Abstract
Translesion synthesis polymerases (TLS Pols) are required to tolerate DNA lesions that would otherwise cause replication arrest and cell death. Aberrant expression of these specialized Pols may be responsible for increased mutagenesis and loss of genome integrity in human cancers. The molecular events that control the usage of TLS Pols in non-pathological conditions remain largely unknown. Here, we show that aberrant recruitment of TLS Polκ to replication forks results in genomic instability and can be mediated through the loss of the deubiquitinase USP1. Moreover, artificial tethering of Polκ to proliferating cell nuclear antigen (PCNA) circumvents the need for its ubiquitin-binding domain in the promotion of genomic instability. Finally, we show that the loss of USP1 leads to a dramatic reduction of replication fork speed in a Polκ-dependent manner. We propose a mechanism whereby reversible ubiquitination of PCNA can prevent spurious TLS Pol recruitment and regulate replication fork speed to ensure the maintenance of genome integrity.
Keywords: genome stability, PCNA ubiquitination, Pol kappa, replication stress, USP1
Introduction
Many DNA mutations that occur in cancer cells arise from the error-generating activities of DNA polymerases (Pols) (Lange et al, 2011). In normal dividing cells, a subtle equilibrium exists between the accurate duplication of the genome and less stringent DNA damage tolerance mechanisms that allow cells to endure DNA damage (Hoffmann and Cazaux, 2010). Translesion synthesis (TLS) is a major DNA damage tolerance mechanism whereby alternative DNA Pols known as TLS Pols are required to bypass bulky DNA lesions that would otherwise cause replication arrest and cell death (Friedberg, 2005). As these TLS Pols are largely inaccurate when replicating undamaged DNA templates, their expression and function need to be carefully monitored and controlled. Dysregulation of these error-prone Pols, including Polκ, can drive genomic instability and tumourigenesis (Bavoux et al, 2005a; Lange et al, 2011). How the cell normally prevents the aberrant usage of these specialized Pols in undamaged cells is presently unknown.
The deubiquitinase (DUB), USP1, is a key regulator of both TLS and the Fanconi Anaemia (FA) crosslink repair/genome stability pathway (Nijman et al, 2005; Huang and D’Andrea, 2006; Huang et al, 2006). First identified in a DUB siRNA screen for the FA pathway (Nijman et al, 2005), USP1 is responsible for antagonizing the monoubiquitination of two FA proteins, FANCD2 and FANCI, involved in DNA crosslink repair. Disruption of the Usp1 gene in mice causes chromosomal instability and produces a phenotype resembling FA mice, implying that dynamic ubiquitin conjugation and deconjugation of FANCD2 and FANCI are critical for efficient DNA repair and for the maintenance of genomic integrity (Kim et al, 2009). However, it is still unclear whether the lack of reversible ubiquitination in the FA pathway, due to the loss of USP1, is indeed the source for genomic instability in mammalian cells (Oestergaard et al, 2007; Kim et al, 2009). Interestingly, knockout of both Usp1 and Fancd2 in mice results in a more severe genomic instability phenotype (Kim et al, 2009). Perhaps, the regulation of additional USP1 substrates is required to protect cells against genomic instability.
USP1 is also responsible for the deubiquitination of proliferating cell nuclear antigen (PCNA), the replication sliding clamp or processivity factor for DNA replication (Huang et al, 2006). Monoubiquitination of PCNA by the ubiquitin E3 ligase RAD18 recruits TLS Pols to sites of DNA damage and stalled replication forks (Kannouche et al, 2004; Watanabe et al, 2004). Most of these specialized enzymes belong to the Y-family Pols, including Polκ, Polη, Polι, and Rev1 (Waters et al, 2009). Importantly, all Y-family Pols possess ubiquitin-binding domains (UBDs) that increase their binding affinity for ubiquitinated forms of PCNA (Bienko et al, 2005). Thus, the recruitment of TLS Pols to the replication fork can be directly regulated by events that activate PCNA ubiquitination. It is currently unknown whether aberrant ubiquitination of PCNA has any detrimental cellular effects during normal S-phase progression. Specifically, it is unknown whether genomic integrity is compromised when PCNA deubiquitination is blocked.
In this study, we set out to determine whether misregulation of TLS was primarily responsible for the genomic instability phenotype observed in USP1-depleted cells. We report that USP1 is required to prevent the aberrant recruitment of Polκ to the replication fork. Failure to do so results in enhanced micronuclei formation (marker of genomic instability) and slower replication fork speed as measured by single-molecule DNA fiber analysis. Overexpression of Polκ by itself can also cause micronuclei formation. Moreover, the direct tethering of Polκ to PCNA can further enhance genomic instability in a manner that is no longer dependent on its ubiquitin-binding function. Based on our findings, we propose a novel replication stress pathway that occurs in the absence of USP1, resulting from elevated PCNA ubiquitination and recruitment of Polκ.
Results
A ubiquitination-defective PCNA mutant can rescue the genomic instability caused by USP1 depletion
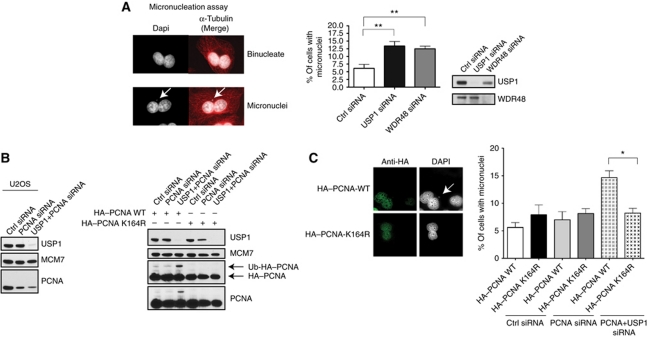
Usp1 knockout mice have increased incidence of perinatal lethality and a strong resemblance to FA mice (small size, infertility, mitomycin C hypersensitivity, and chromosome instability; Kim et al, 2009). However, at the cellular level, it is unclear which genome maintenance pathways are deregulated by the loss of USP1, thereby causing genomic instability. To investigate this further, we employed a micronucleation assay to measure genomic instability in undamaged cells transfected with siRNAs targeting USP1 or UAF1/WDR48 (catalytic cofactor of USP1) (Cohn et al, 2007; Figure 1A). Micronuclei are common in cells undergoing genotoxic or replicative stress and may contain entire chromosome pieces or fragments, making them important and highly sensitive indicators of genomic instability (Utani et al, 2010). After treatment of cells with cytochalasin-B (actin polymerization inhibitor) for 24 h, binucleate cells (cells that have undergone cell division in the absence of cytokinesis) were scored for the presence of micronuclei as a percentage of total cells. We found that depletion of either USP1 or UAF1/WDR48 increased the percentage of cells with micronuclei (Figure 1A). As a positive control, cells were treated with the DNA polymerase inhibitor, aphidicoln (APH), a known inducer of micronuclei formation and genomic instability (Supplementary Figure S1A; Chan et al, 2009; Naim and Rosselli, 2009). These data demonstrate that USP1, along with its catalytic partner UAF1/WDR48, is required for faithful chromosome segregation.
Figure 1.
A ubiquitination-defective PCNA mutant can rescue the genomic instability caused by USP1 depletion. (A) U2OS cells were transfected with siRNAs as indicated and treated with cytochalasin-B for 24 h prior to fixation for micronucleation assay. Representative images of normal and micronuclei-positive binucleate cells. Cells were co-stained for DAPI (grey) and α-tubulin (red). Graph displays percentage (%) of cells with micronuclei. Error bars represent standard deviation of experiment done in triplicate (n=300). (B) Normal U2OS or U20S cells stably expressing siRNA-resistant HA-tagged PCNA WT or K164R mutant were transfected with the indicated siRNAs. (C) Representative images of U2OS cells stably expressing HA-tagged WT or K164R siRNA-resistant PCNA. U2OS cells were transfected with PCNA and USP1 siRNAs and treated with cytochalasin-B for the micronucleation assay. Cells were stained for DAPI (grey) and anti-HA (green). Micronucleation assay was performed in these cells with the indicated siRNAs. Graph displays percentage (%) of cells with micronuclei (only anti-HA positive-stained cells were counted). Error bars represent standard deviation of experiment done in triplicate (n=300). Single asterisk represents P-value <0.05, double asterisks represent P-value<0.01.
In previous work, we and other groups have shown that reversible ubiquitination of PCNA is regulated by USP1 (Huang et al, 2006; Niimi et al, 2008). Therefore, we wanted to examine whether the major effects of USP1 depletion in promoting genomic instability are mediated through PCNA ubiquitination. We generated U2OS cells that stably express HA-tagged wild-type (WT) PCNA, or a K164R mutant that cannot be ubiquitinated. The PCNA cDNA harboured silent mutations that made it refractory to siRNA-directed targeting against endogenous PCNA, using an siRNA sequence that was previously validated (Niimi et al, 2008). We first confirmed that the PCNA siRNA could effectively knockdown endogenous PCNA in untransfected U2OS cells (Figure 1B). Cells depleted of endogenous PCNA but expressing the K164R variant did not accumulate monoubiquitinated PCNA after USP1 knockdown, in contrast to similar cells ectopically expressing WT PCNA (Figure 1B). This was confirmed by western blot by probing for total PCNA (recognizes both HA-tagged PCNA and endogenous PCNA) using an anti-PCNA antibody (Figure 1B). In the context of endogenous PCNA depletion, we found that knockdown of USP1 increased micronuclei formation in the PCNA WT-expressing cells (Figure 1C). Importantly, this effect was prevented in the PCNA K164R mutant-expressing cells (Figure 1C). These data strongly suggest that the genomic instability phenotype observed in USP1-deficient cells is driven by aberrant PCNA ubiquitination.
USP1 depletion contributes to aberrant Polκ recruitment to the replication fork
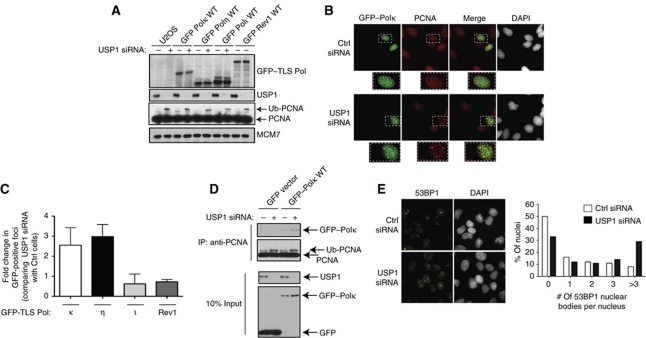
Since PCNA ubiquitination is known to be required for TLS Pol recruitment, we investigated whether USP1 depletion affects the localization of specific TLS Pols. To address this question, we generated stable U2OS cell lines expressing either GFP-tagged TLS Polκ, -η, -ι or -Rev1 (Figure 2A). It has been previously reported that GFP–Polκ forms much less replication foci than that of GFP–Polη in undamaged cells (Ogi et al, 2005). Our immunolocalization results of stably expressing GFP–Polκ and GFP–Polη are consistent with this finding (Figure 2B; Supplementary Figure S2). To determine whether the loss of USP1 results in a further increase in GFP–Pol foci formation above their individual baseline levels, we compared the fold change in the number of cells with GFP–Pol foci formation between control and USP1 siRNA treatments. Interestingly, more Polκ and Polη foci were detected in cells after USP1 depletion while there were very little changes in both Polι and Rev1 foci levels (Figure 2B and C). GFP–Polκ and Polη nuclear foci were also found to partially colocalize with PCNA in USP1-depleted cells (Figure 2B; Supplementary Figure S2). Importantly, ectopic expression of GFP–Polκ can interact preferentially with PCNA in USP1 knockdown U2OS cells (Figure 2D). These data suggest that loss of USP1 in human cells leads to aberrant recruitment or misuse of specific error-prone TLS Pols at the replication fork in undamaged conditions.
Figure 2.
USP1 depletion contributes to aberrant Polκ recruitment to the replication fork. (A) U2OS cells stably expressing GFP–Polκ, -η, -ι or -Rev1 were transfected with control (Ctrl) or USP1 siRNA and western blot analysis was performed and probed with the indicated antibodies. (B) Representative images of GFP–Polκ in cells transfected with indicated siRNAs. Cells were immunostained with DAPI (grey), anti-GFP (green) and anti-PCNA (red) antibodies, with merged images displayed to show colocalization. (C) Graph displays the fold change in cells with GFP-positive foci normalized to Ctrl siRNA-treated cells. GFP-positive cells were counted as cells containing >5 GFP foci (n=600). Experiments were performed in triplicate and error bars represent standard deviation. (D) USP1 siRNA knockdown was followed by transfection with either GFP–Polκ WT or GFP vector control. Cells were lysed and formaldehyde crosslinked as described in Materials and methods. Samples that were not crosslinked (input) were analysed by western blot. Extracts that were crosslinked and solubilized were also immunoprecipitated with anti-PCNA (PC10, Santa Cruz) and probed with the indicated antibodies. (E) Representative images of U2OS cells immunostained with anti-53BP1 antibody (green) and DAPI (Grey). Graph represents the quantification of the number of 53BP1 nuclear bodies per cell for the indicated siRNAs (n>300 for each siRNA knockdown condition).
USP1 depletion elevates 53BP1 nuclear bodies but does not cause replication checkpoint activation or cell-cycle delay
Since mislocalization of TLS Pols may cause elevated endogenous DNA damage or replication stress, we investigated whether replication checkpoint pathways were activated in USP1-depleted cells. In untreated U2OS cells, depletion of USP1 did not increase checkpoint signalling events, as measured by Chk1 and RPA2 phosphorylation (Gatei et al, 2003; Sorensen et al, 2003; Anantha et al, 2007; Vassin et al, 2009), and did not result in cell-cycle delay or arrest (Supplementary Figure 1B and C), even though the monoubiquitination of both FANCD2 and PCNA were elevated (Supplementary Figure S1B). USP1-depleted cells remained checkpoint competent in the presence of the replication inhibitor, hydroxyurea (HU), suggesting that USP1 itself was not required for checkpoint activation (Supplementary Figure S1B). However, unresolved replication stress due to incomplete DNA synthesis during S phase has recently been shown to transmit lingering DNA damage into the G1 phase of successive cell cycles, which are marked by the p53 binding protein-1 (53BP1) (Harrigan et al, 2011; Lukas et al, 2011). These 53BP1 nuclear bodies are sensitive markers of elevated replication stress and serve to prevent loss of chromosome integrity (Harrigan et al, 2011; Lukas et al, 2011). We found that USP1 knockdown resulted in elevated number of 53BP1 nuclear bodies per cell (Figure 2E). Accordingly, the increased number of 53BP1 foci observed in USP1-depleted cells suggests that these cells may possess elevated levels of replication stress, but not above the cellular threshold to cause checkpoint activation and cell-cycle arrest.
Increased micronuclei formation caused by USP1 depletion is Polκ dependent
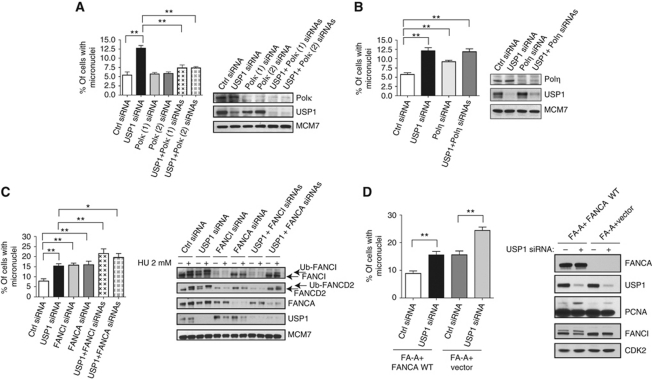
The results in Figure 2C showed that USP1 was required to prevent the mislocalization of Polκ and Polη in undamaged cells. Thus, the misregulation of either Polκ or Polη could be directly responsible for causing elevated levels of genomic instability in USP1-depleted cells. First, we investigated the functional role of Polκ and found that the increase in micronuclei formation caused by USP1 knockdown was prevented by the co-depletion of Polκ (Figure 3A). This effect was observed using either of two different siRNA sequences to knockdown Polκ and also with a second USP1 (2) siRNA sequence (Supplementary Figure S3). In contrast, depletion of Polη was unable to rescue the heightened levels of micronuclei formation occurring after loss of USP1 (Figure 3B). Knockdown of Polη alone, in fact, increased micronuclei formation (Figure 3B). This result is in agreement with a recent study demonstrating a role for Polη in the maintenance of genomic stability during unperturbed S phase (Rey et al, 2009). These findings highlight different functional requirements between Polκ and Polη and suggest that USP1 promotes genomic stability in human cells by selectively inhibiting or limiting Polκ usage during normal DNA replication.
Figure 3.
Increased micronuclei formation caused by USP1 depletion is Polκ dependent. (A, B) U2OS cells were transfected with the indicated siRNAs and assayed for micronucleation (n=600). Western blots were performed with cells from the same transfection to demonstrate efficiency of siRNA knockdown and probed with the indicated antibodies. (C) U2OS cells were transfected with the indicated siRNAs and assayed for micronucleation and western blot analysis. Hydroxyurea (HU) treatment was used as a control for DNA damage-induced monoubiquitination of FANCI and FANCD2 to validate the functional knockdown of the FA pathway. (D) Patient-derived FANCA-deficient (GM6914) fibroblasts retrovirally complemented with either vector or wild-type FANCA cDNA were treated with the indicated siRNAs and analysed by western blot or assayed for micronucleation. Graph displays percentage (%) of cells with micronuclei. Error bars represent standard deviation of experiment done in triplicate (n=300). Single asterisk represents P-value <0.05, double asterisks represent P-value<0.01.
The FA pathway protects cells against genomic instability caused by USP1 depletion
Previous studies have shown that the FA pathway plays an important role in protecting cells undergoing replication stress (Chan et al, 2009; Naim and Rosselli, 2009). Although loss of USP1 results in elevated levels of monoubiquitinated FANCD2 and FANCI even in the absence of DNA damage (Sims et al, 2007; Kim et al, 2009), it is unclear whether this increased monoubiquitination in undamaged cells perturbs the FA pathway to a similar extent as in response to induced DNA damage conditions (Oestergaard et al, 2007; Kim et al, 2009; Murai et al, 2011). As expected, knockdown of either USP1 or UAF1 in U2OS cells causes hyperaccumulation of FANCD2 and FANCI in the chromatin fraction (Supplementary Figure S4A). However, loss of USP1 elevated FANCD2 paired mitotic foci formation, suggesting an active role of the FA pathway in response to USP1-mediated replication stress (Supplementary Figure S4B). Elevated FANCD2 paired mitotic foci are indicative of unresolved replication stress and possibly even fragile site expression (Chan et al, 2009; Naim and Rosselli, 2009). Next, we addressed whether the FA pathway is important for preventing genomic instability in USP1-depleted cells. Knockdown of either FANCI or FANCA alone increased micronuclei formation, in agreement with a previous study that supported a role for the FA pathway in protecting cells undergoing replication stress (Naim and Rosselli, 2009; Figure 3C). Importantly, knockdown of FANCI or FANCA further enhanced micronuclei formation in USP1-depleted cells (Figure 3C). We were also able to confirm this finding in FA patient fibroblasts and show that USP1 knockdown in these cells can increase micronuclei formation, both in the presence and absence of FANCA WT complementation (Figure 3D). Therefore, an intact FA pathway protects cells against genomic instability and does so both in the presence and absence of USP1.
Additionally, we confirmed that RAD18, the major ubiquitin E3 ligase for PCNA in mammalian cells, was required for PCNA ubiquitination after USP1 knockdown (Supplementary Figure S4C). Unexpectedly, the loss of RAD18 alone increased micronuclei formation in U2OS cells, suggesting that RAD18 may play additional roles in the maintenance of genomic stability (Supplementary Figure S4C). RAD18 was recently identified as a positive regulator of the FA pathway (Geng et al, 2010; Williams et al, 2011).
Increase in genomic instability by Polκ overexpression depends on its UBD
Polκ overexpression, which is notably observed in lung cancer, results not only in increased spontaneous mutagenesis, but also in pleiotropic alterations such as DNA breaks, genetic exchanges, and aneuploidy (Ogi et al, 1999; Wang et al, 2001; Bergoglio et al, 2002; Bavoux et al, 2005a, 2005b). We therefore tested whether misregulation of Polκ through overexpression could mimic the genomic instability phenotype observed with USP1 depletion. Indeed, overexpression of GFP-tagged Polκ, but not Polη, enhanced micronuclei formation above the empty vector control (Figure 4A). Together, these data confirm the different functional roles between Polκ and Polη seen with USP1 depletion. Previous studies have shown that PCNA monoubiquitination facilitates Polκ recruitment to stalled replication forks after DNA damage treatment (Bi et al, 2006; Guo et al, 2008). Similarly to the mechanism of recruitment for Polη to PCNA (Bienko et al, 2010), most TLS polymerases possess a UBD to associate with the ubiquitinated form of PCNA (Bienko et al, 2005). We therefore examined whether the UBD of Polκ is important for the increase in micronuclei formation. Significantly, the increase in genomic instability caused by Polκ overexpression required its two ubiquitin-binding zinc-finger domains (UBZs) (Bienko et al, 2005; Guo et al, 2008), because deletion of the Polκ UBZs reduced micronuclei formation to levels similar to that caused by expression of a Polκ catalytic mutant (D198A, E199A) or the GFP vector control (Figure 4A, see schematic diagram). The UBZs of Polκ were previously shown to facilitate the interaction between monoubiquitinated PCNA and Polκ (Guo et al, 2008). Thus, our data strongly suggest that Polκ causes genomic instability through the engagement of its UBZs with ubiquitinated PCNA.
Figure 4.
Increasing the amount of Polκ bound to PCNA can elevate genomic instability. (A) Schematic representation of the structural domains of Polκ and Polη. Domain abbreviations are as follows: polymerase-associated domain (PAD), ubiquitin-binding zinc-finger motif (UBZ), PCNA-interacting region (PIR). U2OS cells were transfected with GFP vector, GFP–Polκ WT, GFP–Polκ CAT mut (catalytic dead, D198A, E199A), GFP–Polκ ΔUBZ (deletion of both UBZ1 and UBZ2) or GFP–Polη WT constructs and scored for micronuclei formation (n=300). Graph displays percentage (%) of cells with micronucleation. Error bars represent standard deviation of experiment done in triplicate. (B) Amino-acid sequences denoting the PIR for Polκ, p21 protein, and the Polκ p21 PIP chimera. Characterization of PIP box affinity is based on the extrapolation of published results (Hishiki et al, 2009). (C) Representative images of U2OS cells transiently expressing the indicated expression constructs. Cells were co-stained for DAPI (grey), GFP (green), and PCNA (red). (D–F) 293T cells were transfected with the plasmids indicated and lysed for co-immunoprecipitation (co-IP) with anti-GFP beads. Whole-cell extracts (WCE) from parallel samples used for co-IP were also analysed. Western blots were probed with the antibodies indicated. In (F), recombinant enzyme from non-specific catalytic domain (DUB-CD) of USP2 was used in ubiquitin deconjugation in-vitro assay to confirm that the slower migrating bands above the PCNA protein were ubiquitin conjugates. (G) Graph displays the percentage (%) of cells with micronuclei. Error bars represent standard deviation of experiment done in triplicate (n=300). Single asterisk represents P-value <0.05, double asterisks represent P-value<0.01.
Tethering Polκ to PCNA enhances micronuclei formation in a ubiquitin binding-independent manner
To further demonstrate the ability of Polκ to promote genomic instability through its direct interaction with PCNA, we developed a novel method to allow tethering of a ‘specific protein of interest’ to PCNA, in which we took advantage of what is currently known about PCNA-interacting domains on high-affinity binding factors. PCNA interacts with a large number of proteins involved in replication, repair, cell cycle, chromatin assembly, and sister chromatid cohesion (Moldovan et al, 2007). Most of these proteins have a conserved sequence, called ‘PCNA-interacting protein box’ or PIP box (Warbrick, 1998). Proteins with a canonical PIP box motif, including the human p21 cell-cycle inhibitor protein, have strong interactions with PCNA (Gulbis et al, 1996). Previous studies have assigned a PCNA-binding sequence in human Polη, Polι, and Polκ, none of which has a canonical PIP box sequence (Haracska et al, 2001a, 2001b, 2001c; Vidal et al, 2004; Ogi et al, 2005). The fact that Y-family TLS Pols do not have a canonical PIP box may be consistent with the notion that TLS Pols, which are intrinsically error-prone, have lower affinity for PCNA than the replicative Pols. A study measuring the binding affinity of PCNA to PIP box peptides derived from canonical and non-canonical PIP box-containing proteins clearly showed that the PIP box of Polκ has a lower affinity for PCNA than those of Polη and Polι, and all three TLS Pol PIP boxes have significantly lower affinity than the canonical p21 PIP box (Hishiki et al, 2009). Furthermore, an extended PCNA interaction surface, termed PCNA-interacting region (PIR), was recently identified on the extreme C-terminus of Polη and encompassed the non-canonical PIP box motif (Bienko et al, 2010). Polκ and other known interactors of PCNA, including p21, also contain a putative PIR on their C-termini (Bienko et al, 2010). To engineer TLS Pols to bind tighter to PCNA, we swapped the PIR of Polκ and Polη for the p21 PIR (see schematic diagram, Figure 4B). To remove the possibility that the p21 PIP box may also function as a degron for the CRL4 (Cdt2) ubiquitin ligase complex (Havens and Walter, 2009), we also changed the critical Arg residue to Ala (shown as an asterisk, Figure 4B) to prevent unscheduled degradation of the chimera protein.
Expression of either GFP-tagged Polκ p21 PIP WT or the Polη p21 PIP WT caused the dramatic formation of punctate nuclear dots that are restricted to S phase (as determined by PCNA staining pattern) (Supplementary Figure S5A and data not shown). The nuclear foci generated by these p21 PIP chimera proteins colocalized with endogenous PCNA, unlike their non-chimera counterparts (Figure 4C). Importantly, deletion of the UBZ domains in the context of Polκ p21 PIP chimera still allowed formation of distinct Polκ foci at levels similar to Polκ p21 PIP WT protein (Figure 4C). In the absence of exogenous DNA damage, expression of the non-chimera Polκ WT and the UBZ mutant constructs did not form robust replication foci (Figure 4C). This may be due to the transitory nature of the Polκ association with the replication fork in the presence of USP1. We also found that both GFP-tagged Polκ PIP WT and Polη p21 PIP WT proteins could robustly interact with both ubiquitinated and unmodified PCNA (Figure 4D and E). Expression of the Polκ WT chimera Pols could stabilize and pull down K164-specific mono- and possibly poly-ubiquitinated forms of PCNA (Figure 4D–F). Recombinant USP2 containing only its catalytic domain (DUB-CD) was added to the anti-GFP immunoprecipitation in order to show that the higher migrating bands that correspond to modified PCNA are indeed ubiquitinated species of PCNA (Figure 4F, right panel). Interestingly, the amount of ubiquitinated versus unmodified form of PCNA captured by the tethered Polκ was influenced by the presence of a functional UBD (Figure 4D). More importantly, expression of the Polκ p21 PIP WT, but not the Polη p21 PIP WT chimera, could dramatically increase micronuclei formation in U2OS cells (Figure 4G). The level of micronuclei formation (both in the percent of cells with micronuclei and the number of micronuclei per cell) caused by the expression of the Polκ p21 PIP WT chimera was significantly above that seen following expression of the Polκ WT non-chimera (Figure 4G; Supplementary Figure S5B). We also show that both the WT and the UBZ-deleted Polκ p21 PIP chimera proteins caused similar levels of elevated micronuclei formation (Figure 4G; Supplementary Figure S5B). Finally, the genomic instability caused by the Polκ p21 PIP protein was still dependent on its catalytic activity, because the Polκ p21 PIP CAT (D198A, E199A) was unable to promote micronuclei formation to the levels of the WT non-chimera protein (Figure 4G). The Polκ p21 PIP CAT was capable of forming nuclear foci at levels similar to that of WT Polκ p21 PIP chimera protein (Supplementary Figure S5C). Collectively, our data strongly suggest that the Polκ ubiquitin-binding function is mainly required for recruitment to the replication fork and is largely dispensable when Polκ is directly tethered to PCNA.
USP1 depletion causes reduced replication fork speed in a Polκ-dependent manner
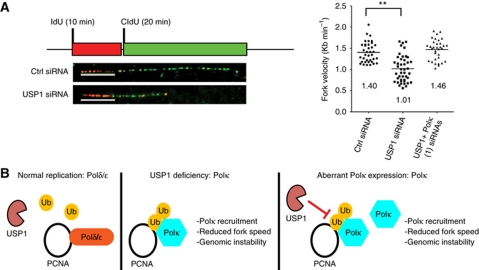
To gain further insight into the molecular basis of how the loss of USP1 contributes to genomic instability, we analysed the replication program of USP1-depleted cells using single-molecule DNA fiber analysis. U2OS cells transfected with different siRNAs were sequentially pulse-labelled with iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU) to determine the polarity of elongating replication forks and replication fork speed (Figure 5A). Following antibody staining and imaging of purified fibers, we observed an average fork speed of 1.44 Kb min−1, consistent with previous studies of asynchronous cells (Berezney et al, 2000) (Figure 5A). In USP1-depleted cells, a significant reduction in the average fork speed (1.10 Kb min−1) was detected (Figure 5A). Importantly, the co-depletion of Polκ in the USP1 knockdown cells rescued the replication fork speed to rates similar to those in control cells (1.45 Kb min−1) (Figure 5A). These results suggest that the misuse of Polκ is primarily responsible for the reduced fork velocity in USP1-depleted cells.
Figure 5.
USP1 depletion reduces the replication fork speed in a Polκ-dependent manner. (A) U2OS cells were transfected with the indicated siRNAs for 72 h and then labelled with IdU and CldU and prepared for single-molecule DNA fiber analysis. Schematic representation of replication pattern on combed DNA molecules. A representative DNA fiber from Ctrl or USP1 siRNA-treated cells. Scale bar=5 Kb. Distribution of replication fork velocities is displayed, showing the mean replication fork velocity for each siRNA condition. (B) Schematic model for the maintenance of genomic stability through the regulation of Polκ recruitment by USP1. In the absence of DNA damage, high levels of USP1 during S phase inhibit PCNA ubiquitination, ensuring replicative polymerases Polδ/ε are maintained at the replication fork (left diagram). In USP1-deficient cells, PCNA ubiquitination is increased, leading to aberrant Polκ recruitment to the replication fork, reduced replication fork speed and genomic instability (middle). Similarly, Polκ overexpression results in increased Polκ recruitment to the replication fork, reduced replication fork speed, and increased genomic instability. Depending on the availability of USP1, genomic instability manifested in these cells by aberrant Polκ expression may vary (right). Double asterisks represent P-value<0.01.
Collectively, our data support a model whereby the loss of USP1 in undamaged cells promotes genomic instability through a novel replication stress mechanism consisting of (1) hyperubiquitination of PCNA, (2) aberrant recruitment of Polκ to ubiquitinated PCNA, (3) uncontrolled engagement of Polκ at the replication fork leading to slower fork speeds, and (4) interference of timely completion of genome-wide DNA synthesis resulting in partially unreplicated genomic regions (expression of common fragile sites) and micronuclei formation after cell division (Figure 5B).
Discussion
Results from our study provide mechanistic insights into how USP1 maintains genomic stability in human cells. Unexpectedly, a major genome stabilizing function of USP1 is to suppress sporadic PCNA ubiquitination during the normal DNA replication process. Failure to reduce the pool of ubiquitinated PCNA leads to the aberrant recruitment of Polκ to the replication fork. Misuse of Polκ during DNA replication results in a slower replication fork, micronuclei formation, and genomic instability. While it is known that the FA pathway can also be regulated by USP1, it is still unclear how USP1 is directly involved in mediating DNA crosslink repair and the maintenance of genomic stability through the function of FA proteins. On the other hand, the FA pathway likely plays a critical role in preventing further replication stress-induced DNA damage in USP1-deficient cells. In summary, our data demonstrate that the genomic instability observed in USP1-depleted cells can be primarily attributed to the dysregulation of the TLS pathway through elevated levels of PCNA ubiquitination and hyperaccumulation of Polκ to the replication fork (model in Figure 5B).
Past studies have strongly hinted at this potentially genome destabilizing function of Polκ. Typically, genetic studies investigating the function of TLS Pols in mammalian systems are based on ‘loss-of-function’ studies (Limoli et al, 2002; Ogi et al, 2002; Dumstorf et al, 2006; Lin et al, 2006; Stancel et al, 2009). However, the inactivation of Polκ, similar to that of Polι, does not result in any dramatic phenotype (Ogi et al, 2002; Stancel et al, 2009). In contrast, the overexpression of Polκ has much more deleterious consequences (Ogi et al, 1999; Bergoglio et al, 2002; Bavoux et al, 2005b). Studies have shown that transient overexpression of Polκ in mouse and human fibroblasts increased the mutation rate at the chromosomal HPRT locus (Ogi et al, 1999; Bergoglio et al, 2002). Ectopic expression of Polκ in Chinese hamster ovary (CHO) cells not only promoted an increase in mutation rate but also DNA breaks and high levels of genetic recombination (both homologous and non-homologous), losses of heterozygosities (LOH), and aneuploidy (Bavoux et al, 2005b). Additionally, excess Polκ in a p53-deficient background favoured tumourigenesis in nude mice. Finally, the role of Polκ in cancer development was highlighted by a study measuring an excess of Polκ in non-small cell lung cancer patients (NSCLC; Wang et al, 2001).
In our study, we showed that overexpression of Polκ, but not Polη, increases micronuclei formation. It was previously shown that Polη overexpression does not elevate the spontaneous mutagenic rate in human cells (King et al, 2005). In contrast, depletion of Polη, but not Polκ, increased micronuclei formation, confirming an important role for Polη in maintaining genomic stability during an unperturbed S phase (Rey et al, 2009). Although the function of Polη during a normal S phase is presently unclear, recent structural analysis suggests that Polη may contain a catalytic domain capable of acting as a wedge to help assist replication through D loop (during homologous recombination) and fragile site structures (Biertumpfel et al, 2010). Together, our data demonstrate that Polκ possesses a ‘gain-of-function’ ability to interfere with the normal replication program, while Polη plays an important DNA repair role during replication to preserve genomic integrity. In the future, it will be important to determine whether the expression, protein turnover, or activity of Polκ is subjected to tighter control than Polη during S-phase progression.
It is presently unclear why the aberrant recruitment of Polκ to PCNA is so detrimental to undamaged cells. Among TLS Pols, Polκ has been reported to have moderate processive DNA Pol activity based on in-vitro primer extension assays despite being error prone, including the generation of frameshift mutations (Ohashi et al, 2000; Zhang et al, 2000; Gerlach et al, 2001). We speculate that Polκ may possess higher affinity for the ubiquitinated forms of PCNA due to its two UBZ domains. Also, once it is loaded onto PCNA, the mutagenic Polκ may remain associated with the replication fork for a much longer time period based on its moderate processivity and ability to generate frameshift mutations in comparison with other TLS Pols. Alternatively, other TLS Pols, including Polι and Rev1, may require additional unknown factors for recruitment to the replication fork. Recently, two Rad5-related ubiquitin E3 ligases, SHPRH and HLTF, have been shown to coordinate the recruitment of different TLS Pols for post-replication repair (Lin et al, 2011). Perhaps, SHPRH is responsible for the selective recruitment of Polκ to the replication fork in the absence of USP1. Structural analysis of Polκ associated with either a damaged or non-damaged template is still lacking and will likely provide important mechanistic clues as to how the misuse of Polκ contributes to genomic instability in unperturbed cells.
We devised a novel PCNA tethering system to enhance our study of the ‘gain-of-function’ property for Polκ. Constructing a Polκ chimera protein harbouring a specific high-affinity PIP box region, we efficiently targeted Polκ to the replication fork to cause elevated PCNA ubiquitination, greater localization to nuclear foci, and enhanced micronuclei formation, as compared with cells ectopically expressing WT Polκ. Interestingly, the tethering of Polη to the replication fork also elevates PCNA ubiquitination and nuclear foci formation, but did not increase genomic instability. This demonstrates that the PCNA tethering system did not introduce grossly artificial functions to the Pols, but rather simply enhanced their intrinsic Pol function due to their prolonged residence on the replication fork. Furthermore, tethering a catalytically dead Polκ mutant to PCNA was unable to elevate micronuclei formation, suggesting that the Polκ-mediated genomic instability was not simply due to blocking or interfering with the recruitment of high-fidelity replicative Pols to the replication fork. We believe that the PCNA tethering system may be a useful tool to study specific ‘gain-of-function’ properties of TLS Pols in an in-vivo setting, especially to determine the natural replicative bypass function of these Pols for damaged and undamaged DNA templates in human cells.
Usp1-deficient mice displayed an FA-like phenotype (Kim et al, 2009). However, the double knockout of Usp1 and Fancd2 resulted in an even more severe phenotype than either single knockout, suggesting that Usp1 may regulate additional genome stability pathways in mice (Kim et al, 2009). Based on the findings presented in this report, it would be of particular interest to determine whether the phenotype linked to chromosomal instability in the Usp1 knockout mice can be partially or fully reversed in animals also containing a Polκ deletion. Importantly, Polκ-deficient mice are viable and do not display obvious phenotypes (Schenten et al, 2002; Stancel et al, 2009). We predict that some of the phenotypic severity of USP1 protein deficiency will be attributable to Polκ misregulation.
We propose that the genomic instability in USP1-depleted cells is due, in part, to the overengagement of Polκ with PCNA, which can ultimately lead to a slow down of replication fork speed. In agreement with our findings, overexpression of Polκ was previously shown to reduce replication fork speed without activation the S-phase checkpoint machinery (Pillaire et al, 2007). In order to compensate for the slow fork, cells overexpressing Polκ were able to promote the activation of additional replication origins (Pillaire et al, 2007). A similar compensatory mechanism was reported by Debatisse and colleagues upon reduction of intracellular nucleotide pools (Anglana et al, 2003). In this study, loss of USP1 did not activate cell-cycle checkpoints nor did it delay S-phase progression, implying that the reduction in replication fork speed may also require a concomitant overall increase in origin firing/initiation in order to complete DNA synthesis in a timely manner. In future studies, it will be interesting to determine whether USP1-deficient cells have shorter inter-origin distances at the genome-wide level.
Precisely, how general slowing of replication fork speeds in human cells can affect genomic stability is still currently not well appreciated. Data from past studies suggest that processes that interfere with replication fork progression are capable of causing genomic instability and fragile site expression. For example, replicative polymerase mutants in budding yeast that affect replication fork progression are capable of inducing chromosomal rearrangements at specific loci reminiscent of common fragile sites (Lemoine et al, 2005). In human cells, elevated fragile site expression has been reported in cells containing a compromised ATR/Chk1 checkpoint signalling pathway (Casper et al, 2002; Durkin et al, 2006) and inhibiting the ATR/Chk1 pathway in an unperturbed S phase has been shown to cause a two-fold reduction in replication fork speed (Petermann et al, 2006, 2010). The use of APH to induce fragile site expression is further evidence linking replication fork speed and genomic instability at fragile sites (Durkin and Glover, 2007). Interestingly, fragile sites may be sensitive to reduced replication forks speeds due to these regions possessing a low abundance of replication initiation events, coupled to the relatively late firing of these origins (Letessier et al, 2011). In the future, it will be interesting to test whether the reduced replication fork speed in USP1-deficient cells is correlated with increased expression of common fragile sites. In summary, our findings unveil a novel mechanism of replication stress in USP1-depleted cells involving reduced replication fork speed due to the misuse of error-prone Polκ.
Materials and methods
Cell culture
HeLa, U2OS, and HEK293 cells (ATCC) were grown in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) with 10% fetal bovine serum (FBS), 1% Pen-Strep, 1% Glutamine at 5% CO2 and 37°C incubator. U2OS cell lines stably expressing GFP–Polκ, -η, -ι and Rev1 were selected for GFP expression by flow cytometry on a MoFlo (Dako, Carpinteria, CA). FA patient-derived FANCA-deficient fibroblasts GM6914 (Coriell Cell Repository) were grown in 15% FBS.
Transfections, DNA constructs, and siRNA oligonucleotides
Transfections with plasmid DNA were performed using Fugene6 transfection reagent (Roche Applied Science) and siRNA oligos were transfected using Hiperfect transfection reagent (Qiagen). The siRNAs used were synthesized by Qiagen. Targeting sequences are human Polκ (1) AACCTCTAGAAATGTCTCATA, (2) AAGATTATGAAGCCCATCCAA; Polη CTGGTTGTGAGCATTCGTGTA; USP1 (1) TCGGCAATACTTGCTATCTTA, (2) TTGGCAAGTTATGAATTGATA; FANCA AAGGGTCAAGAGGGAAAAATA; WDR48 CCGGTCGAGACTCTATCATAA; FANCI CACGGGCATCTGGGAGATATA; ATR AACCTCCGTGATGTTGCTTGA; PCNA GCCGAGAUCUCAGCCAUAUTT and the All-Stars Negative Control siRNA. Mutagenesis was performed by PCR using QuikChange Site-Directed Mutagenesis Kit (Stratagene) with the following primers: PCNA siRNA-resistant forward primer (5′-GGTGAATTTGCACGTATATGCAGGGACTTATCTCATATTGGAGATGCTGTTGTA-3′) reverse (5′-TACAACAGCATCTCCAATATGAGATAAGTCCCTGCATATACGTGCAAATTCACC-3′), Polη p21 PIP forward primer (5′-GAACCTCGAGCTGATCCAAAAAAGAAGAGAAAGGTAATGGCTACTGGACAGGATCGAGTGGTT-3′) reverse (5′-GAACGAATTCTTAGGAGAAGATCAGCCGTGCTTTGGAGTGGTAGAAATCTGTCATGCTGGTCTGCCGCCGTTTTCGGCCTTGATGAGATACGGCAGA-3′), Polκ p21 PIP mutant forward primer (5′-GAACTCGAGCTGATCCAAAAAAGAAGAGAAAGGTAATGGATAGCACAAAGGAGAAGTGTGAC-3′) reverse (5′-GAAGGATCCTTAGGAGAAGATCAGCCGTGCTTTGGAGTGGTAGAAATCTGTCATGCTGGTCTGCCGCCGTTTTCGTGTTCTTGTTACAGCCTTCTG-3′), Polκ UBZ1 deletion forward primer (5′-CAGAGAATTCAGATGACTGTCAGGATGGACCTTCAATCAGTG-3′) reverse (5′-CACTGATTGAAGGTCCATCCTGACAGTCATCTGAATTCTCTG-3′), Polκ UBZ2 deletion forward primer (5′-CCTTACTTATGTGAAGTGAAAACAGGCCAAAATAAAAGTTTTATCCAAGAATTAAGAAAGG-3′) reverse (5′-CCTTTCTTAATTCTTGGATAAAACTTTTATTTTGGCCTGTTTTCACTTCACATAAGTAAGG-3′), Polκ catalytic dead mutant forward primer (5′-GGCCATGAGTCTTGCAGCTGCCTACTTGAATATAACAAAGC-3′) reverse (5′-GCTTTGTTATATTCAAGTAGGCAGCTGCAAGACTCATGGCC-3′).
Western blotting, immunoprecipitation and in-vitro DUB assay
Western blots were performed with whole-cell extracts prepared in SDS sample buffer (0.1 M Tris pH 6.8, 2% (w/v) SDS and 12% (v/v) β-mercaptoethanol). For co-immunoprecipitation studies, cells were lysed in low IP buffer (25 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.2% NP-40) and incubated with anti-GFP agarose (MBL). Protein extracts were separated onto Nupage 3–8% Tris-Acetate or 4–12% Bis-Tris gels (Invitrogen). Immunoblotting was performed as previously described (Sims et al, 2007). The following antibodies were used for western blot analysis: WDR48/UAF1 (Evoquest, Invitrogen), FANCI (Bethyl), PCNA (PC10, Santa Cruz), FANCA (Bethyl), MCM7 (sc-9966, Santa Cruz), Polκ (Bethyl), Polη (Bethyl), GFP (ab290, Abcam), Rad17 (Bethyl), 53BP1 (Ab36823, Abcam), RFC2 (Bethyl), anti-HA (MMS-101R, Covance), and Rad18 (Ab79763, Abcam). Isolation of formaldehyde crosslinked Triton-insoluble and -soluble fractions is as previously described (Huang et al, 2006). In-vitro DUB assay was performed in reaction buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, and 2 mM DTT) at 37°C for 30 min using recombinant USP2 catalytic domain (DUB-CD) (Boston Biochem) and terminated with 2 × SDS loading buffer.
Immunofluorescence and micronucleation assay
Cells were grown and processed in 8-well Lab-Tek® II Chamber Slide™ System slides from Nunc (Naperville, IL). Mitotic FANCD2 staining was performed in U2OS cells fixed with 4% (v/v) paraformaldehyde in PBS. Cells were blocked with PBS containing 1% BSA, 0.2% Triton X-100, and incubated with anti-FANCD2 (1:400, NB 100-182 Novus) for 2 h. GFP TLS Pol nuclear foci formation was detected after methanol fixation and stained with anti-GFP (B-2, Santa Cruz Biotechnology) for 2 h. Micronuclei were measured in cells treated with cytochalasin-B (2 μg ml−1) (Sigma) for 24 h before fixation with methanol. Cells fixed for the micronuceation assay were also stained with mouse anti-α tubulin (Sigma) or DAPI. Secondary antibodies used were Alexa Fluor 488-conjugated goat anti-rabbit, goat anti-mouse IgG (Invitrogen), Alexa Fluor 564-conjugated goat anti-mouse, and goat anti-rabbit IgG (Invitrogen). Slides were mounted in Vectashield with DAPI (Vector Laboratories, Burlingame, CA). Images were deconvolved using Softworx software (Applied Precision). The images were opened and then sized and placed into figures using image J (available at http://rsb.info.nih.gov/ij) or Adobe Photoshop 7.0 Professional (Adobe Systems, Mountain View, CA).
Single-molecule DNA fiber analysis
U2OS cells were labelled for 10 min with 50 μM IdU (Sigma), washed 3 × in PBS and incubated with 200 μM CldU (Sigma) for 20 min. The labelled cells were then treated with 1 mM thymidine for 1 h before they were harvested with trypsin. Genomic DNA was extracted from proteinase K-treated agarose plugs (105 cells per plug). Genomic DNA combed onto the silanated coverslips (Matsunami Glass) as described previously (Michalet et al, 1997). IdU and CldU were detected with monoclonal mouse BrdU (1:10, Becton Dickinson) and rat anti-BrdU antibodies (1:10, Abcam), respectively. Total denatured DNA was also detected to ensure labelled fibers remained intact using Anti-Human ssDNA (MAB3034, Millipore). Secondary antibodies Alexa Fluor 488-conjugated chicken anti-rat IgG (Invitrogen), Alexa Fluor 546-conjugated goat anti-mouse IgG (Invitrogen), and Alexa 647 goat anti-mouse IgG (Invitrogen) were used to detect primary antibodies. Images were captured and distances were measured using a Deltavision personalDV system (Applied Precision, Issaquah, WA) on a base Olympus IX71 microscope and a CoolSnap HQ camera (Photometrics). Fork speed was calculated by dividing the track size in Kb by the labelling time.
Supplementary Material
Acknowledgments
We are grateful to N Hegarat, H Hochegger, and A Murphy for technical assistance with DNA fiber analysis. We thank C Guo, E Friedberg, M Bienko, I Dikic, T Ogi, and A Lehmann for GFP-tagged TLS Pol constructs. We thank A D’Andrea for FANCD2 and USP1 antibodies and J Borowiec for phospho-RPA2 antibody. We greatly appreciate M Bienko and I Dikic for the scientific discussion that contributed to the design of the PCNA tethering system. We thank Y Deng and the Microscopy Core of NYU for assistance with cell imaging. We are also grateful to J Borowiec and G David for their critical reading of the manuscript. We also thank members of the T Huang, M Pagano, D Bar-Sagi, and D Reinberg laboratories for their reagents, technical assistance, and equipment. This work was supported by NIH grant RO1GM084244 and NYU School of Medicine Start-up fund (TTH).
Author contributions: MJ and LC planned and executed the experiments, MJ and TH wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anantha RW, Vassin VM, Borowiec JA (2007) Sequential and synergistic modification of human RPA stimulates chromosomal DNA repair. J Biol Chem 282: 35910–35923 [DOI] [PubMed] [Google Scholar]
- Anglana M, Apiou F, Bensimon A, Debatisse M (2003) Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell 114: 385–394 [DOI] [PubMed] [Google Scholar]
- Bavoux C, Hoffmann JS, Cazaux C (2005a) Adaptation to DNA damage and stimulation of genetic instability: the double-edged sword mammalian DNA polymerase kappa. Biochimie 87: 637–646 [DOI] [PubMed] [Google Scholar]
- Bavoux C, Leopoldino AM, Bergoglio V, O-Wang J, Ogi T, Bieth A, Judde JG, Pena SD, Poupon MF, Helleday T, Tagawa M, Machado C, Hoffmann JS, Cazaux C (2005b) Up-regulation of the error-prone DNA polymerase {kappa} promotes pleiotropic genetic alterations and tumorigenesis. Cancer Res 65: 325–330 [PubMed] [Google Scholar]
- Berezney R, Dubey DD, Huberman JA (2000) Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108: 471–484 [DOI] [PubMed] [Google Scholar]
- Bergoglio V, Bavoux C, Verbiest V, Hoffmann JS, Cazaux C (2002) Localisation of human DNA polymerase kappa to replication foci. J Cell Sci 115: 4413–4418 [DOI] [PubMed] [Google Scholar]
- Bi X, Barkley LR, Slater DM, Tateishi S, Yamaizumi M, Ohmori H, Vaziri C (2006) Rad18 regulates DNA polymerase kappa and is required for recovery from S-phase checkpoint-mediated arrest. Mol Cell Biol 26: 3527–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, Hofmann K, Dikic I (2005) Ubiquitin-binding domains in y-family polymerases regulate translesion synthesis. Science 310: 1821–1824 [DOI] [PubMed] [Google Scholar]
- Bienko M, Green CM, Sabbioneda S, Crosetto N, Matic I, Hibbert RG, Begovic T, Niimi A, Mann M, Lehmann AR, Dikic I (2010) Regulation of translesion synthesis DNA polymerase eta by monoubiquitination. Mol Cell 37: 396–407 [DOI] [PubMed] [Google Scholar]
- Biertumpfel C, Zhao Y, Kondo Y, Ramon-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, Yang W (2010) Structure and mechanism of human DNA polymerase eta. Nature 465: 1044–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper AM, Nghiem P, Arlt MF, Glover TW (2002) ATR regulates fragile site stability. Cell 111: 779–789 [DOI] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID (2009) Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol 11: 753–760 [DOI] [PubMed] [Google Scholar]
- Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, D’Andrea AD (2007) A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell 28: 786–797 [DOI] [PubMed] [Google Scholar]
- Dumstorf CA, Clark AB, Lin Q, Kissling GE, Yuan T, Kucherlapati R, McGregor WG, Kunkel TA (2006) Participation of mouse DNA polymerase iota in strand-biased mutagenic bypass of UV photoproducts and suppression of skin cancer. Proc Natl Acad Sci USA 103: 18083–18088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin SG, Arlt MF, Howlett NG, Glover TW (2006) Depletion of CHK1, but not CHK2, induces chromosomal instability and breaks at common fragile sites. Oncogene 25: 4381–4388 [DOI] [PubMed] [Google Scholar]
- Durkin SG, Glover TW (2007) Chromosome fragile sites. Annu Rev Genet 41: 169–192 [DOI] [PubMed] [Google Scholar]
- Friedberg EC (2005) Suffering in silence: the tolerance of DNA damage. Nat Rev Mol Cell Biol 6: 943–953 [DOI] [PubMed] [Google Scholar]
- Gatei M, Sloper K, Sorensen C, Syljuasen R, Falck J, Hobson K, Savage K, Lukas J, Zhou BB, Bartek J, Khanna KK (2003) Ataxia-telangiectasia-mutated (ATM) and NBS1-dependent phosphorylation of Chk1 on Ser-317 in response to ionizing radiation. J Biol Chem 278: 14806–14811 [DOI] [PubMed] [Google Scholar]
- Geng L, Huntoon CJ, Karnitz LM (2010) RAD18-mediated ubiquitination of PCNA activates the Fanconi anemia DNA repair network. J Cell Biol 191: 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach VL, Feaver WJ, Fischhaber PL, Friedberg EC (2001) Purification and characterization of pol kappa, a DNA polymerase encoded by the human DINB1 gene. J Biol Chem 276: 92–98 [DOI] [PubMed] [Google Scholar]
- Gulbis JM, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J (1996) Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 87: 297–306 [DOI] [PubMed] [Google Scholar]
- Guo C, Tang TS, Bienko M, Dikic I, Friedberg EC (2008) Requirements for the interaction of mouse Polkappa with ubiquitin and its biological significance. J Biol Chem 283: 4658–4664 [DOI] [PubMed] [Google Scholar]
- Haracska L, Johnson RE, Unk I, Phillips B, Hurwitz J, Prakash L, Prakash S (2001a) Physical and functional interactions of human DNA polymerase eta with PCNA. Mol Cell Biol 21: 7199–7206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Johnson RE, Unk I, Phillips BB, Hurwitz J, Prakash L, Prakash S (2001b) Targeting of human DNA polymerase iota to the replication machinery via interaction with PCNA. Proc Natl Acad Sci USA 98: 14256–14261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L (2001c) Interaction with PCNA is essential for yeast DNA polymerase eta function. Mol Cell 8: 407–415 [DOI] [PubMed] [Google Scholar]
- Harrigan JA, Belotserkovskaya R, Coates J, Dimitrova DS, Polo SE, Bradshaw CR, Fraser P, Jackson SP (2011) Replication stress induces 53BP1-containing OPT domains in G1 cells. J Cell Biol 193: 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens CG, Walter JC (2009) Docking of a specialized PIP box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol Cell 35: 93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishiki A, Hashimoto H, Hanafusa T, Kamei K, Ohashi E, Shimizu T, Ohmori H, Sato M (2009) Structural basis for novel interactions between human translesion synthesis polymerases and proliferating cell nuclear antigen. J Biol Chem 284: 10552–10560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann JS, Cazaux C (2010) Aberrant expression of alternative DNA polymerases: a source of mutator phenotype as well as replicative stress in cancer. Semin Cancer Biol 20: 312–319 [DOI] [PubMed] [Google Scholar]
- Huang TT, D’Andrea AD (2006) Regulation of DNA repair by ubiquitylation. Nat Rev Mol Cell Biol 7: 323–334 [DOI] [PubMed] [Google Scholar]
- Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D’Andrea AD (2006) Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol 8: 339–347 [DOI] [PubMed] [Google Scholar]
- Kannouche PL, Wing J, Lehmann AR (2004) Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell 14: 491–500 [DOI] [PubMed] [Google Scholar]
- Kim JM, Parmar K, Huang M, Weinstock DM, Ruit CA, Kutok JL, D’Andrea AD (2009) Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev Cell 16: 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King NM, Nikolaishvili-Feinberg N, Bryant MF, Luche DD, Heffernan TP, Simpson DA, Hanaoka F, Kaufmann WK, Cordeiro-Stone M (2005) Overproduction of DNA polymerase eta does not raise the spontaneous mutation rate in diploid human fibroblasts. DNA Repair (Amst) 4: 714–724 [DOI] [PubMed] [Google Scholar]
- Lange SS, Takata K, Wood RD (2011) DNA polymerases and cancer. Nat Rev Cancer 11: 96–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD (2005) Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell 120: 587–598 [DOI] [PubMed] [Google Scholar]
- Letessier A, Millot GA, Koundrioukoff S, Lachages AM, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M (2011) Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 470: 120–123 [DOI] [PubMed] [Google Scholar]
- Limoli CL, Giedzinski E, Bonner WM, Cleaver JE (2002) UV-induced replication arrest in the xeroderma pigmentosum variant leads to DNA double-strand breaks, gamma -H2AX formation, and Mre11 relocalization. Proc Natl Acad Sci USA 99: 233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JR, Zeman MK, Chen JY, Yee MC, Cimprich KA (2011) SHPRH and HLTF act in a damage-specific manner to coordinate different forms of postreplication repair and prevent mutagenesis. Mol Cell 42: 237–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Clark AB, McCulloch SD, Yuan T, Bronson RT, Kunkel TA, Kucherlapati R (2006) Increased susceptibility to UV-induced skin carcinogenesis in polymerase eta-deficient mice. Cancer Res 66: 87–94 [DOI] [PubMed] [Google Scholar]
- Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Solvhoj Pedersen R, Grofte M, Chan KL, Hickson ID, Bartek J, Lukas J (2011) 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol 13: 243–253 [DOI] [PubMed] [Google Scholar]
- Michalet X, Ekong R, Fougerousse F, Rousseaux S, Schurra C, Hornigold N, van Slegtenhorst M, Wolfe J, Povey S, Beckmann JS, Bensimon A (1997) Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science 277: 1518–1523 [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S (2007) PCNA, the maestro of the replication fork. Cell 129: 665–679 [DOI] [PubMed] [Google Scholar]
- Murai J, Yang K, Dejsuphong D, Hirota K, Takeda S, D’Andrea AD (2011) The USP1/UAF1 complex promotes double-strand break repair through homologous recombination. Mol Cell Biol 31: 2462–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim V, Rosselli F (2009) The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat Cell Biol 11: 761–768 [DOI] [PubMed] [Google Scholar]
- Niimi A, Brown S, Sabbioneda S, Kannouche PL, Scott A, Yasui A, Green CM, Lehmann AR (2008) Regulation of proliferating cell nuclear antigen ubiquitination in mammalian cells. Proc Natl Acad Sci USA 105: 16125–16130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D’Andrea AD, Bernards R (2005) The deubiquitinating enzyme USP1 regulates the fanconi anemia pathway. Mol Cell 17: 331–339 [DOI] [PubMed] [Google Scholar]
- Oestergaard VH, Langevin F, Kuiken HJ, Pace P, Niedzwiedz W, Simpson LJ, Ohzeki M, Takata M, Sale JE, Patel KJ (2007) Deubiquitination of FANCD2 is required for DNA crosslink repair. Mol Cell 28: 798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogi T, Kannouche P, Lehmann AR (2005) Localisation of human Y-family DNA polymerase {kappa}: relatio. J Cell Sci 118: 129–136 [DOI] [PubMed] [Google Scholar]
- Ogi T, Kato T Jr, Kato T, Ohmori H (1999) Mutation enhancement by DINB1, a mammalian homologue of the Escherichia coli mutagenesis protein dinB. Genes Cells 4: 607–618 [DOI] [PubMed] [Google Scholar]
- Ogi T, Shinkai Y, Tanaka K, Ohmori H (2002) Polkappa protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc Natl Acad Sci USA 99: 15548–15553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi E, Bebenek K, Matsuda T, Feaver WJ, Gerlach VL, Friedberg EC, Ohmori H, Kunkel TA (2000) Fidelity and processivity of DNA synthesis by DNA polymerase kappa, the product of the human DINB1 gene. J Biol Chem 275: 39678–39684 [DOI] [PubMed] [Google Scholar]
- Petermann E, Maya-Mendoza A, Zachos G, Gillespie DA, Jackson DA, Caldecott KW (2006) Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol Cell Biol 26: 3319–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Woodcock M, Helleday T (2010) Chk1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci USA 107: 16090–16095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaire MJ, Betous R, Conti C, Czaplicki J, Pasero P, Bensimon A, Cazaux C, Hoffmann JS (2007) Upregulation of error-prone DNA polymerases beta and kappa slows down fork progression without activating the replication checkpoint. Cell Cycle 6: 471–477 [DOI] [PubMed] [Google Scholar]
- Rey L, Sidorova JM, Puget N, Boudsocq F, Biard DS, Monnat RJ Jr, Cazaux C, Hoffmann JS (2009) Human DNA polymerase eta is required for common fragile site stability during unperturbed DNA replication. Mol Cell Biol 29: 3344–3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenten D, Gerlach VL, Guo C, Velasco-Miguel S, Hladik CL, White CL, Friedberg EC, Rajewsky K, Esposito G (2002) DNA polymerase kappa deficiency does not affect somatic hypermutation in mice. Eur J Immunol 32: 3152–3160 [DOI] [PubMed] [Google Scholar]
- Sims AE, Spiteri E, Sims RJ 3rd, Arita AG, Lach FP, Landers T, Wurm M, Freund M, Neveling K, Hanenberg H, Auerbach AD, Huang TT (2007) FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol 14: 564–567 [DOI] [PubMed] [Google Scholar]
- Sorensen CS, Syljuasen RG, Falck J, Schroeder T, Ronnstrand L, Khanna KK, Zhou BB, Bartek J, Lukas J (2003) Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell 3: 247–258 [DOI] [PubMed] [Google Scholar]
- Stancel JN, McDaniel LD, Velasco S, Richardson J, Guo C, Friedberg EC (2009) Polk mutant mice have a spontaneous mutator phenotype. DNA Repair (Amst) 8: 1355–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utani K, Kohno Y, Okamoto A, Shimizu N (2010) Emergence of micronuclei and their effects on the fate of cells under replication stress. PLoS One 5: e10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassin VM, Anantha RW, Sokolova E, Kanner S, Borowiec JA (2009) Human RPA phosphorylation by ATR stimulates DNA synthesis and prevents ssDNA accumulation during DNA-replication stress. J Cell Sci 122: 4070–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal AE, Kannouche P, Podust VN, Yang W, Lehmann AR, Woodgate R (2004) Proliferating cell nuclear antigen-dependent coordination of the biological functions of human DNA polymerase iota. J Biol Chem 279: 48360–48368 [DOI] [PubMed] [Google Scholar]
- Wang JO, Kawamura K, Tada Y, Ohmori H, Kimura H, Sakiyama S, Tagawa M (2001) DNA polymerase kappa, implicated in spontaneous and DNA damage-induced mutagenesis, is overexpressed in lung cancer. Cancer Res 61: 5366–5369 [PubMed] [Google Scholar]
- Warbrick E (1998) PCNA binding through a conserved motif. Bioessays 20: 195–199 [DOI] [PubMed] [Google Scholar]
- Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M (2004) Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J 23: 3886–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Minesinger BK, Wiltrout ME, D’Souza S, Woodruff RV, Walker GC (2009) Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev 73: 134–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SA, Longerich S, Sung P, Vaziri C, Kupfer GM (2011) The E3 ubiquitin ligase RAD18 regulates ubiquitylation and chromatin loading of FANCD2 and FANCI. Blood 117: 5078–5087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Xin H, Wu X, Rajpal DK, Yang D, Wang Z (2000) Human DNA polymerase kappa synthesizes DNA with extraordinarily low fidelity. Nucleic Acids Res 28: 4147–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.