Abstract
In Gram-negative bacteria, type II secretion systems (T2SS) assemble inner membrane proteins of the major pseudopilin PulG (GspG) family into periplasmic filaments, which could drive protein secretion in a piston-like manner. Three minor pseudopilins PulI, PulJ and PulK are essential for protein secretion in the Klebsiella oxytoca T2SS, but their molecular function is unknown. Here, we demonstrate that together these proteins prime pseudopilus assembly, without actively controlling its length or secretin channel opening. Using molecular dynamics, bacterial two-hybrid assays, cysteine crosslinking and functional analysis, we show that PulI and PulJ nucleate filament assembly by forming a staggered complex in the plasma membrane. Binding of PulK to this complex results in its partial extraction from the membrane and in a 1-nm shift between their transmembrane segments, equivalent to the major pseudopilin register in the assembled PulG filament. This promotes fully efficient pseudopilus assembly and protein secretion. Therefore, we propose that PulI, PulJ and PulK self-assembly is thermodynamically coupled to the initiation of pseudopilus assembly, possibly setting the assembly machinery in motion.
Keywords: bacterial protein secretion, membrane proteins, molecular dynamics, pilus assembly, type IV pili
Introduction
Many Gram-negative bacteria use the type II (T2SS) secretion system to secrete folded proteins from the periplasm. Human pathogens like Pseudomonas aeruginosa, Vibrio cholerae and enterotoxigenic Escherichia coli (ETEC) or plant pathogens like Dickeya dadantii use T2SS to secrete toxins and enzymes that damage-specific tissues (Sandkvist, 2001; Cianciotto, 2005). The T2SS machinery is related to the systems involved in bacterial natural transformation (Chen et al, 2005) and in the assembly of type IV pili (T4P) and archaeal flagella (Pelicic, 2008; Albers and Pohlschroder, 2009). All these systems assemble small membrane proteins called pilins into filamentous structures, with the help of a conserved machinery localized to the plasma membrane.
All T2SSs contains five pilins (called PulGHIJK in the Klebsiella oxytoca Pul secretion system; d'Enfert et al, 1987) that have been designated pseudopilins because they do not normally appear on the surface of bacteria. Based on their structural similarities with T4P, it has been postulated that T2SSs assemble periplasmic filaments called pseudopili that promote specific protein transport through the outer membrane (Pugsley, 1993b). In support of this model, the most abundant (major) pseudopilin PulG is assembled into long, surface-exposed pili when overproduced (Sauvonnet et al, 2000). Although necessary, PulG pilus assembly is not sufficient for secretion of the enzyme pullulanase (PulA), the specific substrate of the Pul secreton. Four low abundance (minor) pseudopilins, PulH, PulI, PulJ and PulK, are essential for PulA secretion, although they have not been found in the surface-assembled pili (Sauvonnet et al, 2000; Vignon et al, 2003).
T2SS and T4P pilins are inner membrane proteins composed of a long sigmoidal α helical stem that includes a conserved transmembrane (TM) segment, followed by a variable globular periplasmic domain (Hansen and Forest, 2006; Craig and Li, 2008). Upon membrane insertion via the SRP/Sec pathways (Francetic et al, 2007), a positively charged peptide is removed from the N-terminus of the pilin signal anchor by the prepilin peptidase PulO. Recently, we determined the structure of the PulG pilus at pseudo-atomic resolution using a combination of molecular dynamics (MD)-based modelling and biochemical validation (Campos et al, 2010). We showed that pilus assembly, essential for protein secretion, involves specific electrostatic and hydrophobic contacts between neighbouring pseudopilin subunits. The PulG subunits in the pilus are arranged in a right-handed helix, consistent with the structure of a complex composed of soluble domains of homologous minor pseudopilins GspI, GspJ and GspK from ETEC (Korotkov and Hol, 2008). In this structure, the three pseudopilins are arranged in a right-handed quasi-helix, where GspK (the largest of the minor pseudopilins) caps this complex, suggesting that minor pseudopilins may localize at the tip of the filament.
Many of the T2SS components are similar to those required for T4P assembly (Peabody et al, 2003; Ayers et al, 2010). In the Pul secreton, these components include an assembly platform (Py et al, 2001) composed of a hexameric cytoplasmic ATPase (PulE), inner membrane proteins PulL, PulM and PulF, and an outer membrane channel formed by the secretin PulD, allowing the translocation of the protein substrate. Current models in both T4P and T2SS propose that pili are assembled from the base in the inner membrane. Successive conformational changes of the ATPase would power the membrane assembly platform components to catalyse the addition of pilin monomers, elongating the fiber (Craig et al, 2006; Campos et al, 2010; Misic et al, 2010).
Previous studies have suggested different functions for the components of the pseudopilus ‘tip complex’, including initiation and arrest of pseudopilus polymerization (Sauvonnet et al, 2000; Vignon et al, 2003), its degradation (Durand et al, 2005) or the opening of the secretin channel (Forest, 2008; Korotkov and Hol, 2008). To understand the molecular role of minor pseudopilins, we studied their function in the T2SS of K. oxytoca, functionally reconstituted in E. coli. Our results provide in-vivo, in-vitro and in-silico evidence that pseudopilins PulI and PulJ form a complex in the membrane, to which PulK binds to form a pre-assembled pseudopilus tip. This complex ‘primes’ the initiation of pseudopilus assembly by means of thermodynamic coupling.
Results
Defective pilus assembly in pulI, pulJ and pulK single mutants
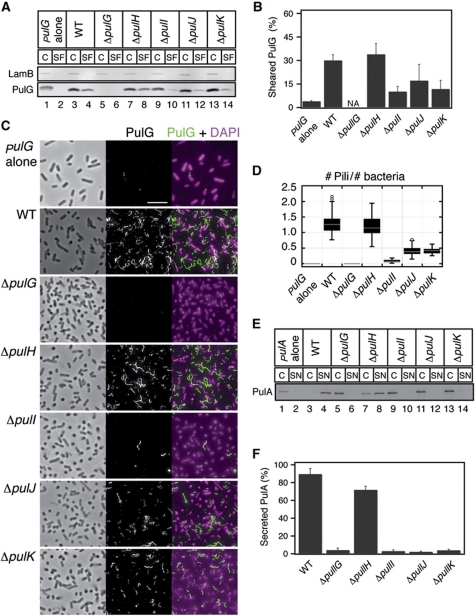
Four minor pseudopilins PulH, PulI, PulJ and PulK are required for efficient pullulanase secreton by the K. oxytoca type II secretion system (Possot et al, 2000). When the genes encoding the Pul secreton are overexpressed in E. coli, the bacteria grown on plates assemble pili on their surface, composed mainly of the major subunit, PulG (Sauvonnet et al, 2000; Kohler et al, 2004). To examine the function of minor pseudopilins in pilus assembly, we tested the effect of single pseudopilin gene deletions in E. coli expressing the pul genes in plasmid pCHAP231 (d'Enfert et al, 1987). Pilus assembly in these bacteria was assessed using immunodetection of the major pilin PulG in bacterial cell and sheared pili fractions (Figure 1A) and the percentage of sheared PulG was quantified (Figure 1B). The absence of PulH did not have a marked effect on the levels of PulG detected in the sheared fraction (Figure 1A, lane 8; Figure 1B). In contrast, in the pulI, pulJ and pulK mutants the amount of PulG in the sheared fraction was reduced (Figure 1A, lanes 10, 12 and 14; Figure 1B).
Figure 1.
Assembly of PulG pili in mutants lacking single minor pseudopilins. (A) PulG immunodetection in 0.005 OD600 nm units of cell and sheared fractions (C, SF) of E. coli carrying the pulG gene in a plasmid (pulG alone), or the pul genes (WT) on plasmid pCHAP231 or its minor pseudopilin gene deletion derivatives. LamB porin is used as a lysis control. (B) PulG percentage in SF (mean+s.d.) from three independent experiments like the one in (A). NA, not applicable. (C) Immunofluorescence (IF) and phase contrast microscopy of strains analysed in (A). PulG staining is in green and DAPI in magenta. Scale bar, 5 μm. (D) Box plot of normalized number of pili observed in (C) from ∼40 randomly selected areas in two independent experiments. (E) PulA immunodetection in cell extracts and supernatants (C, SN) of bacteria carrying pulA on a plasmid (+pulA), or expressing all pul genes on plasmid pCHAP8185 (WT) or its minor pseudopilin single deletion derivatives. (F) Percentage of secreted PulA (mean+s.d.) from five independent experiments like the one in (E). Figure source data can be found in Supplementary data.
To better characterize the phenotypes of minor pseudopilin mutants, we examined the morphology of surface pili by immunofluorescence (IF) microscopy (Figure 1C). Pili in each micrograph were counted and normalized to the number of bacteria (Figure 1D). While the ΔpulH mutant was virtually indistinguishable from WT (Figure 1C and D), the ΔpulJ and ΔpulK mutants had fewer pili (Figure 1C and D). Consistent with the biochemical fractionation results, the strongest defect was observed in the ΔpulI mutant, which assembled very few pili (Figure 1C and D). Overall, the phenotypes of pulI, pulJ and pulK mutants were highly similar, producing a reduced number of filaments. Interestingly, pili produced by the most defective mutant, ΔpulI, were slightly longer than those assembled by the WT (Supplementary Figure S1A). To test whether producing PulG in excess would rescue the piliation defect in these strains, we introduced a second plasmid carrying pulG under placZ control. This led to the assembly of longer pili in pulJ, pulK and pulI mutants compared with WT, but the number of pili remained reduced (Supplementary Figure S1D–I). We conclude that pilus assembly initiation is defective in these mutants, whereas the pilus length appears to be the function of the cellular levels of PulG. All these results suggest a functional interaction between PulI, PulJ and PulK, which is required for efficient pilus assembly.
We also tested secretion of a non-acylated PulA variant in these mutants (Francetic and Pugsley, 2005). PulA secretion was abolished in all pseudopilin mutants, with the exception of pulH, which showed a mild defect (Figure 1E and F), consistent with previously published data (Possot et al, 2000). However, PulH was required for PulA secretion when the copy number of the plasmid carrying the pul genes was reduced by a pcnB mutation (Lopilato et al, 1986; Vignon et al, 2003; Supplementary Figure S1B and C). These results demonstrate a strong correlation between the requirement of the minor pseudopilins PulI, PulJ and PulK for efficient pseudopilus assembly and for PulA secretion.
PulI and PulJ restore pilus assembly in a ΔpulHIJK mutant
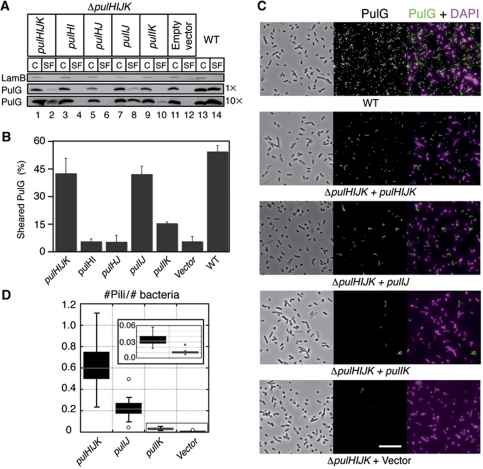
Biochemical in-vitro studies in P. aeruginosa showed that periplasmic domains of all minor pseudopilins, including the PulH homologue XcpU form a quaternary complex (Douzi et al, 2009). To analyse in vivo the function of minor pseudopilins in pilus assembly, we deleted the genes encoding all minor pseudopilins in plasmid pCHAP231 encoding the pul secreton, giving plasmid pCHAP8296 (ΔpulHIJK). E. coli carrying this construct had no PulG in the sheared fraction (Figure 2A, lane 12). Expression of minor pseudopilin genes pulHIJK in trans under placZ control (Figure 2A, lane 2) restored pilus assembly, albeit not to WT levels. This is due to the fact that the ΔpulHIJK mutant and the complemented strain produced less PulG compared with WT (compare Figure 2A, lanes 1 and 13). Quantification of the percentage of sheared PulG showed similar pilus assembly efficiencies (Figure 2B). Surprisingly, further analysis of the ΔpulHIJK mutant by IF microscopy revealed the presence of few pili (Figure 2C, ΔpulHIJK + Vector), which appeared at least as long as pili assembled by the WT and complemented strains (Figure 2C). This further shows that minor pseudopilins are required for efficient pilus initiation, but not for its elongation. Similar results were obtained in E. coli expressing pul genes with a ΔpulGHIJK mutation complemented with either pulG or pulGHIJK genes under placZ control (Supplementary Figure S2).
Figure 2.
PulI and PulJ initiate pilus assembly in the ΔpulHIJK mutant. (A) PulG detection in cell and sheared fractions (C, SF) from WT and the ΔpulHIJK mutant complemented with empty vector or genes pulHIJK, pulHJ, pulHI, pulIJ and pulIK. The equivalent of 0.005 OD600 nm units (1 ×) or 0.05 OD600 nm units (10 ×) of C and SF was analysed. (B) Percentage of PulG in SF (mean+s.d.) from three independent experiments like the one shown in (C) with 0.05 OD600 nm equivalent analysed. (C) IF and phase contrast microscopy of E. coli expressing the pul genes (WT) on a plasmid or its minor pseudopilin deletion derivative (ΔpulHIJK). ΔpulHIJK mutant complemented with empty vector or genes pulHIJK, pulIJ and pulIK. PulG staining is in green and DAPI in magenta. Scale bar, 10 μm. (D) Box plot of normalized number of pili observed in (A) from ∼40 randomly selected areas in two independent experiments. Figure source data can be found in Supplementary data.
Despite its major importance, PulI alone was not sufficient to restore pilus assembly in the ΔpulHIJK mutant (Supplementary Figure S2C and D). Therefore, we tested whether pairs of minor pseudopilins (PulH–PulI, PulI–PulJ, PulI–PulK or PulH–PulJ) could rescue the defect. While the PulH–PulJ, PulH–PulI and PulI–PulK pairs were unable to restore piliation (Figure 2A, lanes 4, 6 and 10), PulI and PulJ together promoted efficient pilus assembly relative to the complemented strain (Figure 2A, lane 8). Interestingly, analysis of 10-fold concentrated samples showed traces of PulG in the ΔpulHIJK mutant complemented with pulIK (Figure 2A, lane 10; and Figure 2B). Consistent with this, IF images of the ΔpulHIJK mutant complemented with pulIJ showed ∼50% of pili compared with the pulHIJK-complemented strain, while co-producing PulI and PulK led to only small numbers of PulG pili (Figure 2C and D).
The minor pseudopilins are not required for opening of the secretin channel
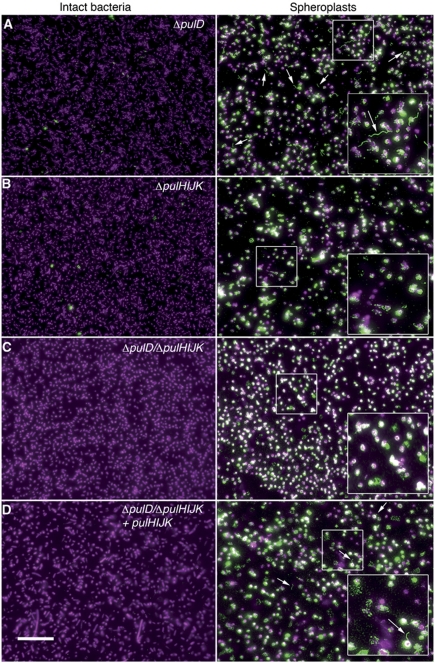
To exclude the possibility that the defect of the ΔpulHIJK mutant was due to inability to open the outer membrane secretin channel (PulD), we examined by IF sphaeroplasts of E. coli transformed with pCHAP231 derivatives carrying pulD, pulHIJK and pulD/pulHIJK knockouts. As expected, IF images of intact bacteria showed no pili on the surface of the ΔpulD mutant (Figure 3A, left panel), while PulG filaments were observed in the sphaeroplasts (Figure 3A right, arrows). However, neither ΔpulHIJK nor the ΔpulD/ΔpulHIJK mutant showed any such filaments, indicating that PulG pili do not assemble in the periplasm in the absence of minor pseudopilins (Figure 3B and C). The co-expression of pulHIJK in trans from a placZ promoter restored the pilus assembly in the periplasm (Figure 3D). Taken together, our results suggest that the initiation of PulG filament assembly is impaired in the ΔpulHIJK mutant, but neither its elongation nor the secretin channel opening is impaired.
Figure 3.
Periplasmic assembly of PulG pili in an outer membrane secretin mutant. IF microscopy showing intact (left) and sphaeroplasted (right) E. coli expressing pul genes with (A) pulD, (B) pulHIJK, (C) pulD/pulHIJK mutations or (D) pulD/pulHIJK mutation complemented with pulHIJK. PulG staining is in green and DAPI in magenta. Arrows indicate PulG pili. Scale bar, 15 μm.
The minor pseudopilins acquire a pre-assembled state in the membrane
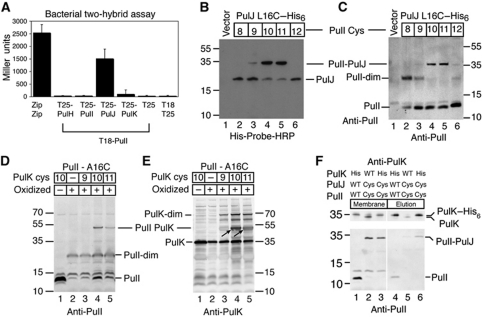
Since soluble versions of PulI and PulJ homologues were shown to be part of a complex (Korotkov and Hol, 2008), we hypothesized that the ability of PulI and PulJ to promote pilus assembly was related to their binding. If this were the case, the interaction between PulI and PulJ should occur in their membrane embedded, unassembled state. To determine whether full-length PulI and PulJ interact in vivo, we used the bacterial two-hybrid assay that allows one to study interactions between membrane proteins (Karimova et al, 1998). As bait, we used PulI that has a central position in the minor pseudopilin complex (Korotkov and Hol, 2008; Douzi et al, 2009). We fused the T18 fragment of the Bordetella pertussis adenylate cyclase toxin (CyaA) to the N-terminus of the mature PulI (Figure 4A). The T25 fragment of CyaA was fused to the N-termini of mature PulH, PulI, PulJ and PulK. The interaction of the two CyaA fragments led to the production of cAMP that was monitored by measuring the β-galactosidase activity in the E. coli cya mutant strain DHT1. Among the minor pseudopilins, only PulJ interacted with PulI (Figure 4A), as indicated by the high β-galactosidase activity in E. coli DHT1 producing T18-PulI and T25-PulJ, comparable to that of the positive control (the yeast protein GCN4 leucine zipper region Zip fused to both T18 and T25). The β-galactosidase activities in strains producing T25-PulH or T25-PulI together with T18-PulI were similar to the negative control (T18 or T18-PulI combined with T25). The activity of the pair T25-PulK and T18-PulI appeared only slightly higher than the negative control. Because strain DHT1 does not contain a functional T2SS, our results show that PulI and PulJ interact in the inner membrane in the absence of other components of the Pul secreton.
Figure 4.
Pre-assembled conformation of minor pseudopilins in the membrane. (A) Interaction of minor pseudopilins analysed by bacterial two-hybrid assay. β-Galactosidase activity (mean+s.d.) of DHT1 bacteria co-producing T18-PulI with T25 PulH, PulI, PulJ and PulK chimera or T25 alone. Zip-T18 and Zip-T25 chimera are used as positive control. (B, C) Position-specific cysteine crosslinking of E. coli co-producing PulJL16C–His6 with PulIV8C, PulIA9C, PulIL10C, PulIV11C, PulIV12C or empty vector. Cell extracts were analysed using His-Probe-HRP (B) or anti-PulI antibody (C). (D, E) Cysteine crosslinking of bacteria co-producing PulIA16C with PulK WT (−), PulKI9C, PulKL10C and PulKA11C. Cell extracts were analysed using an anti-PulI (D) or anti-PulK antibody (E). (F) Pull down of crosslinked PulIL10C and PulJL16C using PulK–His6 and Ni-magnetic beads. Theoretical molecular mass of PulI, 12.9 kDa; PulJ, 21.4 kDa; PulK, 35.2 kDa. Figure source data can be found in Supplementary data.
To gain insight into the conformation of the PulI and PulJ complex in the membrane, we introduced single cysteine substitutions in the TM segments of the two proteins. In a previous study of PulG pilus assembly, we showed that double cysteine substitutions at positions 10 and 16 of PulG led to the formation of covalently crosslinked PulG multimers in the pilus fractions (Campos et al, 2010). Therefore, we introduced single cysteine substitutions at and near positions 10 and 16 in PulI and PulJ, respectively. E. coli co-producing single cysteine variants of PulI and PulJ were chemically oxidized using copper phenanthroline. To detect PulJ, we introduced a hexahistidine tag at its C-terminus and analysed oxidized total cell extracts using His-probe-HRP. Bacteria co-producing single cysteine-substituted variant PulJL16C–His6 together with variants PulIL10C or PulIV11C formed membrane-associated (Supplementary Figure S3C) crosslinked products, migrating with an apparent mass of 35 kDa, corresponding to that of a PulI–PulJ heterodimer (Figure 4B, lanes 4 and 5). This was confirmed by analysing these samples with an anti-PulI antibody (Figure 4C, lanes 4 and 5). However, variants PulIV8C, PulIA9C and PulIV12C formed only homodimers (expected mass, 26 kDa) (Figure 4C, lanes 2, 3 and 6). Homodimers also formed when PulI variants were co-produced with PulJA15C–His6, but crosslinked heterodimers between PulI and PulJ were not observed (Supplementary Figure S3A). This result indicates that the formation of crosslinked heterodimers correlates uniquely with the positions of the cysteine-substituted residues in PulI and PulJ and not with the fact that variants like PulIV8C form homodimers. Similar crosslinking pattern of PulI cysteine-substituted variants with PulJL16C–His6 was observed in the presence of the pul genes with a pulHIJK deletion (Supplementary Figure S3B). Interestingly, after only 10 min of oxidation, variant PulJL16C–His6 formed heterodimers mainly with PulIL10C (Supplementary Figure S3D), suggesting a closer proximity of this residue with PulJL16. All of these results indicate that, if initially the TM segments of monomeric PulI and PulJ reside at similar positions along the membrane plane, binding of PulI to PulJ induces a conformational change, shifting their TM segments by ∼1 nm. This shift between the interacting residues of PulI and PulJ in the membrane is identical to the 1-nm axial rise between major pseudopilins in the assembled PulG filament (Campos et al, 2010). Therefore, we propose that the minor pseudopilins rearrange in the membrane to form a pre-assembled pseudopilus.
Despite the fact that significant interactions were not observed between PulI and PulK in two-hybrid experiments, some PulG pili were assembled in the ΔpulHIJK mutant complemented with pulIK, indicating a weak productive interaction. To test whether these proteins interact in a similar fashion as PulI and PulJ, we introduced single cysteine substitutions at and near positions 16 and 10 in PulI and PulK, respectively. Immunoblot analysis of oxidized E. coli co-producing PulIA16C and PulK cysteine-substituted variants showed that PulKL10C and PulKA11C, but not PulKI9C, crosslinked to PulIA16C, as shown by the presence of a band with an apparent mass corresponding to a PulI–PulK complex (48 kDa) (Figure 4D and E, arrows). Therefore, PulI and PulK do indeed interact in the membrane, and like PulI and PulJ, they might rearrange upon binding to form a staggered array, with a shift corresponding to the axial rise in the PulG filament.
To determine if the full-length minor pseudopilins form a tripartite complex, we performed a pull-down experiment using a hexahistidine-tagged PulK variant in detergent-solubilized membranes. E. coli producing a plasmid-encoded prepilin peptidase PulO (to ensure pseudopilin processing) was transformed with a plasmid encoding WT versions of all pseudopilin genes and PulK–His6, or plasmids encoding variants PulIL10C, PulJL16C and PulK–His6 instead of PulI, PulJ and PulK, respectively. As a control, we used a plasmid encoding PulIL10C, PulJL16C and WT PulK. Although some non-specific binding of PulK was observed to Ni-IDA magnetic beads (Figure 4F, lane 5), PulK–His6 strongly bound to the beads, and the PulI–PulJ crosslinked product appeared specifically in the elution fraction (Figure 4F, lane 6). This result suggests that the interaction of PulI, PulJ and PulK resisted membrane solubilization. Non-crosslinked PulI also eluted with PulK–His6 (Figure 4F, lane 4), although, for lack of specific antibodies, we could not determine whether PulJ was part of this complex. All of these results suggest that the minor pseudopilins interact in the membrane to form a pre-assembled pseudopilus in the absence of other T2SS components.
MD of full-length minor pseudopilin complexes
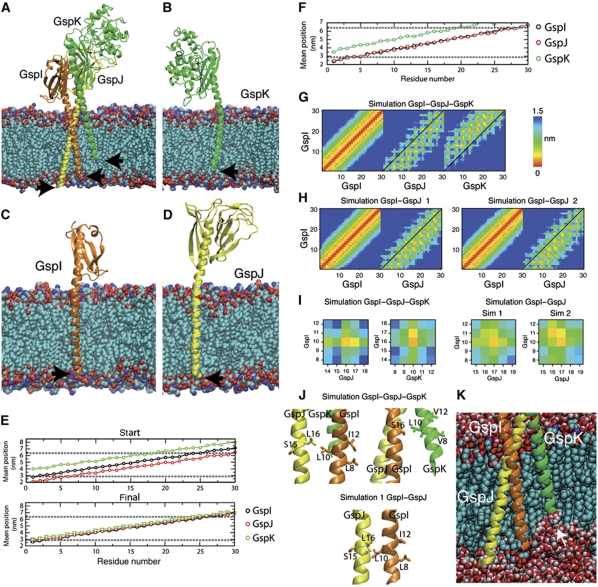
To analyse how PulI, PulJ and PulK might interact in the membrane, we modelled TM segments onto the crystallographic structure of ETEC GspJ–GspI–GspK complex, which showed a quasi-helical register, as reported (Korotkov and Hol, 2008). When introduced into a palmitoyl-oleoyl ethanolamine (POPE) model membrane, their N-termini were placed at different levels of the membrane plane (Figure 5A, arrows). Next, we removed for each protein its two partners and performed MD simulations (Supplementary Video S1). After 20 ns of simulation, all three proteins underwent substantial lateral diffusion and their N-termini adopted similar positions in the membrane plane (Figure 5B–D, arrows). The mean position of each residue with respect to the membrane glycerol backbone atoms in the last 5 ns (Figure 5E) showed that the three proteins reintegrated to a similar extent into the POPE bilayer. These positions could correspond to their initial state upon membrane insertion and maturation, prior to the complex formation.
Figure 5.
Molecular dynamics (MD) of full-length GspI, GspJ and GspK in a model membrane. (A) Initial state of the trimeric complex of GspI, GspJ and GspK after adding POPE lipids with arrows indicating the N-termini. (B–D) Snapshot of the final structure of GspI, GspJ or GspK from MD simulations of each protein alone. (E, F) Position of each residue (mean+s.d.) along the bilayer normal in the starting structure and last 5 ns in GspI, GspJ and GspK alone simulations (E) or the GspI–GspJ–GspK complex simulation (F). Dashed lines show the lipid glycerol backbone atoms position. (G, H) Minimum distance between residues in the TM segments of GspI and itself, GspJ or GspK for the GspI–GspJ–GspK complex simulation (G) and two GspI–GspJ dimer simulations (H). (I) As in (G, H) but for residues targeted for crosslinking experiments. (J) Final position of GspI L10, GspJ L16 and GspK L10 after MD simulations. (K) Snapshot of GspI–GspJ–GspK MD simulation showing the membrane deformation induced by GspK.
We also performed an MD simulation of the three proteins together (Supplementary Video S2). This complex was stable (Supplementary Figure S4) and the mean position of TM segment residues in the membrane showed that GspI and GspJ are completely embedded (Figure 5F). However, GspK TM segment remained partially extracted, with a displacement of ∼1 nm with respect to GspI and GspJ (Figure 5F). This displacement created a severe membrane deformation (Figure 5K). Nevertheless, the 1-nm helical register imposed by the GspI–GspJ–GspK soluble domains was maintained inside the membrane. This is shown in the contact map of GspI and GspJ or GspK where neighbouring residues are shifted from the diagonal, unlike those of GspI with itself (Figure 5G). Interestingly, the main interactions between PulI and PulJ or PulK shown by crosslinking were represented in these maps (Figure 5I and J).
We showed that PulI and PulJ promote PulG pilus assembly and that they interact in the membrane. To examine this interaction, we performed MD simulations of the GspI–GspJ complex with modelled TM segments (Supplementary Video S3). This complex was stable (Supplementary Figure S4) and, as in the GspI–GspJ–GspK simulation, the TM segments remained staggered. In the contact map of GspI and GspJ, neighbouring residues are shifted from the diagonal (Figure 5H). However, only some of the interactions between PulI and PulJ tested by crosslinking were present (Figure 5I and J).
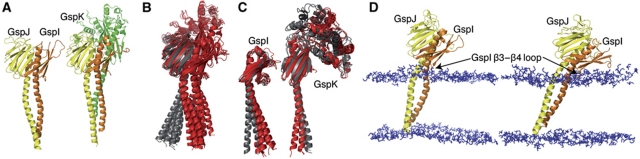
Next, we clustered by similarity all MD simulation-derived structures by aligning their periplasmic α-helical domains and observing the differences in their TM segments. In the representative structures of the main clusters from GspI–GspJ–GspK and two GspI–GspJ simulations, GspJ is curved around glycine 36 (Figure 6A). In contrast, this bending was not present in the main clusters derived from the GspJ alone simulation (Figure 6B). This suggests that this conformational change occurs in the presence of GspI and GspK. GspI and GspK also showed some bending (Figure 6A), although some of the conformations found in the dimer and trimer MD simulations also occurred in the main clusters of the GspI and GspK alone simulations (Figure 6C), suggesting some intrinsic flexibility. One interesting feature is that in both simulations the GspI–GspJ dimer tilted in the membrane (Figure 6D). Apparently, the hydrophobic TM segments maximized their interaction with the phospholipid fatty-acyl chains, bringing the GspI β3-β4 loop close to the phospholipid headgroups (Figure 6D, arrows). This induced tilting could favour the interaction with GspK. Interestingly, a similar in-silico behaviour has been observed for P. aeruginosa PilA (Lemkul and Bevan, 2011).
Figure 6.
Structure of the GspI–GspJ dimer and GspI–GspJ–GspK trimer in the membrane. (A) Representative structure of GspI–GspJ dimer in the main cluster of the two merged simulations (left). Representative structure of GspI–GspJ–GspK in the main cluster of the MD simulation (right). (B) Representative structures of the three main clusters of the GspI–GspJ and GspI–GspJ–GspK simulations (red) and the six main clusters of the GspJ alone simulation (grey), shown in the same view as (A). (C) Representative structures of GspI and GspK in the three main clusters of the GspI–GspJ and GspI–GspJ–GspK simulations (red) and the three main clusters of the GspI or GspK alone simulations (grey). (D) Snapshots of the final structures in two GspI–GspJ MD simulations.
Together, these in-silico results show that (1) the helical register expected for the periplasmic assembled pseudopilus is maintained in the membrane embedded GspI–GspK–GspJ complex, (2) GspJ undergoes conformational changes when bound to GspI and GspK and (3) GspK remains partially extracted from the membrane in this complex, inducing a local membrane deformation.
PulI and PulJ couple their binding energy to pilus assembly initiation
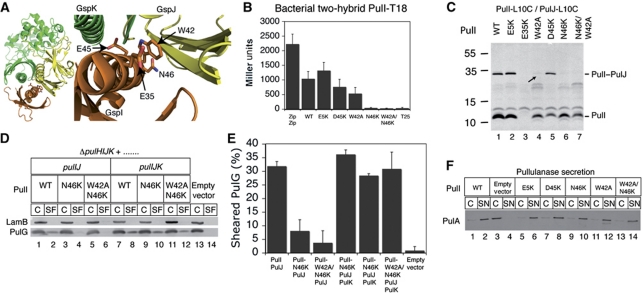
We have shown so far that there is a correlation between the binding of PulI and PulJ and the ability of this complex to restore piliation in the ΔpulHIJK mutant. To examine this correlation, we designed six mutations in pulI that could disrupt PulI binding to PulJ and we tested their impact both on this interaction and on their ability to restore efficient pilus assembly when co-produced with PulJ in the ΔpulHIJK mutant (Figure 7A). The invariant residue E5 of PulG, essential for pilus assembly (Pugsley, 1993a), is also conserved in PulI. However, when substitution E5K was introduced into T18-PulI, its interaction with T25-PulJ was comparable to WT (Figure 7B). Consistent with this result, the double mutant PulI-L10C/E5K crosslinked to PulJ-L16C (Figure 7C, lane 2). Residue E35 of ETEC GspI (Figure 7A) lies close to GspJ (Korotkov and Hol, 2008), but as the substitution E35K in PulI affected protein stability (Figure 7C, lane 3) this variant was not analysed further. The GspI residue W42 (Figure 7A) hides ∼38 Å2 of accessible surface upon GspJ and GspK binding. T18-PulIW42A interacted comparably to WT in the two-hybrid assay (Figure 7B), but the double mutant PulI-L10C/W42A showed only traces of the PulJ-L16C crosslinked product (Figure 7C, lane 4, arrow). Residue E45 in ETEC GspI lies in the interface between GspI, GspJ and GspK (Figure 7A). The substitution of its equivalent (D45K) in T18-PulI had no effect in the two-hybrid assay (Figure 7B), and the double mutant PulI-L10C/D45K crosslinked to PulJ-L16C (Figure 7C, lane 5). Residue N46 (Figure 7A) is highly conserved and is involved in many important contacts with GspJ (Korotkov and Hol, 2008). Substitution N46K in T18-PulI abolished binding to T25-PulJ in the two-hybrid assay (Figure 7B), and the variant PulI-L10C/N46K did not crosslink to PulJ-L16C (Figure 7C, lane 6). Furthermore, PulI-N46K did not restore efficient pilus assembly in the ΔpulHIJK mutant when co-produced with PulJ (Figure 7D, lane 4; Figure 7E). Double-substituted variant PulI-W42A/N46K was similar to PulI-N46K (Figure 7B and C, lane 7), although it initiated pilus assembly even less efficiently (Figure 7D, lane 6; Figure 7E). All these results indicate a strong correlation between the interaction of PulI and PulJ (as measured by the two-hybrid assay and cysteine crosslinking) and pilus assembly initiation in the ΔpulHIJK mutant. Furthermore, they demonstrate that when PulI and PulJ binding is disrupted, the ∼1-nm shift in their TM segments is also affected. This suggests that the binding energy of PulI and PulJ is necessary for the proposed conformational change that would shift their TM segments to acquire a pseudopilus-like structure in the membrane.
Figure 7.
Effect of PulI residue substitutions on pilus assembly. (A) ETEC's GspI–GspJ–GspK complex showing residues E35, W42, E45 and N46 in GspI. (B) Two-hybrid assay of T25-PulJ chimera and WT T18-PulI or indicated mutant variants. Zip-T18 and Zip-T25 are used as positive control and T25 as negative. (C) Cysteine crosslinking of PulJL16C with PulIL10C or its indicated double-substituted variants. (D) Pilus assembly in strains co-producing PulJ or PulJ and PulK with WT PulI or its variants N46K and W42A/N46K. (E) PulG percentage in SF (mean+s.d.) from three independent experiments like the one in (E). (F) PulA immunodetection in cell extracts and supernatants (C, SN) of E. coli expressing all pul genes with a pulI deletion complemented with WT pulI or indicated mutant derivatives. Figure source data can be found in Supplementary data.
Surprisingly, while PulI mutant variants restored PulG pilus assembly to different extents (Supplementary Figure S5) in the ΔpulHIJK mutant, they could all restore full pullulanase secretion in the pulI deletion mutant, even at low pul gene expression levels (Figure 7F). To test whether this was due to the presence of PulK, we produced PulJ and the most defective PulI mutant variants with or without PulK in the ΔpulHIJK mutant. Both PulI-N46K and PulI-W42A/N46K could promote efficient pilus assembly when PulK was present (Figure 7D and E). Presumably, PulK could promote binding of these PulI variants to PulJ to allow PulA secretion.
Discussion
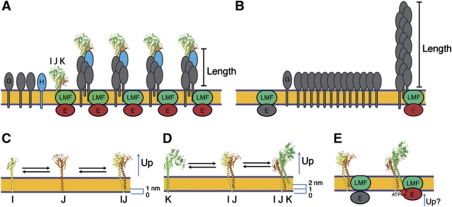
We show here for the first time that the minor pseudopilins PulI, PulJ and PulK interact in vivo in the bacterial inner membrane to initiate PulG filament assembly. IF microscopy analysis revealed highly similar phenotypes of single pulI, pulJ or pulK mutants, all of which assembled reduced number of fibers on their surface compared with WT. The severe defect found in the pulI mutant is consistent with the central position of this protein in the complex formed by the pseudopilin globular domains in vitro (Korotkov and Hol, 2008; Douzi et al, 2009). In contrast, pilus elongation was efficient in mutants lacking PulI, PulJ, PulK, or all minor pseudopilins. The defect in assembly initiation led to the assembly of fewer and longer fibers, which was accentuated by overproducing PulG, with the pulI mutant making very rare and long pili (Supplementary Figures S1 and S2). These observations are consistent with the model in which the pilus length is directly proportional to the size of the PulG pool in the membrane and inversely proportional to the number of active assembly sites (Figure 8A and B). Once the energetic barrier required for the initiation is overcome, pilus assembly continues as a function of the major pseudopilin concentration in the membrane. Thus, in the pulI, pulJ and pulK mutants, the reduced initiation efficiency would lead to pseudopilin accumulation in the membrane, favouring the elongation of the few pili whose assembly had already started. Together, our results suggest that none of the minor pseudopilins play an active role in the pseudopilus length control. Finally, the correlation between the requirement of the minor pseudopilins PulI, PulJ and PulK but not of PulH for efficient pilus assembly and protein secretion, suggests a role for the PulI–PulJ–PulK complex in initiating pseudopilus assembly during protein secretion.
Figure 8.
Pseudopilus assembly model. (A) Length control of pseudopilus based on the number of initiation sites. In WT bacteria, the minor pseudopilin complex (labelled IJK) initiates assembly of H and G pseudopilins into pili of a certain length. (B) In the absence of the minor pseudopilins, spontaneous but sporadic initiation leads to assembly of long pili driven by cell-accumulated major pseudopilins. (C) During the first initiation step, PulI (I) and PulJ (J) interact, leading to a 1-nm shift between their transmembrane segments. (D) PulK (K) is partially extracted from the membrane upon PulI–PulJ binding, forming a pre-assembled trimeric tip complex. (E) The upward movement of the minor pseudopilins upon PulK binding could activate the assembly platform by coordinating structurally the assembly of incoming pseudopilins and by bringing the assembly ATPase in contact with the membrane phopholipids, which is required for the activation of ATP hydrolysis.
The function of the minor pseudopilin PulH appears to be unrelated to the assembly initiation, since the number of pili in the pulH mutant is similar to WT. PulH could instead be involved in the elongation step, at least under physiological conditions where this protein is essential for secretion (Supplementary Figure S1). This is consistent with the fact that the PulH requirement can be partially overcome by overproducing the pul genes. Thus, structural complementarity between the tip complex and the PulG filament is not entirely required for pseudopilus elongation. Therefore, additional factors, like the assembly machinery at the inner membrane, could act downstream of the pseudopilus priming, as discussed below.
We show here that the minor pseudopilins are not required for the opening of the secretin channel. When the pul genes are overexpressed in E. coli grown on agar plates, the pili are surface exposed in single or multiple minor pilin mutants, and rare pili are observed even in the ΔpulHIJK mutant. This suggests that the secretin channel in the outer membrane allows the passage of fibers composed only of PulG. However, in the absence of the secretin channel (ΔpulD mutant), the pseudopilus is formed in the periplasm, as observed previously (Vignon et al, 2003). We found no periplasmic pili in the ΔpulHIJK/ΔpulD mutant, confirming the role of minor pilins in initiating the filament assembly. In addition, the expression of the minor pseudopilin genes in trans restored the periplasmic fiber accumulation. Therefore, the initiation of pseudopilus assembly could be the main function of the PulI–PulJ–PulK complex, although at this point we cannot discard the possibility that it could participate in any event downstream of pseudopilus assembly to regulate secretion in liquid-grown bacteria, where pseudopili do not extend beyond the cell surface. Whether the tip pseudopilins are dispensable or degraded after completing pseudopilus initiation remains an open question. It has been shown recently by surface plasmon resonance that minor pseudopilins interact in vitro with the secreted substrate in P. aeruginosa, suggesting an additional role in protein secretion downstream of pseudopilus initiation (Douzi et al, 2011). The functional significance of these interesting interactions will require further study.
Pilins are synthesized with a positively charged N-terminal signal anchor, which is cleaved at the level of the cytoplasm–membrane interface by prepilin peptidase. Presumably, the cleavage site and the resulting N-termini of the matured pilins lie at approximately the same level in the membrane plane. This is consistent with the results of the MD simulations of PulI, PulJ and PulK monomers, in which the three proteins are positioned at the same level in the absence of their binding partners (Figure 5B–E). However, both MD and cysteine crosslinking suggested that the formation of the native PulI–PulJ–PulK complex in the absence of other components of the Pul secreton leads to rearrangements of their TM segments. As a result of these rearrangements, the interactions between the minor pseudopilin TM segments involve residues that have equivalents in the filament formed by the major pseudopilin PulG. Moreover, the interaction between PulI and PulJ was required for significant pilus assembly initiation in the context of the ΔpulHIJK mutation. Taken together, our results provide strong evidence that the formation of the PulI–PulJ complex in the membrane nucleates the pre-assembled filament-like structure (Figure 8C). Although this complex plays a central and initial role, fully efficient pilus assembly (Figure 1F) and protein secretion require PulK. We showed that PulK bound to the PulI–PulJ complex and that PulK binding to PulI led to the rearrangement of its TM segment. PulK binding to PulI–PulJ in the membrane would lead to their stabilization as suggested by the MD simulations of a complex composed of full-length GspI–GspJ–GspK (Supplementary Video S2). In addition, binding of PulK to PulI–PulJ may induce an upward movement, causing a partial extraction of the PulK TM segment to acquire a pseudopilus-like structure in the membrane (Figure 8D). The tilting of PulI and PulJ complex in the membrane, suggested by the MD, might facilitate this binding, together with the fact that the α-helical N-terminal domain of PulK remains straight. Furthermore, the presence of a kink in the GspJ TM segment is a structural similarity with major pseudopilins, where the highly conserved kink in the α-helical N-terminal domain centred at residue P22 is necessary for efficient assembly of PulG into pili (Campos et al, 2010).
If one considers pseudopilus assembly as a biochemical reaction, then the binding of the minor pseudopilins could have three consequences: kinetic, thermodynamic and structural (or geometrical). The correlation between the binding of PulI to PulJ in the membrane and the pilus assembly supports the idea that the interaction between the minor pseudopilins reduces the kinetic barrier to initiate pseudopilus assembly. The formation of the PulI–PulJ–PulK complex could also have thermodynamic consequences, since tight binding between their globular domains could stabilize the whole pseudopilus, favouring its assembly. In this respect, it is interesting to note that GspK reduced the RMSD of GspI and GspJ in MD simulations (Supplementary Figure S3). Finally, the minor pseudopilin complex formation may have structural consequences. Evidence for this suggestion comes from the fact that GspK maintained its helical register in the GspI–GspJ–GspK complex in MD simulations and that PulI could be crosslinked in the membrane with PulJ and PulK in a staggered manner. Furthermore, GspK binding to GspI–GspJ dimer induced a severe membrane deformation in silico. We propose that the energy used to deform the membrane during MD simulations is directly transduced to the assembly platform, and may lead to its activation. The fact that pilus assembly occurs spontaneously on rare occasions supports this model, suggesting that the tip complex may reduce an energy barrier for a binding event leading to the assembly initiation.
How exactly could the primed conformation of minor pseudopilins initiate the pilus assembly? Although structural information is available for the soluble domains of homologues of PulL, PulF and two domains of the ATPase PulE, the organization of these proteins in the membrane remains unclear, and their interactions with pseudopilins are largely unexplored. Recent studies in V. cholerae show that EpsL interacts with the major pseudopilin EpsG, providing the first direct link between pseudopilins and the assembly platform (Gray et al, 2011). Since EpsE interaction with the membrane phospholipids is required for efficient ATP hydrolysis (Camberg et al, 2007), one plausible model is that the upward movement induced by the minor pseudopilin complex formation is transduced via PulL or PulF to PulE, bringing it close to the membrane to activate ATP hydrolysis. This cascade of conformational changes would set the assembly platform in motion by coordinated ATP hydrolysis, leading to insertion of PulH and PulG into the growing pseudopilus (Figure 8E). Interestingly, the T4P retraction ATPase PilT (Misic et al, 2010) has three main conformations (ready, active and release). If the action of PilT is similar (or the reverse) to that of PulE, then one could speculate that the minor pseudopilins symmetry may correspond to, fit, or lead to these three states.
Type 4a pilus assembly systems contain a set of pilin genes arranged in a similar manner as the pulHIJK genes, and certain features suggest that they share a common function (Forest, 2008; Korotkov and Hol, 2008). First, some of them encode a larger protein (equivalent to PulK) that could cap the tip of the pilus. Second, this protein does not have the conserved residue E5, which would be dispensable at the tip, as it is in PulK (Vignon et al, 2003; Supplementary Figure S6). Third, genetic evidence suggests that in T4P, these minor pilins ‘counteract’ retraction because the pilus assembly defects of single deletion mutants in these genes are suppressed by a deletion of the gene encoding the retraction ATPase PilT (Winther-Larsen et al, 2005; Carbonnelle et al, 2006; Giltner et al, 2010). Therefore, these minor pilins could form a complex that would shift the equilibrium towards pilus assembly by acquiring a pre-assembled conformation. It is also tempting to speculate that their interaction could participate in the switch between assembly and disassembly (Maier et al, 2004), especially if minor pilins were not located exclusively at the tip (Giltner et al, 2010). Therefore, minor pilin binding coupled to partial extraction of TM segments could represent a general mechanism in the initial events of pilus assembly in T4P and other related systems such as DNA competence (Chen et al, 2005), archaeal pili and flagella (Albers and Pohlschroder, 2009).
In conclusion, we propose that the intrinsic binding properties of the minor pseudopilins PulI, PulJ and PulK lead to their self-assembly into a pseudopilus-like complex in the membrane. This event could in turn induce and coordinate key conformational changes of the membrane assembly platform, resulting in its activation to initiate pseudopilus polymerization. The exact sequence and molecular details of this process will be a subject of future studies.
Materials and methods
Bacterial strains, plasmids and molecular biology techniques
The E. coli K12 strains PAP7460 (Possot et al, 2000) and PAP5207 (Campos et al, 2010) were used in this study. Plasmids used in this study are listed in Supplementary Table S1. DNA extraction, plasmid constructions and DNA transformation were performed as described (Maniatis et al, 1982). Site-directed mutagenesis was performed using a modified Quick change method and the Pwo DNA polymerase (Roche). Oligonucleotides, listed in Supplementary Table S2, were synthesized by Sigma Genosys. All plasmids were sequenced by GATC.
Pilus assembly, shearing and fractionation
Shearing assays were performed as described (Sauvonnet et al, 2000). Bacteria carrying derivatives of the plasmid pCHAP231, which contains all the pul genes (Supplementary Table S1) were grown overnight on LB agar containing 100 μg/ml of ampicillin (Ap), 25 μg/ml chloramphenicol (Cm) and 0.4% maltose to induce the expression of the pul genes. Bacteria were collected and resuspended in LB medium at 10 OD600 nm per ml and vortexed (sheared) for 2 min to release pili into the medium. Bacteria were centrifuged at 16 000 g in a table-top centrifuge for 5 min, the pellet was collected and resuspended in the sodium dodecyl sulphate (SDS)–polyacrylamide gel electrophoresis (PAGE) sample buffer (cell fraction). The sheared fraction was further centrifuged for 15 min to remove residual bacteria, the pellet was discarded and the supernatant was precipitated with 10% trichloroacetic acetic acid. After centrifugation for 15 min at 16 000 g, the pellet was washed twice in acetone, air dried and resuspended in SDS–PAGE sample buffer.
Pullulanase secretion assay
Plasmid pCHAP231 carrying the gene encoding a non-acylated variant of PulA pCHAP8185 and its mutant derivatives (Supplementary Table S1) were introduced in strain PAP7460 or its pcnB∷Tn10 derivative PAP5207. Bacteria were grown to late exponential phase in LB media containing appropriate antibiotics and 0.4% maltose. For the pcnB strain, media were supplemented with 1 mM IPTG. Cultures were normalized to OD600 nm of 1 and centrifuged for 5 min at 16 000 g. Bacterial pellets were resuspended in the initial volume of Laemmli sample buffer. The supernatant fractions were transferred to fresh tubes and centrifuged for another 10 min. Samples were taken off top and mixed with an equal volume of 2 × Laemmli sample buffer. The equivalent of 0.05 OD600 nm of cell and supernatant fractions were analysed by SDS–PAGE and immunodetection.
Immunodetection
Immunoblotting was performed as described (Sauvonnet et al, 2000). Proteins were separated by SDS–PAGE in tricine (Schagger and von Jagow, 1987) gels containing 10–15% acrylamide, transferred onto nitrocellulose membranes (Amersham ECL) using semi-dry electrotransfer. Membranes were blocked with 5% milk in TBST (10 mM Tris, 150 mM NaCl, 0.05% Tween-20) and incubated in the specific antiserum (anti-PulG at 1/2000, anti-PulI at 1/500, anti-PulK at 1/1000, anti-PulA 1/2000 or anti-LamB 1/2000) followed by horseradish peroxidase (HRP)-coupled anti-rabbit antibody (1/40 000; Amersham). Membranes were developed by enhanced chemiluminescence ECL-plus (GE-Healthcare) and recorded using the STORM phosphoimager (Molecular Dynamics). Protein quantifications are described in Supplementary data. For detection of proteins containing a hexahistidine tag, nitrocellulose membranes were blocked with 1% BSA in TBST and incubated with a dilution of 1/2000 of His-Probe-HRP (Thermo Scientific). Membranes were developed using West Pico Chemiluminescent Substrate (Thermo Scientific). Anti-PulI and anti-PulK peptide antibodies were produced and purified by Genscript (USA).
Fluorescence microscopy
IF labelling of pili was performed as described (Vignon et al, 2003). Bacteria grown in the same conditions as for pilus assembly assay, were gently resuspended and directly immobilized on poly-L-lysine-coated coverslips. Samples were fixed for 20 min on 3.7% formaldehyde, blocked with 1% BSA in PBS and incubated with anti-PulG antibodies (1:2000) and secondary Alexa Fluor 488-coupled anti-rabbit IgG (Invitrogen). Samples were examined with an Axio Imager.A2 microscope (Zeiss). Images were taken with AxioVision (Zeiss) and processed in ImageJ (Abramoff et al, 2004).
Sphaeroplast preparation
Sphaeroplasts were prepared as described (Randall and Hardy, 1986). Bacteria were resuspended in 0.5 ml of 0.1 M Tris acetate (pH 8.2), 0.5 M sucrose and 5 mM EDTA. Lysozyme was added (0.1 mg/ml) followed by addition of 0.5 ml of ice-cold water. After incubation for 5 min on ice, MgSO4 was added to a final concentration of 18 mM to stabilize the sphaeroplasts. Sphaeroplasting was monitored by light microscopy to verify that bacteria became round (Birdsell and Cota-Robles, 1967). Sphaeroplasts were immobilized onto poly-lysine coverslips, washed with 0.05 M Tris acetate pH 8.2, 0.25 M sucrose, 10 mM MgSO4, and fixed in 3.7% formaldehyde for IF.
MD simulations
Full-length models of GspI, GspJ and GspK were used in MD simulations using the GROMACS package (Hess et al, 2008) version 4.5 (Bjelkmar et al, 2010). The proteins were treated using the CHARMM22/CMAP force field (MacKerell et al, 1998), and the lipid molecules using the new CHARMM36 parameter set (MacKerell et al, 2010). Clustering analysis of each simulation was carried out with GROMACS 4.5, with a neighbour-list cutoff in the pair-wise RMS difference of 0.25 nm. Trajectories were fitted to the backbone atoms of the upper regions of each N-terminal helix (residues 41–53, 36–57 and 29–58 in GspI, GspJ and GspK, respectively). Clustering was performed on the backbone atoms of the bottom regions of each N-terminal helix (residues 5–41, 5–46 and 5–29 in GspI, GspJ and GspK, respectively). The three main clusters of the GspI–GspJ, GspI–GspJ–GspK and GspK alone were present ∼50% of the time in each simulation, while the six main clusters of the GspI alone and GspJ alone were present 50% of the time. Detailed methods for MD simulations are shown in Supplementary data.
Bacterial two-hybrid assay
To produce hybrid proteins with N-terminal T25 and T18 fragments of Bordetella pertussis CyaA, we used plasmids pKT25 and pUT18c (Karimova et al, 1998). Primers are listed in Supplementary Table S2. The cya mutant E. coli strain DHT1 was co-transformed with plasmids containing T18 and T25 chimera. Bacteria were grown for 48 h at 30°C on LB medium, containing 100 μg/ml of ampicillin (Ap) and 25 μg/ml Kanamycin (Km). Randomly picked single colonies were inoculated into LB medium and pre-cultured overnight. Cultures were diluted in medium containing 1 mM isopropyl β-D-1-thiogalactopyranoside, 100 μg/ml of ampicillin (Ap) and 25 μg/ml Kanamycin (Km) for β-galactosidase assays, which were performed as described (Miller, 1972). At least three independent experiments with quadruplicates were performed for each sample.
Copper phenanthroline oxidation
Bacteria carrying cysteine-substituted variants of minor pseudopilins were grown in liquid LB media to OD600 of 1–1.5. Bacteria were harvested, washed with M63 salts (100 mM KH2PO4, 15 mM (NH4)2SO4, 1.7 μM FeSO4·7H2O, pH 7) (Miller, 1972) and incubated 10 min at 30°C to equilibrate. Copper phenanthroline was added to 1.5 mM and incubated for 10–60 min in M63 salts. The reactions were stopped by adding 5 mM EDTA and 25 mM N-ethylmaleimide. Bacteria were washed once in M63 salts and resuspended in SDS–PAGE sample buffer.
PulK–His pull down
Bacteria were oxidized using copper phenanthroline for 1 h and the reaction was stopped with EDTA and N-ethylmaleimide. Bacteria were broken by freeze thaw and sonication. Non-broken bacteria were removed by centrifugation at 4000 g and membrane fractions were then prepared by centrifugation at 112 000 g for 1 h. Membranes were resuspended in 50 mM phosphate buffer, 150 mM NaCl pH 7.4 and solubilized by the addition of 2% Triton X-100. Solubilized membranes were then incubated with nickel-IDA (Ademtech) magnetic beads for 30 min, washed three times in phosphate buffer, and eluted with 0.5 M imidazole in 0.2% Triton X-100.
Supplementary Material
Acknowledgments
We thank Nathalie Nadeau for excellent technical assistance; the Molecular genetics Unit members for helpful discussions; Daniel Ladant and Gouzel Karimova for help and materials for the two-hybrid assays. This work was supported by the Institut Pasteur grant PTR339. DAC was supported by an EMBO long-term fellowship and a Roux fellowship. Simulations were performed using the MareNostrum (Barcelona Supercomputing Center) and Darwin Supercomputer (University of Cambridge High Performance Computing Service), provided by Dell Inc. using Strategic Research, Infrastructure Funding from the Higher Education Funding Council for England. PJB also thanks Thomas J Piggot for advice on CHARMM36 force field.
Author contributions: DAC and OF designed the study and wrote the manuscript, conducted the experiments and with contributions from MC and APP analysed the data. PJB performed molecular dynamics and analysed the data.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abramoff MD, Magelhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophotonics Int 11: 36–42 [Google Scholar]
- Albers SV, Pohlschroder M (2009) Diversity of archaeal type IV pilin-like structures. Extremophiles 13: 403–410 [DOI] [PubMed] [Google Scholar]
- Ayers M, Howell PL, Burrows LL (2010) Architecture of the type II secretion and type IV pilus machineries. Future Microbiol 5: 1203–1218 [DOI] [PubMed] [Google Scholar]
- Birdsell DC, Cota-Robles EH (1967) Production and ultrastructure of lysozyme and ethylenediaminetetraacetate-lysozyme spheroplasts of Escherichia coli. J Bacteriol 93: 427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjelkmar P, Larsson P, Cuendet MA, Hess B, Lindahl E (2010) Implementation of the CHARMM force field in GROMACS: analysis of protein stability effects from correction maps, virtual interaction sites, and water models. J Chem Theory Comput 6: 459–466 [DOI] [PubMed] [Google Scholar]
- Camberg JL, Johnson TL, Patrick M, Abendroth J, Hol WG, Sandkvist M (2007) Synergistic stimulation of EpsE ATP hydrolysis by EpsL and acidic phospholipids. EMBO J 26: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M, Nilges M, Cisneros DA, Francetic O (2010) Detailed structural and assembly model of the type II secretion pilus from sparse data. Proc Natl Acad Sci USA 107: 13081–13086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonnelle E, Helaine S, Nassif X, Pelicic V (2006) A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Mol Microbiol 61: 1510–1522 [DOI] [PubMed] [Google Scholar]
- Chen I, Christie PJ, Dubnau D (2005) The ins and outs of DNA transfer in bacteria. Science 310: 1456–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianciotto NP (2005) Type II secretion: a protein secretion system for all seasons. Trends Microbiol 13: 581–588 [DOI] [PubMed] [Google Scholar]
- Craig L, Li J (2008) Type IV pili: paradoxes in form and function. Curr Opin Struct Biol 18: 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA (2006) Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol Cell 23: 651–662 [DOI] [PubMed] [Google Scholar]
- d'Enfert C, Ryter A, Pugsley AP (1987) Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for production, surface localization and secretion of the lipoprotein pullulanase. EMBO J 6: 3531–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douzi B, Ball G, Cambillau C, Tegoni M, Voulhoux R (2011) Deciphering the Xcp Pseudomonas aeruginosa type II secretion machinery through multiple interactions with substrates. J Biol Chem 286: 40792–40801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douzi B, Durand E, Bernard C, Alphonse S, Cambillau C, Filloux A, Tegoni M, Voulhoux R (2009) The XcpV/GspI pseudopilin has a central role in the assembly of a quaternary complex within the T2SS pseudopilus. J Biol Chem 284: 34580–34589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand E, Michel G, Voulhoux R, Kurner J, Bernadac A, Filloux A (2005) XcpX controls biogenesis of the Pseudomonas aeruginosa XcpT-containing pseudopilus. J Biol Chem 280: 31378–31389 [DOI] [PubMed] [Google Scholar]
- Forest KT (2008) The type II secretion arrowhead: the structure of GspI-GspJ-GspK. Nat Struct Mol Biol 15: 428–430 [DOI] [PubMed] [Google Scholar]
- Francetic O, Buddelmeijer N, Lewenza S, Kumamoto CA, Pugsley AP (2007) Signal recognition particle-dependent inner membrane targeting of the PulG Pseudopilin component of a type II secretion system. J Bacteriol 189: 1783–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francetic O, Pugsley AP (2005) Towards the identification of type II secretion signals in a nonacylated variant of pullulanase from Klebsiella oxytoca. J Bacteriol 187: 7045–7055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giltner CL, Habash M, Burrows LL (2010) Pseudomonas aeruginosa minor pilins are incorporated into type IV pili. J Mol Biol 398: 444–461 [DOI] [PubMed] [Google Scholar]
- Gray MD, Bagdasarian M, Hol WG, Sandkvist M (2011) In vivo cross-linking of EpsG to EpsL suggests a role for EpsL as an ATPase-pseudopilin coupling protein in the Type II secretion system of Vibrio cholerae. Mol Microbiol 79: 786–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JK, Forest KT (2006) Type IV pilin structures: insights on shared architecture, fiber assembly, receptor binding and type II secretion. J Mol Microbiol Biotechnol 11: 192–207 [DOI] [PubMed] [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4: 435–447 [DOI] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA 95: 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler R, Schafer K, Muller S, Vignon G, Diederichs K, Philippsen A, Ringler P, Pugsley AP, Engel A, Welte W (2004) Structure and assembly of the pseudopilin PulG. Mol Microbiol 54: 647–664 [DOI] [PubMed] [Google Scholar]
- Korotkov KV, Hol WG (2008) Structure of the GspK-GspI-GspJ complex from the enterotoxigenic Escherichia coli type 2 secretion system. Nat Struct Mol Biol 15: 462–468 [DOI] [PubMed] [Google Scholar]
- Lemkul JA, Bevan DR (2011) Characterization of interactions between PilA from Pseudomonas aeruginosa strain K and a model membrane. J Phys Chem B 115: 8004–8008 [DOI] [PubMed] [Google Scholar]
- Lopilato J, Bortner S, Beckwith J (1986) Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol Gen Genet 205: 285–290 [DOI] [PubMed] [Google Scholar]
- MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE et al. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102: 3586–3616 [DOI] [PubMed] [Google Scholar]
- MacKerell AD, Klauda JB, Venable RM, Freites JA, O'Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, Pastor RW (2010) Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B 114: 7830–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B, Koomey M, Sheetz MP (2004) A force-dependent switch reverses type IV pilus retraction. Proc Natl Acad Sci USA 101: 10961–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA [Google Scholar]
- Miller JH (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA [Google Scholar]
- Misic AM, Satyshur KA, Forest KT (2010) P. aeruginosa PilT structures with and without nucleotide reveal a dynamic type IV pilus retraction motor. J Mol Biol 400: 1011–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody CR, Chung YJ, Yen MR, Vidal-Ingigliardi D, Pugsley AP, Saier MH Jr (2003) Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149: 3051–3072 [DOI] [PubMed] [Google Scholar]
- Pelicic V (2008) Type IV pili: e pluribus unum? Mol Microbiol 68: 827–837 [DOI] [PubMed] [Google Scholar]
- Possot O, Vignon G, Bomchil N, Ebel F, Pugsley AP (2000) Multiple interactions between pullulanase secretion components involved in stabilization and cytoplasmic membrane association of PulE. J Bacteriol 182: 2142–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley AP (1993a) Processing and methylation of PulG, a pilin-like component of the general secretory pathway of Klebsiella oxytoca. Mol Microbiol 9: 295–308 [DOI] [PubMed] [Google Scholar]
- Pugsley AP (1993b) The complete general secretory pathway in gram-negative bacteria. Microbiol Rev 57: 50–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Py B, Loiseau L, Barras F (2001) An inner membrane platform in the type II secretion machinery of Gram-negative bacteria. EMBO Rep 2: 244–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall LL, Hardy SJ (1986) Correlation of competence for xport with lack of tertiary structure of the mature species: a study in vivo of maltose-binding protein in E. coli. Cell 46: 921–928 [DOI] [PubMed] [Google Scholar]
- Sandkvist M (2001) Type II secretion and pathogenesis. Infect Immun 69: 3523–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvonnet N, Vignon G, Pugsley AP, Gounon P (2000) Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J 19: 2221–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H, von Jagow G (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166: 368–379 [DOI] [PubMed] [Google Scholar]
- Vignon G, Kohler R, Larquet E, Giroux S, Prevost MC, Roux P, Pugsley AP (2003) Type IV-like pili formed by the type II secretion: specificity, composition, bundling, polar localization, and surface presentation of peptides. J Bacteriol 185: 3416–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winther-Larsen HC, Wolfgang M, Dunham S, van Putten JP, Dorward D, Lovold C, Aas FE, Koomey M (2005) A conserved set of pilin-like molecules controls type IV pilus dynamics and organelle-associated functions in Neisseria gonorrhoeae. Mol Microbiol 56: 903–917 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.