriptional events are observed, which are separable from senescence-associated effects. Yet, depleting HELLS in asynchronously growing T98G cells did not alter the RNA levels of the E2F-targets CDC6, MCM6 or MCM4 (Figure 3A) using quantitative real-time PCR (qRT–PCR). By western blotting, no substantial changes in the expression of the E2F-target p107 or the tested E2Fs were detected (Figure 3B). Likewise, the expression of the other pocket proteins (pRB, p130) were unaffected by HELLS-depletion. The absence of E2F3 has similarly limited effects on E2F-targets in asynchronous cells, but evokes difficulties upon cell-cycle entry (Humbert et al, 2000). For this reason, we used the shHELLS or control-infected T98G cells, synchronized these by serum deprivation and permitted S-phase re-entry upon addition of serum. In accordance with previous results (Raabe et al, 2001), HELLS increases upon cell-cycle re-entry in control cells (Figure 3C) as determined by qPCR. Concomitant with HELLS-depletion, we detected a severely reduced and delayed induction of the E2F-targets CDC6, E2F1, CCNA2 and MCM4, while the ribosomal S18 remained constant. That loss of HELLS impeded the induction of E2F-targets on a transcript level is consistent with a possible role of HELLS in aiding E2F-dependent gene activation. We completed the analysis of cell-cycle regulators by assessing possible disturbances of the cell-cycle machinery at the G1/S transition. The induction of Cyclin D1, as well as the levels of pRB phosphorylated at Serine 780, were unchanged in HELLS-depleted cells (Supplementary Figure S3), except for a slight decrease of pRB in HELLS-depleted cells at peak S-phase. This is likely due to the fact that RB1 itself, like p107, is an E2F-target (Ren et al, 2002). Thus, it appears that the HELLS activity is not critical in asynchronously growing cells but is essential for the induction of E2F-targets upon the immediate cell-cycle entry after quiescence.
Figure 3.
HELLS is dispensable for E2F-target gene expression under asynchronous growth conditions but impairs E2F-targets upon cell-cycle re-entry. (A) Quantitative real-time PCR analysis (qPCR) of lentivirally infected T98G cells using two shRNA sequences against HELLS (shHELLS S1; shHELLS S2). Neither expression of MCM6, nor CDC6, nor MCM4 were deregulated in HELLS-depleted cells in comparison to control infections (shCtrl). The expression of all RNAs was normalized to β-actin and the RNA in control-infected cells was set to 1. (B) Western blot analyses after HELLS-depletion as in (A). The expression of cell cycle-related proteins were analysed using antibodies recognizing E2F1, pan-E2F3 (against both E2F3 isoforms), E2F4, pRB, p130 and p107. α-Tubulin served as a loading control. (C) Knockdown of HELLS using sh-lentivirus (as in A) leads to altered target gene expression. All cells were serum-starved, reintroduced into cell cycle by addition of 20% serum and the expression of the indicated genes was measured by qPCR. S18 served as a control gene. Figure source data can be found in Supplementary data.
Loss of HELLS activity cannot be overcome by forced E2F3-expression
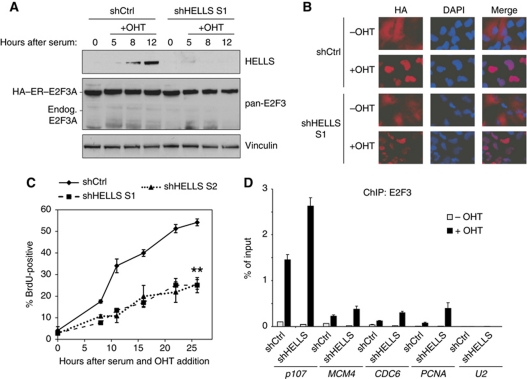
Since we showed that HELLS activity at promoters appears sensitive to the presence or absence of E2F3, we tested if a strong E2F3 overexpression could compensate for an absence of HELLS to permit E2F-target gene induction. We addressed this by constitutively expressing a 4-hydroxytamoxifen (OHT)-inducible HA–ER–E2F3A chimera in T98G cells. The oestrogen receptor (ER) allows a rapid translocation of HA–ER–E2F3A into the nucleus after OHT-treatment. This system was used to demonstrate that E2Fs recruit TIP60 to promoters of E2F-targets (Taubert et al, 2004). These T98G–HA–ER–E2F3 cells were depleted of HELLS using sh-lentivirus, serum-deprived and then induced with OHT in addition to serum (Figure 4A). As expected (Figure 3C), we were only able to observe an induction of HELLS in control-infected cells after serum addition, but not if HELLS is depleted. The exogenous HA–E2F3A was abundantly present (Figure 4A) and only nuclear upon OHT/serum induction. This translocation was observed within only 1 h of OHT-induction and was not perturbed in either the presence or absence of HELLS (Figure 4B). More interestingly, we found that the depletion of HELLS in these E2F3A-overexpressing cells still led to a statistically highly significant delay in entry into S-phase as measured by BrdU-labelling (Figure 4C). Since, the HELLS activity is essential for immediate E2F-target gene induction and also S-phase induction, even after an exposure to OHT/serum for 26 h, we questioned if E2F3A needs HELLS to appropriately bind to its target promoters. As anticipated, endogenous E2F3 (i.e., E2F3A/B) was detected at target promoters in the absence of OHT of control-infected cells (Figure 4D). Performing ChIPs in the presence of OHT/serum resulted in a massive surge of promoter-bound E2F3 (i.e., exogenous E2F3A). Moreover, depleting HELLS led to a similar or even slightly increased levels of E2F3 (i.e., E2F3A) at the promoters of the investigated E2F-target genes. The inability of exogenous E2F3A to compensate for the loss of HELLS despite its strong overexpression and proficient chromatin binding is surprising. The lack of induction of MCM4 or CDC6 (Figure 3C), despite detectable chromatin affinity (Figure 4D) is remarkable and we conclude that only the activating function of E2F3 is strictly dependent on the presence of HELLS, but not its recruitment to chromatin.
Figure 4.
Cell-cycle re-entry critically depends on the activity of HELLS. (A) Stable T98G HA–ER–E2F3A cells were infected with either shHELLS S1 or shCtrl, serum-starved and either untreated or treated with a combination of 20% serum and 300 nM OHT. (A) Infected T98G HA–ER–E2F3A cells were induced with OHT/serum and harvested at the given time points. An even exogenous E2F3A expression was verified by western blotting. Endogenous (endog.) E2F3A is hardly detectable. The induction or knockdown of HELLS was analysed with a HELLS antibody. Vinculin served as a loading control. (B) The same cells of (A) were used to observe the proper nuclear translocation of the HA–ER–E2F3A protein by immunofluorescence using an antibody recognizing the (HA) epitope tag. Nuclei were counterstained with DAPI and the overlap of both stainings is shown (Merge). (C) shHELLS S1, shHELLS S2 or shCtrl-infected cells were assayed for BrdU incorporation after serum stimulation and OHT-treatment. The data points represent the average ±s.e.m. from three independent experiments (**P<0.01, Student's t-test for both shRNAs). (D) The cells were treated similar to (A) but were processed for ChIP using pan-E2F3 antibody to investigate the recruitment of E2F3 after 1 h of OHT/serum (+OHT) or ethanol/serum (–OHT) treatment as solvent control. The relative binding to the E2F-target genes p107, MCM4, CDC6 or PCNA was determined by qPCR. The U2 promoter served as a negative control in this experiment. Figure source data can be found in Supplementary data.
HELLS and E2F3 cooperate in prostate cancer
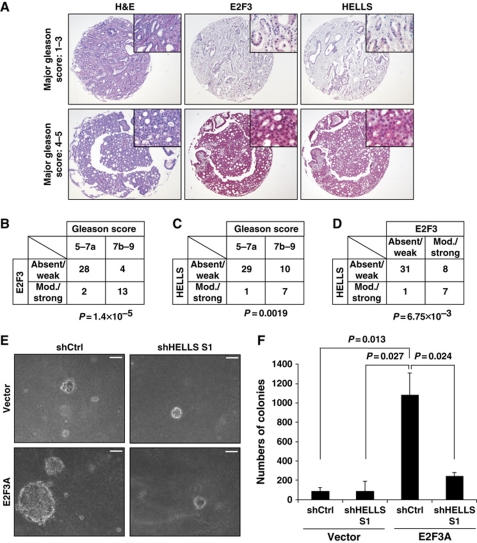
Perhaps the most striking example that inappropriate levels of E2F3 provoke human tumours is prostate cancer. Here, high E2F3-expression independently predicts a poor overall survival or clinical outcome for patients (Foster et al, 2004). To elucidate if the oncogenic E2F3 function also critically depends on HELLS in vivo, we screened biopsies from a total of 47 prostate cancer patients for E2F3 and HELLS expression. The separate patient tumour tissue cores were assembled into prostate cancer tissue arrays to allow multiplex histological analysis. These arrays were screened with specific antibodies recognizing either pan-E2F3 or HELLS. Next, a grading of the staining intensities was categorized according to the Gleason score, the standard score for prostate cancers (Figure 5A). In accordance with the previous results (Foster et al, 2004), we detected a weak or absent E2F3 staining intensity in tumours with a low Gleason score of 5–7a and a moderate-to-strong E2F3 staining in the tumours with a high Gleason score of 7b–9 (Figure 5B). When analysed in a semiquantitative manner, we were able to detect a moderate-to-strong E2F3 staining in only two samples (6%) of the 30 tumours with low Gleason score. In contrast, 13 (76%) of the 17 tumours with high Gleason score showed a moderate-to-strong E2F3 signal. Strikingly, we observed the same correlation for HELLS with weak or absent staining in 29 (97%) out of 30 tumours with low Gleason score and a moderate or strong HELLS staining in 7 (41%) of 17 for the more aggressive tumours with a Gleason score of at least 7b (Figure 5C). Furthermore, the co-expression of HELLS and E2F3 in the respective tumours is statistically significant (Figure 5D). There is a general trend for E2F3 and HELLS co-expression in specific other solid tumour entities that we tested including bladder, retina, lung and ovary (Supplementary Figure S4A and B), but the most significant positive correlation is still detected in the prostate cancers, where the intensity of E2F3 and HELLS co-staining is linked to the most aggressive tumour stages. These results imply that the activity of the E2F3:HELLS complex promotes tumourigenesis and aggravates prostate cancer malignancy. To understand why HELLS is elevated in advanced human prostate tumours, we assessed if the HELLS gene itself might be an E2F-dependent transcriptional target. Indeed, in MEFs that were derived from pRB-deficient embryos, Hells is elevated as compared with WT MEFs, which also translates into higher amounts of the protein (Supplementary Figure S4C and D). In silico, we identified E2F-sites at the promoter of Hells and performed ChIPs revealing that E2F1, E2F3 and E2F4 are all recruited to the Hells promoter (Supplementary Figure S4E and F). That Hells is an E2F-target, was recently corroborated for E2F1 (Niu et al, 2011) and since pRB/E2F is deregulated in many human tumours (Hanahan and Weinberg, 2011), this likely has far reaching consequences. Our results prompted us to assess if a reduction of the HELLS activity could serve as a future therapeutic measure. To do so, we depleted HELLS in a prostate cancer cell line, DU145, and performed soft agar assays. This type of assay measures anchorage independent survival and is considered a stringent test for the malignant transformation of cells. The strong lentiviral knockdown of HELLS had no effect on the E2F3A expression (Supplementary Figure S4G) and had surprisingly little effect on the colony forming capacity of the cells (Figure 5E). Consistent with its oncogenic function, the lentiviral-transduced E2F3A strongly promoted growth in the semisolid environment, as indicated by the number of large colonies in soft agar (Figure 5E and F). More importantly, the concomitant HELLS-depletion reversed the capacity of E2F3 to promote transformation. Combined, this suggests that the HELLS activity is a critical component of E2F3's ability to promote the malignant progression of prostate tumours.
Figure 5.
Increased E2F3 and HELLS expression correlate with the aggressiveness in prostate cancer. (A) Representative examples of patient biopsies of prostate cancer (PCA) assembled into a tissue microarray. Representative sections were analysed by haematoxylin and eosin (H&E) and also used for immunohistochemistry to detect pan-E2F3 or HELLS. The staining was developed with the Fast Red substrate and counterstained with haematoxylin. (B) Summary of the semiquantitative expression of E2F3, and of HELLS (C) with the Gleason score of the respective PCA. The less aggressive tumours were grouped in 5–7a and the most aggressive ones in 7b–9. This distinction correlated with the respective expression of E2F3 and HELLS. The P-value was calculated using the Fisher's exact test. (D) Mutual relationship between the semiquantitative E2F3 and HELLS expression of all tumours. (E, F) The DU145 PCA line was retrovirally infected with E2F3a or infected with an empty virus (Vector). The ensuing lines were both infected with lentiviral shHELLS S1 or shCtrl (scramble control). All cell lines were grown in soft agar for 2–3 weeks. (E) Phase contrast pictures of representative colonies are shown. (F) The ensuing colonies of a minimum diameter of 100 μm were counted. The P-values were calculated with the Student's t-test and are displayed above the bar graph.
HELLS and E2F3 are enriched at a large fraction of promoters
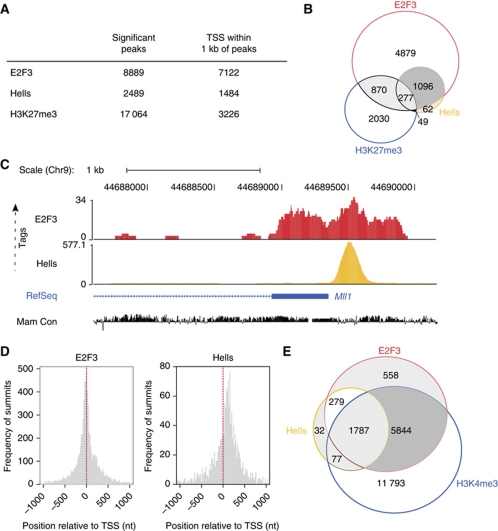
The joint regulation of cell-cycle entry by E2F3 and HELLS and their co-expression in human tumours are strong evidence that these proteins share functional overlaps. To reveal the full extent of the genome-wide chromatin association of E2f3 and Hells, we applied ChIP-Seq in mouse fibroblasts. Since Hells was previously reported to localize to heterochromatin (Yan et al, 2003) and enforce DNA methylation and silencing of selected Hox genes (Xi et al, 2007), we expected to detect Hells also at inactive, repressed loci. Such regions are also targets of the polycomb group-mediated repression and characterized by trimethylation of histone H3 at lysine 27, H3K27me3 (Sawarkar and Paro, 2010). Thus, the genome-wide distribution of Hells and E2f3 was assessed in WT MEFs and compared with that of the polycomb-associated, repressive H3K27me3 mark. The ChIP-Seq data were mapped to the genome and subjected to a bi-modal transcription factor type peak analysis to identify enriched regions (Supplementary data). The highest number of significant peaks within 1 kb of RefSeq TSSs (TSS) were found for E2f3 (Figure 6A; Supplementary Tables SII and SIII). Importantly, only 22% of the Hells targets coincide with H3K27me3, but 93% of all Hells-associated TSS coincide with E2f3-bound TSS (Figure 6B). These findings indicate that Hells is more likely to co-bind E2f3 targets than the H3K27me3-enriched, presumably ‘repressed’ promoters. For a graphical overview of E2f3 and Hells peaks, we chose the chromosomal context of Mll1 (Figure 6C) because for Hells it is the highest-ranking region according to the ChIP-Seq data (Supplementary Table SIII). The ChIP-Seq data confirm our conventional ChIP analysis that Hells and E2f3 co-occupy overlapping sets of promoters. Knowing that E2Fs have strong preference for genomic regions closest to the TSS (DeGregori and Johnson, 2006), we assessed the global positional preference of E2f3 and Hells (Figure 6D). As anticipated, E2F3 sharply concentrates at TSS. Also for Hells, the ChIP-Seq summits clustered closely to the TSS, but with a preference towards a region of about 120 nucleotides downstream of the TSS. Having observed that there is a global overlap of Hells and E2F3-bound targets, we determined if the co-bound target genes are likely to be active or repressed by assessing the overlap with previously mapped regions that are enriched for histone H3 trimethylated at lysine 4, H3K4me3 (Mikkelsen et al, 2007). As expected, the majority of E2f3 targets are also enriched for H3K4me3 (Figure 6E). Importantly, 86% of the Hells targets are likely to be actively transcribed, since they also show H3K4me3 enrichment at their promoter.
Figure 6.
ChIP-sequencing reveals that E2f3 and Hells share many common target genes. (A) Results of the ChIP-sequencing (ChIP-Seq) for WT MEFs. Genome-wide counts of significant ChIP-Seq peaks for E2f3, Hells and H3K27me3. The number of transcriptional start sites (TSS) within 1 kb of peaks is also given. (B) Venn diagram depicting the contingency between the number of TSS within 1 kb of E2f3, Hells or H3K27me3 ChIP-Seq peaks in WT MEFs. (C) Tag profiles of E2f3 and Hells in WT MEFs in a screenshot from the UCSC genome browser displaying number of ChIP-Seq tags mapping to the Mll1 genomic context. Tag profiles were created by extending each read to a length of 100 bases, and normalizing to 10 million mapped reads. RefSeq transcripts and mammalian sequence conservation (Mam Con) is also shown. (D) Positional preference of the ChIP-Seq peaks for E2f3 and Hells in WT MEFs. Positions are given in nucleotides (nt), and negative values correspond to locations upstream of the TSS. Counts are binned in 20 nucleotide windows. Red-dashed lines indicate the positions of the TSS relative to the peaks. (E) Venn diagram depicting the overlap of E2f3 and Hells-bound promoters (RefSeq) with promoters that are enriched for the trimethylated histone H3 on Lysine 4 (H3K4me3) in WT MEFs.
The activities of HELLS and E2F3 are essential to induce MLL1 and other targets to create a proliferative circuitry
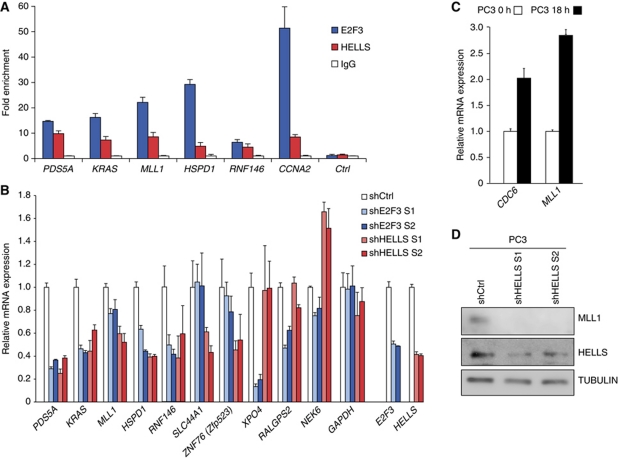
Next, confirmatory ChIP analysis was performed in DU145 prostate cancer cells, since here the E2F3:HELLS activity is physiologically relevant. Consistently, both E2F3 and HELLS were enriched at the promoter of MLL1 and other ChIP-Seq targets such as PDS5A, KRAS, HSPD1 and RNF146 (Figure 7A; Supplementary Figure S5). We used the established E2F-target CCNA2 that was previously shown to depend on HELLS (Figure 3C), as a positive control. Also in the mouse fibroblasts, Hells ChIP-Seq reads were found at the Ccna2 promoter (Supplementary Figure 6C). Next, we used human prostate carcinoma cell lines again to determine if in this context the HELLS activity would affect the expression of MLL1 or any of the other targets (Supplementary Table SII). Either HELLS or E2F3 was acutely depleted by lentiviral shRNA and the knockdown of E2F3 or HELLS was verified using qRT–PCR. Importantly, HELLS-depletion led to a reduction of MLL1 and also of PDS5A, KRAS, HSPD1, RNF146, SLC44A1, ZNF76 (Mus musculus Zfp523), XPO4 and RALGPS2. Only NEK6 expression was elevated upon HELLS loss (Figure 7B). This is in stark contrast to the effect of Hells loss on MCM4/6 or CDC6 shown before (Figure 3A). Importantly, also E2F3 is needed for normal MLL1 levels and also regulates the expression of most of the other targets, except SLC44A1. Since we also detected a diminished expression of MLL1 and other HELLS targets in T98G cells after HELLS-depletion (Supplementary Figure S7A–C), we assessed whether the expression pattern of MLL1 resembles that of typical E2F-targets. In synchronized human prostate cancer cells, MLL1 was induced more strongly than CDC6 upon serum-induced cell-cycle re-entry (Figure 7C). Given that the expression of the MLL1 RNA not always correlates with the protein level (Liu et al, 2007), we tested the effect of HELLS-depletion upon the expression of the MLL1 protein in the same prostate cells and clearly found MLL1 to be reduced (Figure 7D). Furthermore, the Mll1 RNA and protein also declined in E2f3-deficient mouse cells, where it is associated with a reduction of H3K4me3. In accordance to the ChIP-Seq data, the Mll1 promoter is indeed bound by Hells in both WT and E2f3-deficient cells (Supplementary Figure S7D–F). Combined, our data potently support that the top-ranking ChIP-Seq targets of HELLS, among them MLL1, are HELLS targets in vivo. Importantly, Mll1, Kras and Rnf146 are Hells-regulated targets in both mouse and human cells. Lastly, both E2F3 and HELLS are necessary for proper activation of MLL1 and other targets.
Figure 7.
E2F3 and HELLS regulate many common targets, most notably MLL1. (A) Confirmatory ChIP analyses of HELLS target genes in DU145 prostate carcinoma cells. The enrichment of E2F3 and HELLS was assessed using PDS5A, KRAS, MLL1, HSPD1 and RNF146 primers spanning the genomic regions around the TSS. CCNA2 is an E2F-dependent promoter and U2 served as a negative promoter (Ctrl) and IgG served as antibody control. (B) qRT–PCR (qPCR) analysis depicting the relative expression of representative HELLS target genes in human prostate carcinoma cells (PC3) after lentiviral depletion of E2F3 or HELLS using two independent sequences each. The knockdown was verified by qPCR analysis of E2F3 and HELLS. The transcriptional regulation of PDS5A, KRAS, MLL1, HSPD1, RNF146, SLC44A1, ZNF76 (M. musculus Zfp523), XPO4, RALGPS2 and NEK6 was analysed. GAPDH served as a control. (C) MLL1 was analysed by qRT–PCR in synchronized PC3 cells, which were serum-arrested and harvested at 0 or 18 h after serum addition. CDC6 is a S-phase-specific control. (D) Effects of depleting HELLS in PC3 cells determined by western blots for MLL1, HELLS and TUBULIN, using two independent hairpins (shHELLS S1 or S2) or control infections (shCtrl). Figure source data can be found in Supplementary data.
Hells still binds to targets in the absence of E2f3
Since either E2f3 or Hells is essential for proper gene activation and the Hells association to certain E2F-targets seem to decline (Figure 2G), one hypothesis would be that the Hells association to E2F-targets depends on E2F3. Thus, to address this question, Hells ChIP-Seq were performed in E2f3−/− MEFs (Supplementary Tables SII and SIII). Even in the absence of E2F3, Hells was found to localize to a large set of TSS (Supplementary Figure S5A). The number of Hells targets increased in the E2f3−/− MEF ChIP-Seq data, as compared with WT MEFs. However, the lack of further data precludes a statement about the relative numbers of Hells targets in the two cell types. Importantly, 70% of the TSS bound in the WT condition are also found in the mutant condition (Supplementary Figure S5B). Differently from E2F3, which exhibited many targets with high ChIP-Seq peaks, only few Hells targets showed peaks that yielded high statistical significance. This holds true for Hells targets in both WT and E2f3−/− MEFs.
This finding and the low enrichment for Hells in ChIP (Figure 2G) suggest that Hells may bind chromatin differently than E2f3, which binds sequence specifically (Supplementary Table SIV). Thus, we reanalysed the data by determining normalized tag density relative enrichment to IgG (Supplementary Figure 7G), confirming that Hells exhibited enriched tag densities around the TSS. Upon loss of E2f3, the relative tag density enrichment of Hells appears shifted towards a location that is more downstream of the TSS. This shift may explain why in the absence of E2f3, we detected a minor decrease of Hells recruitment using amplicons that best fitted the E2f3 positioning (Figure 2G; Supplementary Figure S6). Combined these results suggest, in accordance with our prior findings, that Hells associates with regions around the TSS at specific target promoters even in the absence of E2f3, but may shift its position at some promoters.
Discussion
In studies using knockout mice, E2f3 is considered the most critical E2f for the control of cellular proliferation, the induction of target genes but also survival (Humbert et al, 2000; Tsai et al, 2008). Our screen for E2F3-interactors adds a new aspect: the SNF2-like helicase HELLS is specifically complexed to E2F3 in vivo but not to other E2Fs (Figure 1G), thus loss of HELLS, like E2F3, creates cell-cycle defects. We show that Hells binds well to either the E2F3A or the E2F3B isoform (Figure 2A and C), which is congruent with both isoforms contributing to G1/S-specific gene expression and proliferation (Danielian et al, 2008; Chong et al, 2009). The E2F3 isoforms have different N-termini and are differentially regulated during the cell cycle (Leone et al, 2000), but they have identical CC-domains and they have largely overlapping functions in vivo. HELLS depends on this E2F3-CC-domain for complex formation (Figure 1G). Murine cells deficient for E2f3a and E2f3b, as well as Hells mutant cells show inappropriate up-regulation of the p19Arf–p53–p21Cip1 network (Fan et al, 2003; Aslanian et al, 2004). Although initially E2f3b was regarded to be the sole repressor of p19Arf, this now seems incorrect (Danielian et al, 2008). In this study, E2f3 ChIP-Seq data established with an antibody recognizing E2f3a and E2f3b show a large overlap with Hells-bound promoters. Importantly, this led to the identification of unanticipated, physiologically relevant E2f3:Hells ChIP-Seq targets (e.g., Kras or Mll1). By conventional ChIP and in the tag density representation (Supplementary Figure S6), Hells was also detectable at classical E2f-targets (e.g., CCNA2), at mostly E2f3b-controlled promoters (p19ARF and other CDK-inhibitors; Aslanian et al, 2004) as well as some of the classical Hells targets (as Hoxa5). The poor ChIP quality of E2f3 isoform-specific antibodies has precluded a more detailed analysis of isoform-specific target genes (Aslanian et al, 2004; own observation).
Here, we propose that Hells largely aids E2f3a in the activation of specific target genes (Figures 3C and 7B). This is supported by the S-phase expression of Hells (Figure 3C), the ability to form ternary complexes with E2F3:DP2 (fostering the ability to bind to DNA), the largely overlapping target set, the presence of an E2F-like T[CG][CG]CGC motif in 74% of all Hells-bound promoters and the fact that a massive surge of E2F3A at E2F-promoters cannot compensate for the loss of HELLS in target gene and S-phase induction (Figure 4C and D). Our findings contrast that Hells acts solely as a repressor involved in long-term silencing of heterochromatic regions (Huang et al, 2004) or Hox genes (Xi et al, 2007). This is supported by the fact, that HELLS overexpression does not hinder cell-cycle re-entry (Supplementary Figure S8). A positive role for Hells in aiding transactivation is also supported by the fact that most Hells-bound promoters overlap with the active H3K4me3 and not the repressive H3K27me3 histone mark. Intriguingly, HELLS binds via its N-terminus to E2F3 (Figure 1D), the same domain that is responsible for recruitment of the DNA methyltransferase DNMT3B, which in turn is complexed with HDAC1/2 and DNMT1 to silence specific genomic targets (Zhu et al, 2006; Myant and Stancheva, 2008; Myant et al, 2011). The fact that they bind to the same region of HELLS, may lead under physiological conditions to competition between E2F3 and the DNMT/HDAC complex for the available pools of HELLS for the co-recruitment to genes that are to be activated by E2F3A. This view is supported by the fact that E2F3 is hardly detected at certain HELLS/DNMT3B-bound Hox genes (Xi et al, 2007) by ChIP assays (Supplementary Table SIII) and that HDAC1 can only be detected in quiescent cells at E2F-regulated promoters in a pocket protein-dependent manner (Rayman et al, 2002). Since E2f3-loss might shift the equilibrium, we monitored Hells chromatin recruitment in cells devoid of E2f3 activity. In these cells, Hells still bound to a large set of promoters, but we did not observe a larger overlap to H3K27me3 as compared with WT cells. Importantly, we rather counted more than less Hells-bound promoters with the E2f3-like motif, 95% instead of 74% (Supplementary Table SIV). These findings are consistent with the idea that Hells chromatin association, for example, at classic Hox-regulatory elements, is not restrained by E2f3. The ChIP-Seq data support that Hells binds chromatin E2f3 independently, but we have indications suggesting that its chromosomal localization at some promoters is altered in the absence of E2f3 and/or loses part of the signal at the TSS in the case of Kras and Rnf146 (Supplementary Figure S7G). If Hells in conjunction with E2f3 changes its localization in a context-dependent manner, this could lend further support to the idea of Hells acting as a chromatin remodeller. In the future, it will be important to determine if and what kind of Hells-shifts can be found at specific promoters.
We are not adverse to the idea that Hells also acts as a repressor, since the ability to function as activator and repressor of transcription is also found for other SWI/SNF molecules (Bell et al, 2011). Here, we imply a model based on the interplay between Hells and E2f3a/b in which the interaction of E2f3a/b and the promoter depends on its sequence (containing the E2f3-binding site) and this process coincides with Hells recruitment, facilitating transcriptional activation. It remains to be seen if Hells recruitment leads to a nucleosome sliding or if specific histone variants are involved to explain how transcription of genes such as Mll1 might be initiated within the cell cycle.
An important insight of our study is that E2F3:HELLS leads to sustained proliferation in human tumours. Consistent with this finding, we show that the elevated level of E2F3 is mirrored by high HELLS expression in specific human tumour entities (Supplementary Figure S4A and B), likely owing to the fact that HELLS itself is an E2F-target (Supplementary Figure S4C–F). Since the full E2F3-driven transformation capacity of prostate tumour cells also depends on HELLS, it is consistent with the high levels of E2F3:HELLS found marking the most advanced stages of prostate tumours. Combined, we infer that the E2F3:HELLS connection fuels the progression of tumours (Figure 5), making HELLS an attractive therapeutic target. Interestingly, HELLS was recently identified as a progression marker in a rare human tumour (Waseem et al, 2010). All our results combined, we present HELLS as a novel member of the pRB/E2F-pathway. The activity of HELLS plays a surprising positive role on E2F3-promoters to allow transcriptional activation and proliferation. This analysis is the first to link HELLS with E2F-controlled processes that are critical to establish a proliferative tumour circuitry. This study highlights that ChIP-Seq is the method of choice to obtain an unambiguous picture of the crosstalk between chromatin-associated factors. Among the many newly identified E2F3:HELLS target genes, a target such as MLL1 may provide a mechanistic explanation into the transformation and growth phenotypes of E2F3. Lastly, identifying HELLS as a partner of E2F3 might prove helpful in the development of new targets for future drug developments against specific E2F3-propelled malignancies.
Materials and methods
Cell culture and serum starvation
All cells were cultured in DMEM plus 1% Pen/Strep and 10% FCS. For serum starvation experiments, cells were kept without serum for 48–72 h and re-stimulated with medium containing 20% serum.
GST-pulldowns and co-immunoprecipitations
For a GST-pulldown experiment, 250–400 μg nuclear extract was incubated with 2 μg GST protein bound to GSH sepharose for 2 h at 4°C. The beads were washed four times with HEGN150, boiled in SDS sample buffer and separated by SDS–PAGE. For GST-pulldown coupled to mass spectrometry 4.5 mg of nuclear extract and 40 μg of GST protein were used. GST-IVT 1 μl from the 35S-Methionine-labelled IVT reaction (TNT Translation System, Promega) was used in a volume of 500 μl HEGN150 supplemented with 0.5 mg/ml BSA and detected by autoradiography after SDS–PAGE. Immunoprecipitations were performed with HEGN buffer containing 1% NP-40. In all, 250–600 μg of nuclear extracts was incubated on a rotating wheel overnight with 4 μg antibody. The next day, 20 μl of Protein A sepharose was added for 1 h and the beads were washed three times with HEGN150 (containing 1% NP-40).
Immunofluorescence and BrdU assay
Cells were plated on glass coverslips and fixed the next day with 4% PFA, 2% sucrose. Cells were washed three times with PBS permeabilized for 10 min with PBS +0.5% Triton X-100. After washing, the cells were blocked for 30 min with blocking solution with 5% BSA. The coverslips were incubated with the primary antibody diluted in blocking solution at 4°C. Next day, the coverslips were washed and incubated with the secondary antibody for 30 min at room temperature, washed again, counterstained with DAPI and mounted. For BrdU assays, the cells were pulse-labelled with 50 μM BrdU for 2 h and processed as described above, with minor modifications as the primary antibody was incubated at 37°C and the blocking solution was supplemented with 3 mM MgCl2 and 100U DNase I/ml.
ChIP and ChIP-Seq data
The procedures of the ChIP and Re-ChIP assays are found in the Supplementary data. The ChIP-sequencing data from this publication have been submitted to the Gene Expression Omnibus database http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi and assigned the identifier GSE33791.
Supplementary Material
Acknowledgments
We thank our colleagues from the Hagemeier, Helin, JA Lees, Nevins, Eilers and Gaubatz laboratories for reagents. Our special thanks go to Svetlana Lebedeva and Wei Chen. The Protein Sample Production Facility at the MDC is funded by the Helmholtz Association of German Research Centres. We thank J Tischer and T Dornblut for excellent technical assistance. This projected was supported by a KKP-Charité grant (C/09/04). UZ was funded by a Marie Curie Excellence Grant (FP6-2658 ES-TRAP).
Author contributions: All authors contributed to the work presented in this paper. NR and UZ supervised the project. BvE designed and performed the experiments, analysed the data and wrote an initial draft; MDT built initial constructs and validated them in vitro. AS created constructs and performed the interaction studies; SM, KM and JM designed and performed the experiments; CL graded and screened human tumours. AO performed the mass spectroscopic analysis. JM and NR analysed all ChIP-Seq data. UZ designed the experiments, analysed the data and wrote the paper. All authors reviewed the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aslanian A, Iaquinta PJ, Verona R, Lees JA (2004) Repression of the Arf tumor suppressor by E2F3 is required for normal cell cycle kinetics. Genes Dev 18: 1413–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asp P, Acosta-Alvear D, Tsikitis M, van Oevelen C, Dynlacht BD (2009) E2f3b plays an essential role in myogenic differentiation through isoform-specific gene regulation. Genes Dev 23: 37–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell O, Tiwari VK, Thomä NH, Schübeler D (2011) Determinants and dynamics of genome accessibility. Nat Rev Genet 12: 554–564 [DOI] [PubMed] [Google Scholar]
- Blais A, Dynlacht BD (2007) E2F-associated chromatin modifiers and cell cycle control. Curr Opin Cell Biol 19: 658–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart DL, Sage J (2008) Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer 8: 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JL, Wenzel PL, Sáenz-Robles MT, Nair V, Ferrey A, Hagan JP, Gomez YM, Sharma N, Chen HZ, Ouseph M, Wang SH, Trikha P, Culp B, Mezache L, Winton DJ, Sansom OJ, Chen D, Bremner R, Cantalupo PG, Robinson ML et al. (2009) E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature 462: 930–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Friesenhahn LB, Faust AM, West JC, Caron AM, Bronson RT, Lees JA (2008) E2f3a and E2f3b make overlapping but different contributions to total E2f3 activity. Oncogene 27: 6561–6570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGregori J, Johnson DG (2006) Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr Mol Med 6: 739–748 [DOI] [PubMed] [Google Scholar]
- Dennis K, Fan T, Geiman T, Yan Q, Muegge K (2001) Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev 15: 2940–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan T, Yan Q, Huang J, Austin S, Cho E, Ferris D, Muegge K (2003) Lsh-deficient murine embryonal fibroblasts show reduced proliferation with signs of abnormal mitosis. Cancer Res 63: 4677–4683 [PubMed] [Google Scholar]
- Feber A, Clark J, Goodwin G, Dodson AR, Smith PH, Fletcher A, Edwards S, Flohr P, Falconer A, Roe T, Kovacs G, Dennis N, Fisher C, Wooster R, Huddart R, Foster CS, Cooper CS (2004) Amplification and overexpression of E2F3 in human bladder cancer. Oncogene 23: 1627–1630 [DOI] [PubMed] [Google Scholar]
- Foster CS, Falconer A, Dodson AR, Norman AR, Dennis N, Fletcher A, Southgate C, Dowe A, Dearnaley D, Jhavar S, Eeles R, Feber A, Cooper CS (2004) Transcription factor E2F3 overexpressed in prostate cancer independently predicts clinical outcome. Oncogene 23: 5871–5879 [DOI] [PubMed] [Google Scholar]
- Halstrom TC, Nevins JR (2003) Specificity in the activation and control of transcription factor E2F-dependent apoptosis. Proc Natl Acad Sci USA 100: 10848–10853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Huang J, Fan T, Yan Q, Zhu H, Fox S, Issaq HJ, Best L, Gangi L, Munroe D, Muegge K (2004) Lsh, an epigenetic guardian of repetitive elements. Nucleic Acids Res 32: 5019–5028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert PO, Verona R, Trimarchi JM, Rogers C, Dandapani S, Lees JA (2000) E2f3 is critical for normal cellular proliferation. Genes Dev 14: 690–703 [PMC free article] [PubMed] [Google Scholar]
- Hurst CD, Tomlinson DC, Williams SV, Platt FM, Knowles MA (2008) Inactivation of the Rb pathway and overexpression of both isoforms of E2F3 are obligate events in bladder tumours with 6p22 amplification. Oncogene 27: 2716–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong LJ, Chang JT, Bild AH, Nevins JR (2007) Compensation and specificity of function within the E2F family. Oncogene 26: 321–327 [DOI] [PubMed] [Google Scholar]
- Lang SE, McMahon SB, Cole MD, Hearing P (2001) E2F transcriptional activation requires TRRAP and GCN5 cofactors. J Biol Chem 276: 32627–32634 [DOI] [PubMed] [Google Scholar]
- Leone G, Nuckolls F, Ishida S, Adams M, Sears R, Jakoi L, Miron A, Nevins JR (2000) Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol Cell Biol 20: 3626–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Cheng EH, Hsieh JJ (2007) Bimodal degradation of MLL by SCFSkp 2 and APCCdc 20 assures cell cycle execution: a critical regulatory circuit lost in leukemogenic MLL fusions. Genes Dev 21: 2385–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, Desai A, Dorrestein PC, Glass CK, Rosenfeld MG (2010) PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 466: 508–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD (1998) The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94: 363–374 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myant K, Stancheva I (2008) LSH cooperates with DNA methyltransferases to repress transcription. Mol Cell Biol 28: 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myant K, Termanis A, Sundaram AY, Boe T, Li C, Merusi C, Burrage J, de Las Heras JI, Stancheva I (2011) LSH and G9a/GLP complex are required for developmentally programmed DNA methylation. Genome Res 21: 83–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Nnez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW (2003) Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113: 703–716 [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, Kouzarides T (2001) Rb targets histone H3 methylation and HP1 to promoters. Nature 412: 561–565 [DOI] [PubMed] [Google Scholar]
- Niu J, Chen T, Han L, Wang P, Li N, Tong T (2011) Transcriptional activation of the senescence regulator Lsh by E2F1. Mech Ageing Dev 132: 180–186 [DOI] [PubMed] [Google Scholar]
- Oeggerli M, Tomovska S, Schraml P, Calvano-Forte D, Schafroth S, Simon R, Gasser T, Mihatsch MJ, Sauter G (2004) E2F3 amplification and overexpression is associated with invasive tumor growth and rapid tumor cell proliferation in urinary bladder cancer. Oncogene 23: 5616–5623 [DOI] [PubMed] [Google Scholar]
- Parisi T, Yuan TL, Faust AM, Caron AM, Bronson R, Lees JA (2007) Selective requirements for E2f3 in the development and tumorigenicity of Rb-deficient chimeric tissues. Mol Cell Biol 27: 2283–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raabe EH, Abdurrahman L, Behbehani G, Arceci RJ (2001) An SNF2 factor involved in mammalian development and cellular proliferation. Dev Dyn 221: 92–105 [DOI] [PubMed] [Google Scholar]
- Rayman JB, Takahashi Y, Indjeian VB, Dannenberg JH, Catchpole S, Watson RJ, te Riele H, Dynlacht BD (2002) E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev 16: 933–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD (2002) E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev 16: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra HI, Maiti B, Timmers C, Altura R, Tokuyama Y, Fukasawa K, Leone G (2003) Inactivation of E2F3 results in centrosome amplification. Cancer Cell 3: 333–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawarkar R, Paro R (2010) Interpretation of developmental signaling at chromatin: the polycomb perspective. Dev Cell 19: 651–661 [DOI] [PubMed] [Google Scholar]
- Simon M, Köster G, Menon AG, Schramm J (1999) Functional evidence for a role of combined CDKN2A (p16-p14ARF)/CDKN2B (p15) gene inactivation in malignant gliomas. Acta Neuropatho 98: 444–452 [DOI] [PubMed] [Google Scholar]
- Sun LQ, Lee DW, Zhang Q, Xiao W, Raabe EH, Meeker A, Miao D, Huso DL, Arceci RJ (2004) Growth retardation and premature aging phenotypes in mice with disruption of the SNF2-like gene, PASG. Genes Dev 18: 1035–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Rayman JB, Dynlacht BD (2000) Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev 14: 804–816 [PMC free article] [PubMed] [Google Scholar]
- Taubert S, Gorrini C, Frank SR, Parisi T, Fuchs M, Chan HM, Livingston DM, Amati B (2004) E2F-dependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol Cell Biol 24: 4546–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SY, Opavsky R, Sharma N, Wu L, Naidu S, Nolan E, Feria-Arias E, Timmers C, Opavska J, de Bruin A, Chong JL, Trikha P, Fernandez SA, Stromberg P, Rosol TJ, Leone G (2008) Mouse development with a single E2F activator. Nature 28: 1137–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S, Chabes AL, Wysocka J, Herr W (2007) E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell 27: 107–119 [DOI] [PubMed] [Google Scholar]
- van den Heuvel S, Dyson NJ (2008) Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol 9: 713–724 [DOI] [PubMed] [Google Scholar]
- Waseem A, Ali M, Odell EW, Fortune F, Teh MT (2010) Downstream targets of FOXM1: CEP55 and HELLS are cancer progression markers of head and neck squamous cell carcinoma. Oral Oncol 46: 536–542 [DOI] [PubMed] [Google Scholar]
- Xi S, Zhu H, Xu H, Schmidtmann A, Geiman TM, Muegge K (2007) Lsh controls Hox gene silencing during development. Proc Natl Acad Sci 104: 14366–14371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Cho E, Lockett S, Muegge K (2003) Association of Lsh, a regulator of DNA methylation, with pericentromeric heterochromatin is dependent on intact heterochromatin. Mol Cell Biol 23: 8416–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Geiman TM, Xi S, Jiang Q, Schmidtmann A, Chen T, Li E, Muegge K (2006) Lsh is involved in de novo methylation of DNA. EMBO J 25: 335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebold U, Lee EY, Bronson RT, Lees JA (2003) E2F3 loss has opposing effects on different pRB-deficient tumors, resulting in suppression of pituitary tumors but metastasis of medullary thyroid carcinomas. Mol Cell Biol 23: 6542–6552 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.