Abstract
Intracellular deoxyribonucleoside triphosphate (dNTP) pools must be tightly regulated to preserve genome integrity. Indeed, alterations in dNTP pools are associated with increased mutagenesis, genomic instability and tumourigenesis. However, the mechanisms by which altered or imbalanced dNTP pools affect DNA synthesis remain poorly understood. Here, we show that changes in intracellular dNTP levels affect replication dynamics in budding yeast in different ways. Upregulation of the activity of ribonucleotide reductase (RNR) increases elongation, indicating that dNTP pools are limiting for normal DNA replication. In contrast, inhibition of RNR activity with hydroxyurea (HU) induces a sharp transition to a slow-replication mode within minutes after S-phase entry. Upregulation of RNR activity delays this transition and modulates both fork speed and origin usage under replication stress. Interestingly, we also observed that chromosomal instability (CIN) mutants have increased dNTP pools and show enhanced DNA synthesis in the presence of HU. Since upregulation of RNR promotes fork progression in the presence of DNA lesions, we propose that CIN mutants adapt to chronic replication stress by upregulating dNTP pools.
Keywords: checkpoints, DNA damage, dNTPs, hydroxyurea, replication stress
Introduction
The genome of eukaryotic cells is particularly at risk during the S-phase of the cell cycle. Indeed, replication forks are fragile structures that are prone to collapse and have been implicated in cancer development through the induction of chromosome breaks (Halazonetis et al, 2008; Branzei and Foiani, 2010). To ensure the accurate duplication of their chromosomes, eukaryotic cells have developed complex surveillance pathways called intra-S-phase checkpoints. In budding yeast, arrested forks and DNA breaks are signalled by distinct pathways called the DNA replication checkpoint (DRC) and the DNA damage checkpoint (DDC), respectively (Tourriere and Pasero, 2007). Both pathways are activated by Mec1 (ATR in human), a sensor kinase that is recruited to stalled forks and chromosome breaks through its interaction with the ssDNA-binding factor RPA. Mec1 activates in turn the effector kinase Rad53 (CHK1 in human), which orchestrates the multiple cellular responses to genotoxic stress (Alabert et al, 2009; Barlow and Rothstein, 2009; Zegerman and Diffley, 2009). Rad53 activation is mediated at DNA breaks by the adaptor protein Rad9 and at stalled forks by Mrc1 and RFCCtf18 (Alcasabas et al, 2001; Gilbert et al, 2001; Crabbe et al, 2010). The DRC pathway promotes the maintenance and the recovery of stalled forks (Lopes et al, 2001; Tercero and Diffley, 2001; Cobb et al, 2005) and prevents the activation of late replication origins (Santocanale and Diffley, 1998; Lopez-Mosqueda et al, 2010; Zegerman and Diffley, 2010).
Faithful duplication of the genome also depends on a balanced supply of deoxyribonucleoside triphosphates (dNTPs). In Saccharomyces cerevisiae, dNTP pools increase by about three-fold upon entry into S-phase relative to G1 levels (Chabes et al, 2003). Since dNTP pools are sufficient to replicate only a small fraction of the genome, high ribonucleotide reductase (RNR) activity is maintained throughout the S-phase (Kumar et al, 2010). This activity must be tightly coordinated with DNA synthesis. Indeed, increased dNTP pools interfere with initiation and induce mutagenesis in yeast (Chabes and Stillman, 2007; Kumar et al, 2010). Moreover, reduced or imbalanced pools impede fork progression and promote tumourigenesis in human cells (Bester et al, 2011; Chabosseau et al, 2011). However, S. cerevisiae cells exposed to high doses of the RNR inhibitor hydroxyurea (HU) are able to complete S-phase by slowing down the execution of their replication programme (Alvino et al, 2007). The consequences of deregulated dNTP pools on DNA replication in eukaryotic cells remain therefore poorly understood.
Besides DNA replication, DNA lesions also induce an upregulation of dNTP pools. In budding yeast, dNTP levels show a three- to five-fold increase in response to DNA damage relative to a normal S-phase, through the checkpoint-dependent induction of RNR genes, the allosteric regulation of RNR activity and the degradation of the Rnr1 inhibitor Sml1 (Zhao et al, 1998; Zhao and Rothstein, 2002; Chabes et al, 2003). In mammalian cells, RNR is directly recruited to DNA damage sites in a Tip60-dependent manner (Niida et al, 2010a). It has been proposed that expansion of dNTP pools helps cells survive DNA damage by promoting the repair and/or the bypass of DNA lesions (Mathews, 2006; Sabouri et al, 2008; Niida et al, 2010b). In budding yeast, deletion of the SML1 gene is essential for the viability of mec1Δ, rad53Δ and mrc1Δrad9Δ mutants, suggesting that increased dNTP pools promote survival in the absence of intra-S-phase checkpoints (Zhao et al, 1998; Alcasabas et al, 2001, Corda et al, 2005). Whether checkpoint-dependent mechanisms regulate dNTP pools in order to promote S-phase progression under replication stress is an important question that remains to be addressed.
Here, we have used a combination of genome-wide and single-molecule approaches to monitor the kinetics of DNA replication in the presence of altered dNTP pools. We show that upregulation of RNR activity accelerates fork speed, indicating that DNA precursors are normally limiting for DNA synthesis. We also show that HU-treated cells undergo a sharp transition from a regular- to a slow-replication mode when dNTP pools drop below a critical level. Interestingly, this transition is primarily determined by dNTP pools present in G1, as upregulation of RNR activity drastically increased fork progression in HU-treated cells. We also report that a collection of chromosomal instability (CIN) mutants including sgs1Δ, rrm3Δ, rtt101Δ, asf1Δ, rad52Δ, ctf4Δ, ctf18Δ and elg1Δ cells replicate faster than wild-type cells in the presence of HU. These mutants also display a constitutive activation of the DDC pathway and a concomitant increase of dNTP pools. Since increased levels of DNA precursors promote both initiation and elongation, we propose that CIN mutants compensate for increased fork instability by upregulating RNR activity. This view is supported by the fact that expansion of dNTP pools also facilitates elongation in the presence of exogenous DNA damage. In conclusion, we propose that dNTP levels are key determinants of replication fork speed and that cells adapt to replication stress by upregulating dNTP pools.
Results
BrdU-IP-chip analysis of replication dynamics in HU-treated cells
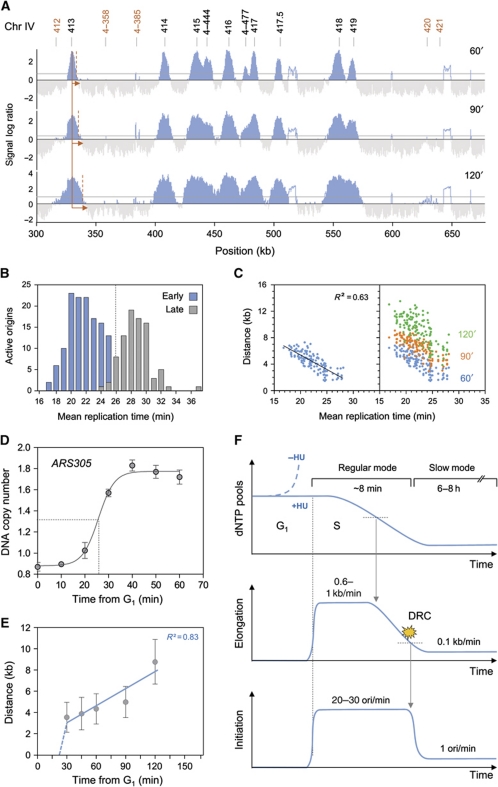
We have recently used a microarray-based assay to dissect the checkpoint pathways that repress late replication origins in S. cerevisiae cells exposed to HU (Crabbe et al, 2010). Here, we have adapted this assay to determine how low-dNTP conditions affect the execution of the DNA replication programme. To this end, wild-type cells were synchronized in G1-phase with α-factor and were released into S-phase in the presence of 200 mM HU. Newly synthesized DNA was labelled with bromodeoxyuridine (BrdU) and cells were harvested 60, 90 and 120 min after release from the G1 arrest. BrdU-labelled DNA was immunoprecipitated and hybridized to high-resolution tiling arrays. Representative replication maps are shown in Figure 1A and Supplementary Figure S1A and whole-genome maps are shown in Supplementary Figure S2. Analysis of DNA content by flow cytometry confirmed that replication progresses at a slow but constant pace in HU-treated wild-type cells (Supplementary Figures S1B and S3A and B), as described previously (Alvino et al, 2007). An automated process was used to determine the rates of initiation and elongation from 60 to 120 min in HU, as described in Materials and methods. After 60 min, we detected the activation of 194 origins, which represents ∼40% of all the yeast origins. This number gradually increased after 90 and 120 min (246 active origins), giving an initiation rate of one origin firing per minute (Supplementary Figure S1C). In contrast, all the origins fired simultaneously in rad53-11 cells (Supplementary Figure S1A and C), which is consistent with the fact that the Mec1–Rad53 pathway represses late origins firing in the presence of HU (Santocanale and Diffley, 1998).
Figure 1.
Replication dynamics in low-dNTP conditions. (A) Replication profiles of a region of chromosome 4 in wild-type cells (PP872). Cells were synchronized in G1 with α-factor, and released into medium containing 200 mM HU and 400 μg/ml BrdU for 60, 90 or 120 min. After DNA extraction and fragmentation, BrdU-labelled DNA was immunoprecipitated and hybridized on high-resolution tiling arrays. Enrichment of replicated DNA fragments relative to a whole-genome sample (signal log ratio) is shown. Significant peaks are filled in blue, horizontal grey lines indicate the threshold used for peak calling (50% of signal range). Empty areas correspond to non-significant peaks or repeated sequences. Black and red numbers indicate early and late origins, respectively. (B) Bimodal distribution of active replication origins plotted relative to the mean replication time in a normal S-phase in wild-type cells (Yabuki et al, 2002). (C) Scatter plot of the distance covered by replication forks versus the mean replication time in a normal S-phase. Early origins are plotted as blue dots for the 60-min time point. The distance covered by individual forks after 60, 90 and 120 min in HU is shown as blue, red and green dots, respectively. (D) Kinetics of ARS305 sequence duplication monitored by DNA copy number change using qPCR. Cells were synchronized in G1 with α-factor and released into medium containing 200 mM HU. DNA amount was normalized to a negative region unreplicated at 60 min. (E) Mean distances covered by individual replication forks over time in HU-treated wild-type cells. Distances are derived from genome-wide replication profile at a given time (see Materials and methods). (F) Schematic representation of DNA replication parameters (dNTP pool, elongation rate expressed in kb/min, initiation rate expressed in number of origins activated per minute) over time in wild-type cells during a replicative stress (+HU). DRC indicates activation of the DNA replication checkpoint. Arrows indicate a causal link. When dNTP levels drop below a critical threshold, forks slowdown and accumulate ssDNA, which activates in turn the DRC and delays the temporal programme of origin activation.
To further characterize the kinetics of DNA replication in HU-treated cells, we compared the pattern of origin usage in HU-treated cells to their mean replication time during a normal S-phase. Since the exact time of origin firing is not known in budding yeast, we used the time of 50% replication along the origin sequence defined by Yabuki et al (2002) as best approximation for origin firing time. As illustrated in Figure 1B, origins are replicated from 17 to 33 min during a normal S-phase and show a biphasic distribution, with a transition between early and late origins occurring at t=25 min. Next, we plotted the distance covered by individual replication forks in HU relative to the mean replication time of the corresponding origins during a normal S-phase (Figure 1C). This analysis revealed that BrdU track lengths in HU correlate with the mean replication time of origins (R2=0.63). For instance, forks activated at t=17 min during a normal S-phase replicated >6 kb in the presence of HU while forks generated 8 min later covered <1.5 kb, resulting in a mean elongation rate of 0.6 kb/min. Analysis of later time points (90 and 120 min) indicated that all forks progressed at a reduced rate of 0.1 kb/min, regardless of their initiation time (Figure 1C). This is consistent with single-molecule analyses of fork progression in cells exposed for up to 6 h to 200 mM HU (80 bp/min; Supplementary Figure S3C and D). This slow fork progression was not observed in the absence of Rad53 activity (Supplementary Figure S1D), which explain why DNA content does not increase in HU-treated rad53-11 cells despite the derepression of late origins (Supplementary Figure S1B and C).
To determine elongation rates within the first minutes of S-phase in HU-treated cells, we first analysed the duplication of ARS305, an efficient early-firing origin. Wild-type cells were synchronized in G1-phase with α-factor and were released into S-phase in the presence of 200 mM HU. Cells were harvested every 10 min during 60 min. Analysis of DNA copy number by quantitative PCR indicated that the ARS305 locus is duplicated ∼25 min post-G1 release, which is comparable to normal S-phase kinetics (Figure 1D). Next, to measure fork rates, wild-type cells were synchronized in G1-phase with α-factor and were released in S-phase in the presence of 200 mM HU and BrdU. Samples were collected from 30 to 120 min as indicated (Figure 1E). This analysis revealed a transition from a regular (∼0.5 kb/min) to a slow-replication mode (0.1 kb/min) ∼30 min after release from the G1 arrest. Taken together, these data indicate that HU-treated cells display two distinct modes of DNA replication (Figure 1F). During the first minutes, cells use G1 dNTP pools to progress normally through S-phase. However, after activation of 40% of the origins and duplication of 10–15% of the genome, dNTP pools become critically limiting and cells elicit a checkpoint response that impedes further initiation events. Then, cells change to a slow-replication mode using decreased dNTP pools to complete S-phase. This slow mode is characterized by a 10-fold reduction of the elongation rate and a 25-fold reduction of the initiation rate. Analysis of initiation and elongation profiles indicates that this transition occurs ∼8 min after entry into S-phase and that slow replication persists for at least 8 h before S-phase completion (Supplementary Figure S3A and B).
Physiological dNTP levels are limiting for DNA replication
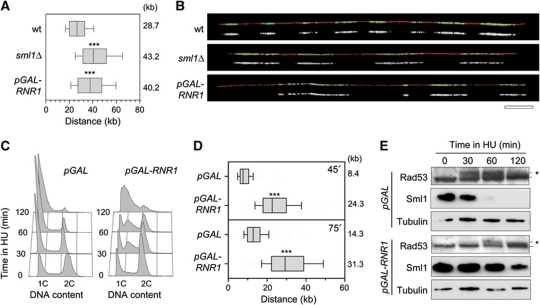
Our data suggest that the initial kinetics of DNA replication in the presence of HU is determined by the level of preexisting dNTP pools. To address this possibility, we next monitored the effect of increased RNR activity on replication fork progression, in cells exposed or not to HU. We first measured the effect of the RNR inhibitor Sml1 (Zhao et al, 1998), on the rate of fork progression. Deletion of the SML1 gene leads to a 2.5-fold increase in dNTP levels (Zhao et al, 1998). Untreated exponentially growing wild-type and sml1Δ cells were pulse labelled for 30 min with BrdU and the length of BrdU tracks was measured along individual DNA fibres stretched by DNA combing. We found that BrdU tracks were significantly longer in sml1Δ mutants (43.2 kb) than in wild-type cells (28.7 kb; Figure 2A and B) and that sml1Δ cells progressed slightly faster through S-phase than their wild-type counterparts (Supplementary Figure S4A). To confirm that higher dNTP pools increase fork speed, we overexpressed the RNR1 gene with a pGAL-RNR1 construct (Chabes and Stillman, 2007). Analysis of fork progression performed as above revealed a 35% increase of BrdU track length in pGAL-RNR1 cells (40.2 kb) compared with control pGAL cells (28.7 kb; Figure 2A and B). Although the pGAL-RNR1 construct increased dNTP pools to a much higher level than the deletion of the SML1 gene (Chabes and Stillman, 2007), this did not further increase fork speed. These data suggest that other factors become rate limiting for elongation above a two-fold increase in dNTP levels.
Figure 2.
Increased dNTP pools accelerate fork progression. (A) Exponentially growing wild-type (PP872) and sml1Δ (PP924) cells were pulse labelled with 400 μg/ml BrdU for 30 min. Prior to BrdU labelling, pGAL-RNR1 (PP918) cells were incubated for 2 h in the presence of 2% galactose. DNA fibres were analysed by DNA combing. Graph depicts the distribution of BrdU tracks length. Box and whiskers indicate 25–75 and 10–90 percentiles, respectively. Mean BrdU tracks length is indicated in kb. Asterisks indicate the P-value of the statistical test (Mann–Whitney rank sum t-test, ***P-value <0.0001). (B) Representative images of DNA fibres are presented. Green: BrdU, red: DNA. Bar, 50 kb. (C–E) Exponentially growing pGAL (PP917) and pGAL-RNR1 (PP918) cells were cultured for 2 h with 2% galactose before addition of 100 mM HU. (C) Flow cytometry profiles of pGAL and pGAL-RNR1 cells during the time course in HU. (D) After 30 min in HU, BrdU was then added for 45 or 75 min. DNA fibres were analysed by DNA combing. Box and whiskers indicate 25–75 and 10–90 percentiles, respectively. Mean BrdU tracks length is indicated in kb. (E) Protein extracts were collected during the time course and subjected to SDS–PAGE followed by an immunoblotting with antibodies against Rad53, Sml1 and tubulin. Asterisks indicate the hyperphosphorylated form of Rad53. Figure source data can be found in Supplementary data.
Expansion of dNTP pools promotes DNA synthesis during replicative stress
Next, we tested whether increased dNTP pools also increase DNA synthesis in the presence of HU. Asynchronous cultures of control cells (pGAL) and cells overexpressing RNR1 (pGAL-RNR1) were grown in galactose-containing medium before addition of 100 mM HU for 2 h. Analysis of dNTP levels indicates that although pools decreased over time when both strains are exposed to HU, they remained higher in pGAL-RNR1 cells compared with control cells (Supplementary Figure S4B). Flow cytometry analysis of DNA content revealed that pGAL-RNR1 cells continue to synthesize DNA for at least 2 h after HU addition. In contrast, control cells (pGAL) accumulated in early S after 30 min in HU (Figure 2C). Finally, analysis of fork progression by DNA combing confirmed that pGAL-RNR1 cells replicate 2.9-fold more DNA than control cells after 45 min in HU (Figure 2D). Interestingly, high-dNTP levels also impeded the activation of Rad53 and the Rad53-dependent degradation of Sml1 (Figure 2E). We therefore conclude that expansion of dNTP pools delays the inhibition on fork progression by HU and the subsequent activation of the DRC pathway.
Increased dNTP pools promote replication of damaged DNA templates
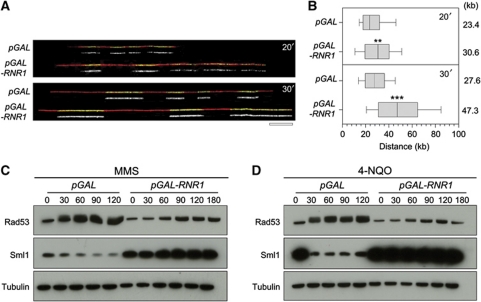
Since high-dNTP levels promote fork progression in HU-treated cells, we reasoned that increased RNR activity could also facilitate DNA replication in other types of replication stress situation. To address this possibility, we have monitored the effect of RNR upregulation on fork progression in the presence of MMS, a DNA alkylating agent inducing replication fork arrest. Asynchronous cultures of control cells (pGAL) and cells overexpressing RNR1 (pGAL-RNR1) were grown in galactose-containing medium before addition of 0.033% MMS for 3 h. Strikingly, analysis of fork progression by DNA combing revealed that forks progress faster in the presence of MMS upon overexpression of RNR1 (Figure 3A and B). In addition, this faster replication in MMS-treated cells expressing pGAL-RNR1 occurs without detectable activation of Rad53 or degradation of Sml1 (Figure 3C). Similar results were obtained in the presence of the UV-mimetic agent 4-NQO (Figure 3D). It is worth noting that the absence of Rad53 activation by MMS has already been reported in the presence of a much higher level of dNTPs (Chabes and Stillman, 2007). Altogether, these data support the view that a moderate increase in dNTP pools protect cells from replication stress and promotes fork progression without activating the DRC.
Figure 3.
High-dNTP pools improve tolerance to DNA lesions in vivo. Exponentially growing pGAL (PP917) and pGAL-RNR1 (PP918) cells were incubated for 2 h in the presence of 2% galactose prior to the addition of 0.033% MMS during 30 min. Then, cells were pulse labelled with 400 μg/ml BrdU for 20 and 30 min. DNA fibres were analysed by DNA combing. (A) Representative images of DNA fibres are presented. Green: BrdU, red: DNA. Bar, 50 kb. (B) Graph depicts the distribution of BrdU tracks length. Box and whiskers indicate 25–75 and 10–90 percentiles, respectively. Mean BrdU tracks length is indicated in kb. Asterisks indicate the P-value of the statistical test (Mann–Whitney rank sum t-test, **P-value <0.001; ***P-value <0.0001). (C, D) Protein extracts were collected at the indicated times in the presence of 0.033% MMS (C) or 0.2 μg/ml 4-NQO (D) and subjected to SDS–PAGE followed by an immunoblotting with anti-Rad53, anti-Sml1 and anti-tubulin antibodies. Note that basal levels of Sml1 are higher in pGAL-RNR1 due to the upregulation of RNR. Figure source data can be found in Supplementary data
dNTP levels modulate origin usage in HU-treated cells
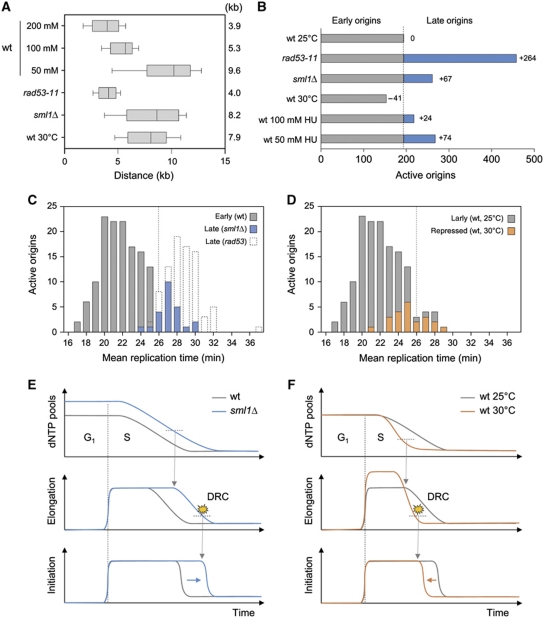
Our data indicate a transition of HU-treated cells to a slow-replication mode characterized by Rad53 activation when dNTP pools drop below a critical level (Figure 1F). Since expansion of dNTP pools delays Rad53 activation in HU-treated cells (Figure 2E), we reasoned that it could also affect the execution of the DNA replication programme. To address this possibility, we monitored origin usage in cells showing different levels of DNA precursors. Replication profiles were first analysed in sml1Δ cells. This analysis revealed a two-fold increase of fork rate in sml1Δ cells (Figure 4A) and the activation of 67 late origins, even though sml1Δ mutants are proficient to activate the DRC pathway (Figure 4B; Supplementary Figure S5).
Figure 4.
Available dNTP pools determine the temporal programme of origin activation. (A, B) BrdU-IP-chip analysis of distances covered by individual replication forks and the number of active origins in the indicated strains exposed to 200 mM HU. (A) Box and whiskers indicate 25–75 and 10–90 percentiles, respectively. Mean distance is indicated in kb. (B) Grey and blue bars indicate early and late origins, respectively. The number of late origins is indicated relative to wild-type cells grown at 25°C. (C) Distribution of active origins in wild-type, rad53-11 and sml1Δ cells exposed for 60 min to 200 mM HU relative to their mean replication time during a normal S-phase (Yabuki et al, 2002). Grey boxes indicate origins that are active in all strains (early origins). Late origins repressed by Rad53 in the presence of HU are indicated with empty boxes. Late origins that fire in sml1Δ cells are indicated in blue. (D) Distribution of active origins in HU-treated wild-type cells grown at either 25 or 30°C. Early origins fired at 25°C but not at 30°C are indicated in orange. (E, F) Schematic representation of dynamic changes in dNTP pools, elongation and initiation rates in the experiments shown in (A–D). DRC indicate the onset of the DRC activation. See text and Figure 1F legend for details.
We next analysed origin usage in wild-type cells grown at either 25 or 30°C (Figure 4B; Supplementary Figure S5). We found that the distance covered by replication forks were much larger at 30°C than 25°C (7.9 versus 3.9 kb; Figure 4B), which is consistent with the fact that replication forks are faster at 30°C than at 25°C in the absence of HU (Supplementary Figure S6C). Remarkably, we also found that 41 origins that are normally active at 25°C were repressed at 30°C (Figure 4B; Supplementary Figure S6A and B). Since dNTP pools do not change with temperature (Supplementary Figure S6D), these data suggest that faster exhaustion of dNTP pools at 30°C activates the DRC pathway prematurely. Finally, we measured a proportional increase of fork rate and origin activity at lower doses of HU (50 or 100 mM; Figure 4A and B), which supports the view that dNTP levels determine both initiation and elongation rates in the presence of HU.
To check whether dNTP pools determine the transition from regular- to slow-replication mode in HU-treated cells, we next analysed the mean replication time of the origins that are specifically affected in sml1Δ cells and in wild-type cells grown at 30°C. This analysis revealed that increased dNTP pools allow the activation of the earliest of the late origins in sml1Δ cells (Figure 4C; P<0.0001). Conversely, faster consumption of DNA precursors at 30°C prevents the activation of the latest of the early origins (Figure 4D; P<0.0001). Collectively, these data indicate that the difference between early and late origins in HU-treated cells is determined by the size of preexisting dNTP pools and the kinetics of dNTP consumption by replication forks (Figure 4E and F).
Replication mutants show increased dNTP levels and fork progression in HU
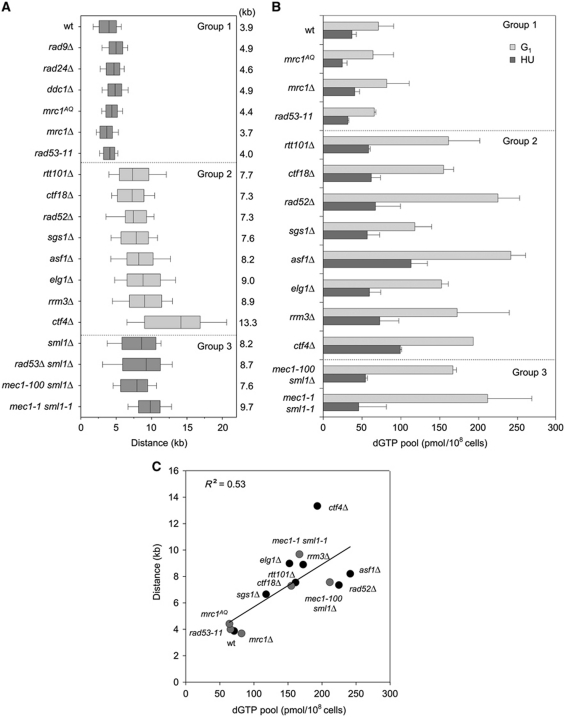
To further dissect the mechanisms that determine fork progression in HU-treated cells, we next monitored fork progression in a collection of mutants showing spontaneous CIN. This includes mutants affected in intra-S-phase checkpoints (mec1-1 sml1-1, mec1-100 sml1Δ, rad53Δ sml1Δ, rad53-11), the DRC pathway (mrc1Δ, mrc1AQ, ctf18Δ), the DDC pathway (ddc1Δ, rad24Δ, rad9Δ), homologous recombination (rad52Δ), chromatin assembly (asf1Δ) and in the maintenance of fork integrity (sgs1Δ, ctf4Δ, elg1Δ, rtt101Δ, rrm3Δ). This analysis identified three groups of mutants showing normal (group 1) or wider (groups 2 and 3) BrdU tracks (Figure 5A). Groups 2 and 3 differ in the status of the SML1 gene, which is mutated in group 3 strains. Remarkably, deletion of the SML1 gene in wild-type and rad53 cells increased the distance covered by replication forks by two-fold (8.2 and 8.7 kb, respectively), a figure comparable to mec1-1 sml1-1 cells (9.7 kb; Figure 5A; Supplementary Figure S7). These data indicate that SML1 mutation can be accounted for the wider BrdU tracks of group 3 mutants.
Figure 5.
Increased dNTP pools promote fork progression in CIN mutants. (A, B) Analysis of fork progression and dGTP concentration in G1 and in 200 mM HU in wild-type (PP872), mrc1AQ (PP679), mrc1Δ (PP913), rad53-11 (PP37), rtt101Δ (PP285), ctf18Δ (PP907), rad52Δ (PP501), sgs1Δ (PP35), asf1Δ (PP889), elg1Δ (PP1013), rrm3Δ (PP1039), ctf4Δ (PP908), mec1-100 sml1Δ (PP685) and mec1-1 sml1-1 (PP276) cells. (A) Cells were released into YPD containing 200 mM HU and BrdU for 60 min. Box and whisker graph depicts the distribution of distances covered by individual replication forks in the indicated strains. Distances were measured on the whole-genome BrdU-IP-chip replication profiles. Box and whiskers indicate 25–75 and 10–90 percentiles, respectively. Mean distance is indicated in kb. (B) dGTP concentration was measured in G1-arrested cells and after 60 min in HU. (C) Mean fork distances in kb plotted relative to the dGTP pool size in pmol/108 cells in HU in the indicated strains. Filled black circles: DRC-proficient mutants. Filled grey circles: DRC-deficient mutants. Correlation coefficient (R2) is indicated for a linear regression taking into account all the strains.
Since SML1 deletion and RNR1 overexpression increase fork progression in HU by upregulating dNTP pools, we next measured dNTP pools in representative mutants of the three groups. Strikingly, we found that all mutants that show enhanced DNA synthesis in HU also had increased dNTP levels in G1, which were maintained after 60 min in HU (Figure 5B; Supplementary Figure S8). To determine whether faster forks correlate with increased dNTP levels in these mutants, we plotted BrdU tracks length relative to the amount of dGTP, the most limiting of the four dNTPs (Kumar et al, 2010). This analysis revealed a good correlation between dNTP levels and fork progression (R2=0.53; Figure 5C). These data indicate that G1 dNTP levels are key determinants of fork progression in HU. They also indicate that for equivalent dNTP levels, forks are slower in DRC or HR mutants, presumably because fork collapse more frequently when these mutants are exposed to HU.
Spontaneous DNA damage increases dNTP production
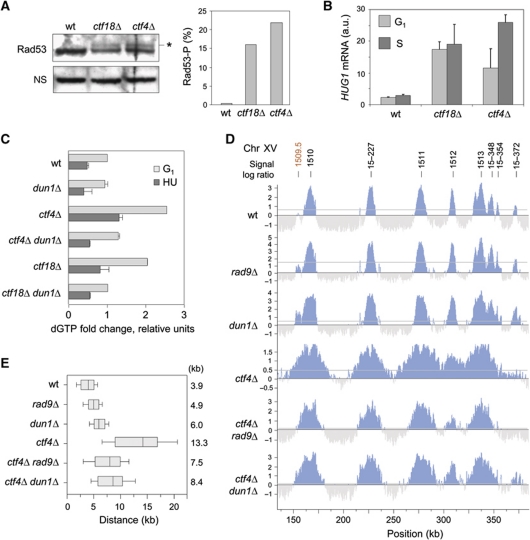
RNR activity is tightly controlled by several mechanisms, including inhibition by the Sml1 protein. Sml1 levels decrease during S-phase and become undetectable after DNA damage (Zhao and Rothstein, 2002). The DDC-dependent activation of RNR induces a three- to five-fold increase of dNTP levels relative to a normal S-phase (Chabes et al, 2003). Interestingly, we noticed that group 2 mutants have all in common an unstable genome and/or a perturbed replication programme. For instance, ctf4Δ and ctf18Δ cells present spontaneous Rad52 foci and depend on a functional homologous recombination pathway for viability (Alvaro et al, 2007). We therefore asked whether elevated dNTPs levels in the absence of Ctf4 or Ctf18 result from spontaneous DDC activation. We measured a weak but reproducible activation of Rad53 in asynchronous cultures of untreated ctf4Δ and ctf18Δ mutants (Figure 6A) and a sharp induction of HUG1 transcription (Figure 6B), which are both indicative of a chronic activation of the DDR pathway (Basrai et al, 1999).
Figure 6.
Spontaneous DNA damage increases dNTP production in ctf4Δ and ctf18Δ mutants. (A) Protein extracts from asynchronous wild-type, ctf18Δ and ctf4Δ cells were collected and subjected to SDS–PAGE followed by an immunoblotting anti-Rad53. Star indicates Rad53 phosphorylated form. The histogram is a quantification of Rad53-P levels relative to Rad53. (B) Relative mRNA levels of HUG1 in wild-type, ctf18Δ and ctf4Δ cells in G1 and after 40 min in S-phase. Error bars indicate standard deviation from three independent experiments. (C) Relative dGTP levels in wild-type (PP872), dun1Δ (PP1387), ctf4Δ (PP908), ctf4Δdun1Δ (PP1322), ctf18Δ (PP907) and ctf18Δ dun1Δ (PP1321) strains in G1 and after 60 min in 200 mM HU. (D, E) Cells were synchronized in G1 with α-factor, and released into medium containing 200 mM HU and 400 μg/ml BrdU for 60 min to perform BrdU-IP-chip. (D) Replication profiles of a representative region of chromosome 15 in wild-type (PP872), rad9Δ (PP914), dun1Δ (PP1387), ctf4Δ (PP908), ctf4Δrad9Δ (PP851) and ctf4Δdun1Δ (PP1322) strains. Significant peaks are filled in blue, horizontal grey lines indicate the threshold used for peak calling (50% of signal range). Early and late origins are labelled in black and red, respectively. (E) Distribution of the distances covered by individual replication forks in the indicated strains. Box and whiskers indicate 25–75 and 10–90 percentiles, respectively. Mean BrdU tracks length is indicated in kb. Figure source data can be found in Supplementary data
In response to DNA damage, the Mec1–Rad9–Rad53 pathway activates Dun1, a checkpoint kinase that triggers the degradation of the RNR inhibitor Sml1 (Zhao and Rothstein, 2002). To evaluate the implication of the DDC pathway in enhanced DNA synthesis in the presence of HU in ctf4Δ and ctf18Δ mutants, we next monitored the contribution of the checkpoint mediator Rad9 and the Dun1 kinase in the increased fork progression and dNTP levels in these mutants. DUN1 inactivation significantly reduced dNTP levels in ctf4Δ and ctf18Δ cells (Figure 6C). We also measured a proportional reduction of the length of replicated tracks in both ctf4Δ dun1Δ and ctf4Δ rad9Δ cells, indicating that enhanced replication in HU depends at least in part on Rad9 and Dun1 (Figure 6D and E). Deletion of the RAD9 and DUN1 genes did not significantly decrease fork rate in ctf18Δ cells (Supplementary Figure S9A and B). We assume that this difference reflects the fact that unlike ctf4Δ mutants, ctf18Δ cells are unable to fully activate Rad53 in response to HU (Crabbe et al, 2010). However, deletion of the DUN1 gene significantly increased the HU sensitivity of ctf18Δ cells, this effect being partially relieved by sml1Δ (Supplementary Figure S9C). Collectively, our results suggest that chronic replication stress upregulates dNTP pools through a mechanism that depends at least in part on Rad9 and Dun1. Expansion of dNTP pools in these mutants promotes fork progression and/or viability in the presence of HU.
Discussion
In optimal growth conditions, haploid S. cerevisiae cells synthesize up to 500 kb of DNA per minute and complete the replication of their ∼14 Mb genome within <30 min. This tremendous task depends on the sequential activation of hundreds of replication origins and on the unimpeded progression of twice as many replication forks (Raghuraman et al, 2001; Yabuki et al, 2002). Efficient DNA replication also depends on a constant supply of dNTPs, which are synthesized throughout the length of the S-phase. Levels of DNA precursors are tightly controlled as unbalanced or deregulated dNTP pools are deleterious for genome integrity (Chabes and Stillman, 2007; Kumar et al, 2010). This control is mostly exerted at the level of RNR, an enzyme regulating a rate-limiting step in dNTP biosynthesis.
In this study, we have modulated RNR activity by different means to monitor the effect of altered dNTP pools on the dynamics of DNA replication in budding yeast. We first deleted SML1, a gene encoding an allosteric repressor of RNR, to induce a 2.5-fold increase in dNTP levels (Zhao et al, 1998). Remarkably, this modest change in nucleotide pools induced a 66% increase in fork rate, indicating that dNTP levels are normally limiting for elongation. Next, we overexpressed the RNR subunit Rnr1 to further increase dNTP levels by 10-fold (Chabes and Stillman, 2007). This did not further increase fork rate, suggesting that other factors, such as newly synthesized histones, become limiting for elongation in high-dNTP conditions.
Remarkably, we also observed that overexpression of Rnr1 induced an ∼3-fold increase and fork rate in the presence of high levels of MMS, a potent DNA alkylating agent that impedes fork progression in triggers a robust replication stress response (Tourriere and Pasero, 2007). This 10-fold increase in dNTP levels also prevented activation of the DRC in cells exposed to either MMS or 4-NQO, a UV-mimetic agent interfering with fork progression. Together, these data indicate that increased dNTP pools protect cells from replication stress, by promoting lesion bypass (Chabes et al, 2003; Sabouri et al, 2008) or through a mechanism that remains to be determined.
In HU-treated cells, it has been reported that the rate of dNTP production decreases dramatically to residual levels and S-phase extends over 8–10 h (Koc et al, 2004; Alvino et al, 2007). Here, we have used a combination of BrdU-IP-chip and DNA combing approaches to determine how this very slow S-phase is regulated. Our data indicate that cells released synchronously into S-phase in the presence of HU undergo a rapid transition from a regular- to a slow-replication mode when dNTP pools drop below a critical level. This transition is characterized by an ∼10-fold reduction of fork speed and a 25-fold reduction of initiation rate. In wild-type cells exposed to 200 mM HU, it occurs after activation of ∼40% of the origins and duplication of 10–15% of the genome. This transition was not reported by Alvino et al (2007), presumably because the density shift assay used in this study is not adapted to detect rapid changes in fork rate.
Interestingly, we observed that subtle changes in the size or the consumption rate of dNTP pools have a direct effect on fork speed and on origin usage in HU-treated cells, supporting the view that the transition from regular to slow DNA replication is primarily determined by the size of intracellular dNTP pools before the addition of HU. This observation has major implications for the interpretation of mutant phenotypes. For instance, we and others have reported that mec1-1 mutants, but not rad53-11 cells, show much longer BrdU tracks in HU (Feng et al, 2009; Supplementary Figure S7). At first sight, this difference could reflect the fact that Mec1 regulates fork progression independently of Rad53. However, we show here that it only reflects the status of SML1, which is mutated in mec1-1 cells but not in rad53-11 mutants. Deletion of the SML1 gene in rad53-11 cells increased fork speed in HU to a level comparable to mec1-1 sml1-1 mutants. These data suggest therefore that the distance covered by replication forks in HU-treated cells is primarily determined by dNTP levels and not checkpoint kinases.
In this study, we have also screened a large collection of DNA replication and checkpoint mutants for their ability to replicate in low-dNTP conditions. Unexpectedly, we found that mutants with unstable replication forks or impaired fork repair pathways, such as elg1Δ, ctf4Δ, ctf18Δ, sgs1Δ, rad52Δ, asf1Δ, rtt101Δ and rrm3Δ, also show increased dNTP pools and enhanced DNA synthesis in HU. Most of these mutants exhibit spontaneous replication defects, activate the DDR pathway, accumulate in G2/M in a Rad9-dependent manner and show increased levels of spontaneous HR foci (Versini et al, 2003; Luke et al, 2006; Schmidt and Kolodner, 2006; Alvaro et al, 2007; Davidson and Brown, 2008; Tanaka et al, 2009; Crabbe et al, 2010). Since the DDR pathway induces the expansion of dNTP pools in response to DNA damage (Chabes et al, 2003), we propose that chronic DNA damage in these mutants promotes fork progression through the upregulation of dNTP pools. This view is supported by the fact that inactivation of the DDC mediator Rad9 and of the checkpoint kinase Dun1 reduces both dNTP levels and fork progression in HU-treated ctf4Δ cells. Interestingly, upregulation of dNTP pools in elg1Δ mutants also depends on the transcriptional activator Swi4 and on Ccr4, a component of the cytoplasmic mRNA deadenylase complex, indicating that Dun1-independent mechanisms contribute to the upregulation of dNTP pools in response to chronic DNA damage (Davidson et al, 2011). Taken together, these data indicate that CIN mutants upregulate dNTP levels through Rad9/Dun1-dependent and -independent mechanisms in order to facilitate DNA replication and adapt to chronic replication stress.
Alike yeast CIN mutants, oncogene-activated human cells suffer from chronic replication stress and display a constitutive activation of the DDC pathway (Halazonetis et al, 2008). Whether cancer cells upregulate dNTP pools as an adaptation to replication stress is an attractive possibility that remains to be addressed. It has been recently reported that alteration in dNTP pools promotes tumourigenesis through the deregulation of the Rb pathway or the inactivation of the BLM helicase (Bester et al, 2011; Chabosseau et al, 2011). Moreover, earlier studies have shown that abnormally low or imbalanced dNTP pools in mammalian cells can be complemented by the addition of exogenous nucleotide precursors to restore normal fork progression (Anglana et al, 2003; Courbet et al, 2008). However, unlike in yeast, genotoxic agents do not induce global changes in intracellular dNTP pools in mammalian cells, but rather promote the recruitment of RNR to sites of DNA damage. Whether RNR is also recruited to replication sites in the presence of genotoxic agents is currently unclear. Further work is therefore needed to decipher the fascinating connections between dNTP pools, replication stress and tumourigenesis in human cells.
Materials and methods
Yeast strains and plasmids
All strains are derivatives of W303 (MATa, ade2-1 trp1-1 can1-100 leu2,3,112 his3-11,15 ura3-52) (Supplementary Table SI). YEP medium was supplemented with 2% glucose or galactose. MATa cells were synchronized in G1 by adding α-factor (5 μg/ml) for 170 min at 25 or 30°C. To arrest cells in early S-phase, cells were released from G1 block by addition of pronase (50 μg/ml) into medium containing 0.2 M HU and 400 μg/ml BrdU for 60 min (genome-wide experiments).
Flow cytometry
Samples were prepared as previously described (Haase and Lew, 1997). Data were acquired on a FACScalibur (Becton Dickinson) and analysed with Cell Quest software.
Protein extracts and western blotting
TCA precipitation was performed as previously described (Longhese et al, 1997). Extracts were resolved by SDS–PAGE (Invitrogen). Rad53 phosphorylation shift was detected with a rabbit polyclonal antibody and tubulin was detected with the YOL1/34 antibody (Abcam; #ab6161). Sml1 was detected with a rabbit polyclonal antibody (Agrisera AB, Sweden, #AS10 847).
DNA copy number analysis by qPCR
For DNA extraction, 50 ml of yeast cells at 1 × 107 cells/ml were shaken five times 2 min in NIB buffer (17% (v/v) glycerol, 50 mM MOPS buffer, 150 mM potassium acetate, 2 mM magnesium chloride, 500 μM spermidine and 150 μM spermine, pH 7.2) with Zirconium beads on a Vibrax (VXR basic, Ika) at 4°C. DNA was isolated using QiagenGenomic DNA extraction kit. For qPCR, extracted DNA was diluted to 0.1 ng/ml and amplified using Roche Diagnostics LightCycler480 PCR system. Enrichment values at ARS305 (305F: 5′-AGCCTTCTTTGGAGCTCAAGTG-3′; 305R: 5′-TTTGAGGAATTTCTTTTGAAGAGTTG-3′) were calculated against an unreplicated region on chromosome V confirmed by BrdU-IP-chip (NEGV; NEGV-F: 5′-GCACTTAATTGGCGTAAGCTG-3′; NEGV-R: 5′-TCGCAGGAGCATATTTCGTA-3′). Enrichment values fluctuate between 1 (unreplicated DNA region) and 2 (fully replicated DNA region).
RNA extraction and RT
Cytoplasmic RNAs were extracted from G1, S-phase and 200 mM HU-treated yeast cells using Phenol:Chloroform 5:1 (Sigma; P-1944). In all, 1.5 μg of RNA dissolved in RNAse-free water was treated with DNAse I (1 U/μl; Promega) for 30 min at 37°C, followed by 10 min at 67°C for enzyme denaturation. RNA solution was purified using the mini kit RNeasy (Qiagen). RNA integrity was checked by agarose electrophoresis prior reverse transcription at 42°C for 1 h in a total volume of 20 μl reaction buffer (50 mM Tris pH 8.3, 30 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol) containing 50 U of SuperScript II RT (Invitrogen), 100 pM of oligo(dT)20 and 40 U RNaseOUT inhibitor (Invitrogen).
dNTP quantification
dNTP quantification was performed as previously described (Chabes et al, 2003).
BrdU-IP-chip, peak identification and measurement of fork progression
Signals from two to four biological replicates were normalized with TAS 1.1.02 (Affymetrix) using quantile normalization. Analysis results were stored in Log2 scale for signal intensity and in −10Log10 for P-values. Probe signals were analysed using perfect matches only with a bandwidth of 300. P-value intervals were generated using the following parameters: cutoff: 10−5, max gap: 80 and min run: 40. Significant intervals were scored as active origins when signal intensity was above a threshold arbitrarily determined as 50% of the signal range and when BrdU tracks were >1 kb. Replication profiles (signal log ratio) were displayed with IGB software (Nicol et al, 2009). BrdU peaks overlapping with replication origins referenced in OriDB (Nieduszynski et al, 2007) were scored with Galaxy (Blankenberg et al, 2010; Goecks et al, 2010). BrdU peaks that are not listed in OriDB as a known replication origin were assigned a name corresponding to the chromosome number followed with the position of the peak in kb. BrdU peaks that overlap with repetitive sequences such as transposons and subtelomeric repeats were not used in this study. Peaks that do not grow in size over time in time course experiments were also eliminated. Distances covered by forks in HU were determined as the distance between origin initiation sites and the edge of BrdU intervals at a subset of 129 individual forks using the P-value data calculated as described above. This subset was determined as all the active forks in HU-treated wild-type cells that are located at sufficient distance from the neighbouring origin in order to avoid BrdU tracks fusions. The time of 50% replication along origin sequences was used as best approximation for origin firing time (Yabuki et al, 2002).
DNA combing
DNA combing was performed as described (Tourriere et al, 2005). Single-stranded DNA was detected with a mouse monoclonal antibody (Chemicon; clones 16–19) and a goat anti-mouse antibody coupled to Alexa 546 (Invitrogen). BrdU was detected with a rat monoclonal antibody (Abcys; clone BU1/75) and a goat anti-rat antibody coupled to Alexa 488 (Invitrogen). Images were recorded on a Leica DM6000 microscope equipped with a CoolSNAP HQ CCD camera (Roper Scientific) and were processed as described (Pasero et al, 2002). Fork speed (S in kb/min) was determined as S=(Lt2−Lt1)/2 × T, where Lt2 is the median length of BrdU tracks measured at t2, Lt1 is the median length of BrdU tracks measured at t1T is n min (t2−t1).
Accession codes
Microarray data presented in this article can be obtained from Gene Expression Omnibus with the accession number GSE33686 and GSE21014.
Supplementary Material
Acknowledgments
We thank O Aparicio for plasmids. We thank E Schwob and the DNA combing facility of Montpellier for providing silanized coverslips and the Montpellier RIO Imaging microscopy facility. We thank G Brown for sharing information before publication. We thank H Tourrière and YL Lin for critical reading of the manuscript. JP was supported by a fellowship from the French ‘Ministère de la Recherche et de l’Enseignement Supérieur’. This work was supported by FRM (Equipe FRM) and ANR to PP and by the Swedish Foundation for Strategic Research, the Swedish Research Council and the Swedish Cancer Society to AC.
Author contributions: JP, LC, AC, AL and PP designed the experiments. JP, OT, LC, AK, VP and AL performed the experiments. JP, AL and PP wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alabert C, Bianco JN, Pasero P (2009) Differential regulation of homologous recombination at DNA breaks and replication forks by the Mrc1 branch of the S-phase checkpoint. EMBO J 28: 1131–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ (2001) Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol 3: 958–965 [DOI] [PubMed] [Google Scholar]
- Alvaro D, Lisby M, Rothstein R (2007) Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet 3: e228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvino GM, Collingwood D, Murphy JM, Delrow J, Brewer BJ, Raghuraman MK (2007) Replication in hydroxyurea: it's a matter of time. Mol Cell Biol 27: 6396–6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglana M, Apiou F, Bensimon A, Debatisse M (2003) Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell 114: 385–394 [DOI] [PubMed] [Google Scholar]
- Barlow JH, Rothstein R (2009) Rad52 recruitment is DNA replication independent and regulated by Cdc28 and the Mec1 kinase. EMBO J 28: 1121–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basrai MA, Velculescu VE, Kinzler KW, Hieter P (1999) NORF5/HUG1 is a component of the MEC1-mediated checkpoint response to DNA damage and replication arrest in Saccharomyces cerevisiae. Mol Cell Biol 19: 7041–7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bester AC, Roniger M, Oren Yifat S, Im Michael M, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach Donna S, Kerem B (2011) Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145: 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J (2010) Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol Chapter 19: 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M (2010) Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11: 208–219 [DOI] [PubMed] [Google Scholar]
- Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L (2003) Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112: 391–401 [DOI] [PubMed] [Google Scholar]
- Chabes A, Stillman B (2007) Constitutively high dNTP concentration inhibits cell cycle progression and the DNA damage checkpoint in yeast Saccharomyces cerevisiae. PNAS 104: 1183–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabosseau P, Buhagiar-Labarchède G, Onclercq-Delic R, Lambert S, Debatisse M, Brison O, Amor-Guéret M (2011) Pyrimidine pool imbalance induced by BLM helicase deficiency contributes to genetic instability in Bloom syndrome. Nat Commun 2: 368. [DOI] [PubMed] [Google Scholar]
- Cobb JA, Schleker T, Rojas V, Bjergbaek L, Tercero JA, Gasser SM (2005) Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev 19: 3055–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corda Y, Lee SE, Guillot S, Walther A, Sollier J, Arbel-Eden A, Haber JE, Geli V (2005) Inactivation of Ku-mediated end joining suppresses mec1Δ lethality by depleting the ribonucleotide reductase inhibitor Sml1 through a pathway controlled by Tel1 kinase and the Mre11 complex. Mol Cell Biol 25: 10652–10664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courbet S, Gay S, Arnoult N, Wronka G, Anglana M, Brison O, Debatisse M (2008) Replication fork movement drives remodeling of chromatin loops and origin choice in mammalian cells. Nature 455: 557–560 [DOI] [PubMed] [Google Scholar]
- Crabbe L, Thomas A, Pantesco V, De Vos J, Pasero P, Lengronne A (2010) Analysis of replication profiles reveals key role of RFC-Ctf18 in yeast replication stress response. Nat Struct Mol Biol 17: 1391–1397 [DOI] [PubMed] [Google Scholar]
- Davidson MB, Brown GW (2008) The N- and C-termini of Elg1 contribute to the maintenance of genome stability. DNA Repair (Amst) 7: 1221–1232 [DOI] [PubMed] [Google Scholar]
- Davidson MB, Katou Y, Keszthelyi A, Sing TL, Xia T, Ou J, Vaisica JA, Thevakumaran N, Marjavaara L, Myers CL, Chabes A, Shirahige K, Brown GW (2011) Endogenous DNA replication stress results in expansion of dNTP pools and a mutator phenotype. EMBO J (advance online publication, 10 January 2012; doi:; DOI: 10.1038/emboj.2011.485) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Bachant J, Collingwood D, Raghuraman MK, Brewer BJ (2009) Centromere replication timing determines different forms of genomic instability in Saccharomyces cerevisiae checkpoint mutants during replication stress. Genetics 183: 1249–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert CS, Green CM, Lowndes NF (2001) Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol Cell 8: 129–136 [DOI] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase SB, Lew DJ (1997) Flow cytometric analysis of DNA content in budding yeast. Methods Enzymol 283: 322–332 [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J (2008) An oncogene-induced DNA damage model for cancer development. Science 319: 1352–1355 [DOI] [PubMed] [Google Scholar]
- Koc A, Wheeler LJ, Mathews CK, Merrill GF (2004) Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J Biol Chem 279: 223–230 [DOI] [PubMed] [Google Scholar]
- Kumar D, Viberg J, Nilsson AK, Chabes A (2010) Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res 38: 3975–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese MP, Paciotti V, Fraschini R, Zaccarini R, Plevani P, Lucchini G (1997) The novel DNA damage checkpoint protein ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. EMBO J 16: 5216–5226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M (2001) The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412: 557–561 [DOI] [PubMed] [Google Scholar]
- Lopez-Mosqueda J, Maas NL, Jonsson ZO, DeFazio-Eli LG, Wohlschlegel J, Toczyski DP (2010) Damage-induced phosphorylation of Sld3 is important to block late origin firing. Nature 467: 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Versini G, Jaquenoud M, Zaidi IW, Kurz T, Pintard L, Pasero P, Peter M (2006) The Cullin Rtt101p promotes replication fork progression through damaged DNA and natural pause sites. Curr Biol 16: 786–792 [DOI] [PubMed] [Google Scholar]
- Mathews CK (2006) DNA precursor metabolism and genomic stability. FASEB J 20: 1300–1314 [DOI] [PubMed] [Google Scholar]
- Nicol JW, Helt GA, Blanchard SG Jr, Raja A, Loraine AE (2009) The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25: 2730–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieduszynski CA, Hiraga S, Ak P, Benham CJ, Donaldson AD (2007) OriDB: a DNA replication origin database. Nucl Acids Res 35: D40–D46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida H, Katsuno Y, Sengoku M, Shimada M, Yukawa M, Ikura M, Ikura T, Kohno K, Shima H, Suzuki H, Tashiro S, Nakanishi M (2010a) Essential role of Tip60-dependent recruitment of ribonucleotide reductase at DNA damage sites in DNA repair during G1 phase. Genes Dev 24: 333–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida H, Shimada M, Murakami H, Nakanishi M (2010b) Mechanisms of dNTP supply that play an essential role in maintaining genome integrity in eukaryotic cells. Cancer Sci 101: 2505–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasero P, Bensimon A, Schwob E (2002) Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev 16: 2479–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL (2001) Replication dynamics of the yeast genome. Science 294: 115–121 [DOI] [PubMed] [Google Scholar]
- Sabouri N, Viberg J, Goyal DK, Johansson E, Chabes A (2008) Evidence for lesion bypass by yeast replicative DNA polymerases during DNA damage. Nucleic Acids Res 36: 5660–5667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF (1998) A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395: 615–618 [DOI] [PubMed] [Google Scholar]
- Schmidt KH, Kolodner RD (2006) Suppression of spontaneous genome rearrangements in yeast DNA helicase mutants. PNAS 103: 18196–18201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Katou Y, Yagura M, Saitoh K, Itoh T, Araki H, Bando M, Shirahige K (2009) Ctf4 coordinates the progression of helicase and DNA polymerase α. Genes Cells 14: 807–820 [DOI] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF (2001) Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553–557 [DOI] [PubMed] [Google Scholar]
- Tourriere H, Pasero P (2007) Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair 6: 900–913 [DOI] [PubMed] [Google Scholar]
- Tourriere H, Versini G, Cordon-Preciado V, Alabert C, Pasero P (2005) Mrc1 and tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell 19: 699–706 [DOI] [PubMed] [Google Scholar]
- Versini G, Comet I, Wu M, Hoopes L, Schwob E, Pasero P (2003) The yeast Sgs1 helicase is differentially required for genomic and ribosomal DNA replication. EMBO J 22: 1939–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki N, Terashima H, Kitada K (2002) Mapping of early firing origins on a replication profile of budding yeast. Genes Cells 7: 781–789 [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JFX (2009) DNA replication as a target of the DNA damage checkpoint. DNA Repair 8: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JFX (2010) Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature 467: 474–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Muller EG, Rothstein R (1998) A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell 2: 329–340 [DOI] [PubMed] [Google Scholar]
- Zhao X, Rothstein R (2002) The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. PNAS 99: 3746–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.