Abstract
Cell adhesion organizes the structures of tissues and mediates their mechanical, chemical, and electrical integration with their surroundings. Here, we describe a strategy for chemically controlling cell adhesion using membrane anchored single-stranded DNA oligonucleotides. The reagents are pure chemical species prepared from phosphoramidites synthesized in a single chemical step from commercially available starting materials. The approach enables rapid, efficient, tunable cell adhesion, independent of proteins or glycans, by facilitating interactions with complementary labeled surfaces or other cells. We demonstrate the utility of this approach by imaging drug-induced changes in the membrane dynamics of non-adherent human cells while chemically immobilized on a passivated glass surface.
Keywords: cell patterning, self-assembly, cell-cell interactions, lipid-DNA, cytoskeleton, imaging, lymphocyte, membrane dynamics, bottom-up
Cells interact with cells and materials through adhesion molecules expressed on their surfaces. These interactions physically couple cells to their surroundings and alter cellular behavior by triggering intracellular signaling cascades. Decoupling the mechanics of cell adhesion from intracellular signaling remains a challenge that limits the study of many fundamental questions in cell biology.1
Chemical strategies for controlling cell adhesion have attempted to address this challenge in two ways. First, chemical ligands specific for the native adhesion machinery have been used to direct adhesion with high temporal and spatial resolution2–5. Second, artificial adhesion molecules6 have been used to direct the physical interactions between cells and substrates independent of the adhesion machinery. Promising examples of this latter approach use cell-surface grafted oligonucleotides7–11 as artificial adhesion molecules. Indeed, nucleic acids offer several advantages, including combinatorial encoding of interactions, ease of synthesis and modification, and minimal cross reactivity with the suite of other molecules typically found at the cell surface. DNA-programmed adhesions are also strong, form rapidly, and are reversible upon addition of DNAse12.
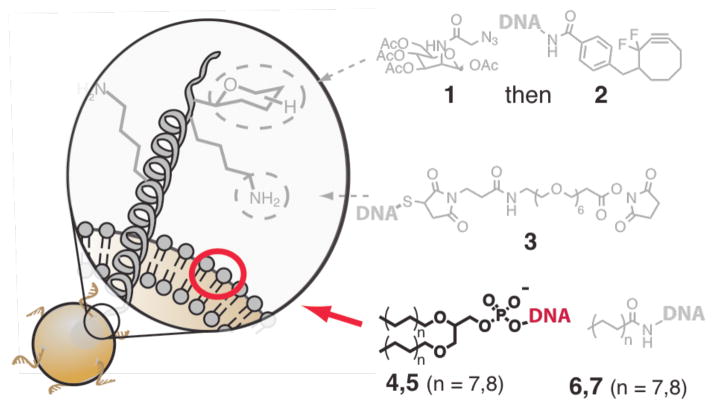
Cell surfaces have been modified with DNA by engineering the glycocalyx9 or by both non-covalent7,8 and covalent10,13 modification of cell surface proteins (Figure 1). These strategies offer an effective means of programming adhesion. However, they may also inadvertently engage the adhesion machinery, perturb the cytoskeleton, or activate cell surface receptors due to the somewhat indiscriminate nature of the modification process. Their overall utility would also benefit by increasing the speed, generality, and cost effectiveness of the labeling process. We therefore sought an alternative means for programming cell adhesion using DNA that avoids covalent reactions with cell surface proteins and glycans, thus facilitating new experimental applications while increasing the method’s overall feasibility.
FIGURE 1.
DNA-conjugates used in this study. Cell surface glycans are targeted for chemical remodeling following a three-day incubation in azido sugar 1 and subsequent covalent modification with DIFO 2 conjugated DNA. Protein lysine side chains are conjugated to NHS-ester modified DNA 3. Fatty acid amides 6, 7 and dialkylphosphoglyceride-modified oligonucleotides 4, 5 (bold) target the lipid bilayer non-covalently.
Previous reports have shown that lipid bilayers can be modified with oligonucleotides incorporating hydrophobic molecules at their 3′ or 5′ ends.14–20 Here, we apply this approach to programming cell adhesion to provide significant advantages over previous strategies including (i) diffusion of the oligonucleotides independent of cell surface proteins and glycans; (ii) adhesion decoupled from the cytoskeleton; and (iii) attachment of oligonucleotides at a common physical site relative to the cell membrane. These improvements enable experiments requiring programmed cell adhesion while avoiding activation of adhesion-dependent signaling cascades.
We anticipated three barriers to implementing this approach for chemically programming cell adhesion. First, the tendency of amphipathic DNA conjugates towards self-aggregation may interfere with their partitioning into the cell surface. Second, steric hindrance from a 20+ nm thick glycocalyx may prevent access of membrane bound oligonucleotides to complementary labeled surfaces. Third, improper control of the extent of labeling may alter cellular viability, growth, or behavior by disrupting membrane integrity or mechanics.
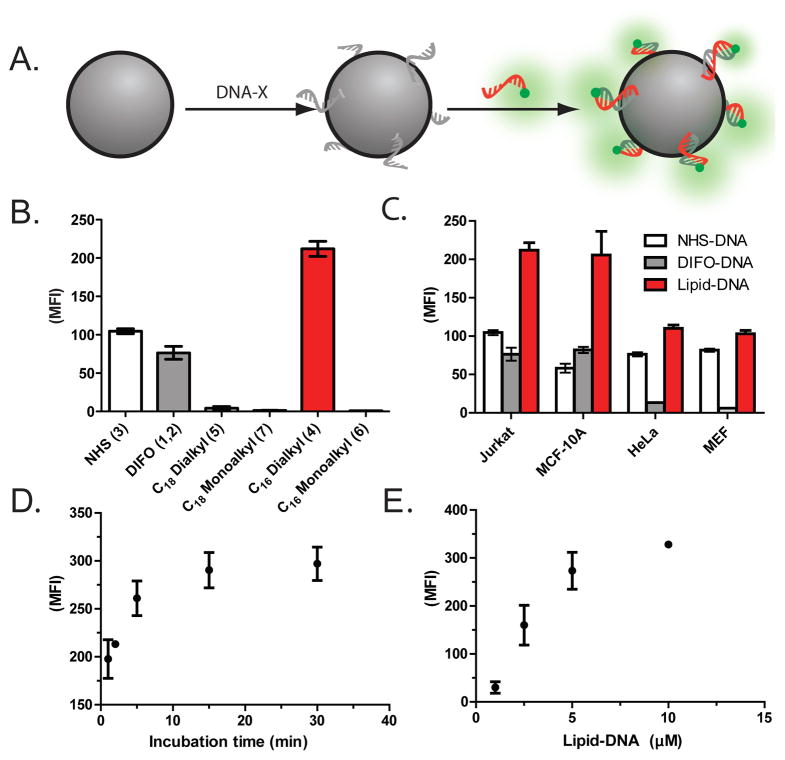
To address these concerns, we first prepared oligonucleotides conjugated to a C18 dialkylphosphoglyceride (5, n=8) and assayed their ability to partition into Jurkat T-lymphocyte cell membranes using flow cytometry (Figure 2a). This modification was previously reported to be stable in liposomes and is easily coupled to the 5′ end of oligonucleotides via the phosphoramidite (Scheme S1)17. Although we observed some oligonucleotides incorporating into cell surfaces, the extent of incorporation was insufficient to mediate cell-cell or cell-surface adhesion (Figure 2b). Neither increasing the concentration of modified DNA, the temperature, nor duration of incubation resulted in substantial increases in membrane incorporation (Figure S1).
FIGURE 2.
Incorporation of oliognucleotides to cell surfaces. (A) Scheme for labeling and selectively quantifying cell surface oligonucleotides by flow cytometry. (B) Median Fold Fluorescence Increase (MFI) of Jurkat cells labeled with DNA. (C) Extent of cell surface modification by 1/2, 3, and 4 (n=7) across four different cell types. (D) Time course for cell surface modification with 4 (n=7). (E) Concentration dependence of cell surface labeling by 4 (n=7).
We hypothesized that the low efficiency of cell surface modification was due to the anticipated slow exchange between aggregated and solvated forms of the lipid-modified oligonucleotides. However, attempts to destabilize aggregates by adding lipid-DNA solutions containing 50% ethanol or lacking salt did not lead to increased labeling (Figure S1). We also explored oligonucleotides 6 or 7 incorporating single acyl chains, as they are predicted to exchange more rapidly between solution and membrane-bound forms.16 However, these oligonucleotides appeared unstable on the cell surface as we were unable to detect the DNA conjugates by flow cytometry (Figure 2b).
In examining the physical properties of diacylphosphoglycerides, structurally similar C18 and C16 phospholipids differ significantly with respect to their kinetics of membrane exchange,21 suggesting that even slightly shorter alkyl chains might dramatically improve the rate of partitioning out of aggregates without significantly affecting retention in cell membranes. Indeed, several reports have described C16 lipids or unsaturated C18 lipids with similar physical properties as effective modifications for directing the partitioning of polymers into cell surfaces.22,23
Consistent with these studies, we found that C16 dialkylphosphoglyceride-modified oligonucleotides 4 rapidly incorporated into Jurkat cell membranes (Figure 2B). Even after repeated washing with PBS containing 1% serum we observed only minor reductions in cell-surface 4-DNA. Trypsin treatment did not affect the degree of modification, a significant advantage of this approach when working with adherent cells such as HeLa, MEF, or MCF-10A lines (Figure 2C). Additionally, labeling was rapid (Figure 2D) and dose dependent (Figure 2E). Finally, we assayed the lifetime of the cell surface DNA by flow cytometry and found that on Jurkat cell surfaces, DNA decayed to 86% of its initial concentration by 160 minutes at 25 °C and to 67% of its initial intensity over the same time period at 37 °C. These values were similar to those measured for protein or glycan-modified cell surfaces (Figure S3).
Oligonucleotides modified with 2 and 3 react with functionality present throughout the cell surface. However, 4 modified oligonucleotides are predicted to only localize proximal to the cell membrane, where steric hindrance from the glycocalyx may limit accessibility to complementary labeled surfaces. We therefore compared the ability of 3 and 4 modified cell surfaces to adhere to complementary labeled surfaces. We first prepared DNA-modified glass by reacting 5′-amino-terminated oligonucleotides with aldehyde-coated microscope slides by reductive amination. Subsequent reduction of unreacted aldehyde with NaBH4 and passivation with Sigmacote then Pluronic F108 provided a non-adhesive surface. We then prepared populations of Jurkat cells labeled with a complementary 20mer strand anchored to proteins through 3 or to the cell membrane through 4. Finally, we compared the ability of 3- and 4-labeled cells to adhere to the DNA-labeled glass. Cells labeled with the 4-conjugated 20mer strand did not significantly adhere to the glass surface whereas 3-labeled cells showed significant adhesion despite similar absolute numbers of oligonucleotides at the cell surface (Figure 3A).
FIGURE 3.
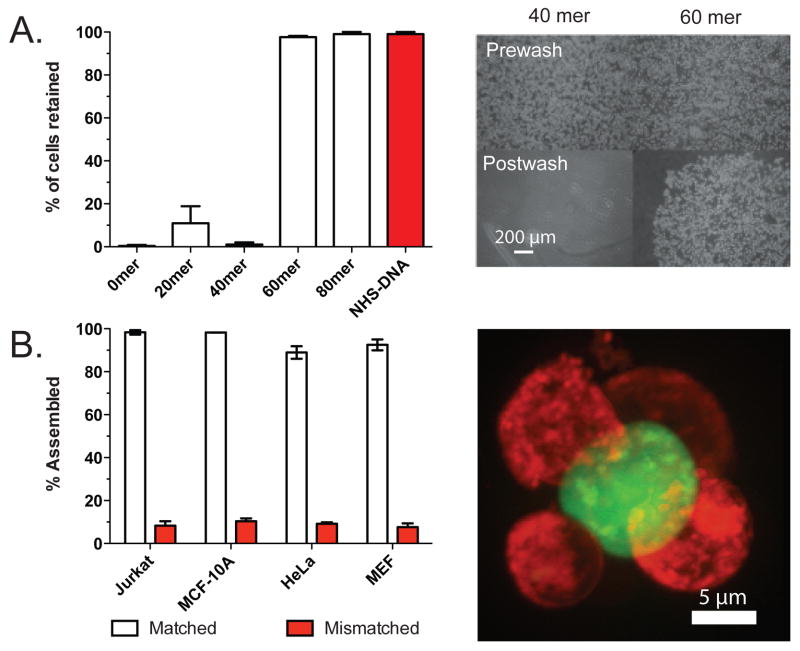
Chemical control of cellular adhesion by 4-DNA. (A) Jurkat cells bearing a 60- or 80mer poly(dT) linker had considerably more cell-surface adhesion than cells with shorter linkers. (B) The indicated cell type bearing 4-modified DNA incorporating 80mer poly(dT) linkers was assembled with a 100x excess of a complementary-labeled population of Jurkat cells and then analyzed by flow cytometry. Assembly efficiency is reported as the fraction of minority cells associated with at least one majority cell. A FACS purified cell cluster imaged by confocal fluorrescence microscopy is shown to the right.
We attributed these differences in cell adhesion to steric hindrance caused by the cells’ dense glycocalyx. We therefore reasoned that the addition of poly(dT) linkers between the 20mer sequence and the lipid anchor, a modification that is easily introduced on a DNA synthesizer, might extend the adhesive 20mer region beyond the glycocalyx thereby increasing the adhesive capacity of labeled cells. To test this idea, we synthesized a series of oligonucleotides incorporating 20, 40, 60, or 80 thymine nucleotides between the 20mer sequences and the lipid anchor and tested their ability to direct adhesive interactions between labeled cells and complementary labeled surfaces. We found a sharp transition in adhesive capacity between the oligonucleotides incorporating 40 and 60 thymine nucleotides (Figure 3a). Several reports have estimated the contribution of each single dT to the contour length of poly(dT) to be, on average, 5.2 Å.24 This would be consistent with the notion that the glycocalyx reduces access to 20mer sequences that are incapable of extending between 20 and 30 nm above the cell membrane.
Utilizing the 80mer poly(dT) linker, we also measured the degree to which 4-modified DNA could program the self-assembly of complementary labeled populations of Jurkat cells. We observed near quantitative formation of chemically programmed cell-cell adhesions after 10 minutes of mixing (Figure 3b).12 Similar results were obtained with adherent MCF-10A, MEF, and HeLa cells.
To be useful in subsequent experiments, populations of DNA-labeled cells must also retain high viability and normal proliferative capacity. We found that the viability of Jurkat cells labeled with 4-modified 100mer oligonucleotides approached or exceeded 90% -- similar to controls treated with PBS alone and to cells labeled by glycan engineering or direct conjugation to lysine side chains (Figure S4A). We also measured cell proliferation in labeled cell populations using the resazurin assay. Jurkat cells showed no differences in their rate of proliferation after being modified with 4. Similar results were obtained with MCF-10A, HeLa and MEF cells (Figure S4C).
As described previously, an advantage of this approach to programming cell adhesion is the potential to decouple adhesion from both the cytoskeleton and cell surface proteins and glycans. This facilitates imaging of non-adherent cells that undergo rotations and translation in the x, y, and z directions, but that change their morphology and polarity upon interaction with adhesive surfaces1. To image the membrane dynamics of non-adherent cells, we envisioned a strategy in which 4-modified cells were chemically immobilized on passivated glass surfaces by grids of 5–7 μm spots bearing complementary DNA sequences. Based on previous reports, we anticipated that chemical immobilization through such small DNA patches would be minimally perturbing3 but still facilitate imaging and allow for the addition and removal of small molecule drugs.
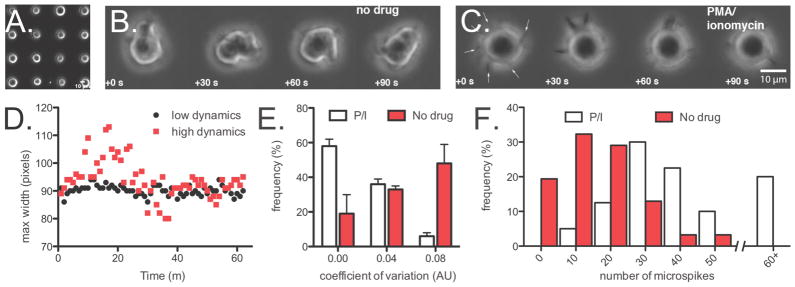
To test the feasibility of the approach, we first confirmed that the morphology of 4-modified cells was similar to unmodified cells when imaged in suspension over glass (supplementary movies S5). We next prepared small, 5–7 μm patches of DNA on passivated glass surfaces using a Bioforce Nano eNabler and confirmed the presence of DNA on the surface by annealing a FITC-labeled complementary strand (Figure S6). We also confirmed that Jurkat cells were unable to interact with the passivated glass surface (Supplementary movies S7). Jurkat cells bearing 4-modified complementary sequences were immobilized on these patches by DNA hybridization (Figure 4A). Time-lapse imaging revealed a heterogeneous population of cells with respect to cell shape that was changing on the minute timescale (Figure 4B, D, and E and supplementary movie S8). Interestingly, small molecule T-cell activators PMA and ionomycin (P/I) seemed to reduce the cell shape heterogeneity within the population (Figure 4E). At higher magnification, we noticed that many cells also extended membrane microspikes from the cell surface (Figure 4C and supplementary movie S9). These appeared to be distinct from the gross morphological changes observed at lower magnification, as they were enhanced rather than reduced by P/I treatment (Figure 4F).
FIGURE 4.
Imaging the membrane dynamics of T-lymphocytes. (A) A representative field of Jurkat cells arrayed on 5–7 μm spots. (B) Timelapse images illustrating the dynamics of a single immobilized Jurkat Cell. (C) P/I treated Jurkat cells are more homogeneous with respect to cell shape but have increased membrane microspikes (arrows). (D) Trajectories of maximum cell width for representative cells of high and low membrane dynamics. (E) Population-level analysis of maximum width for cells treated with or without PMA/ionomycin. (F) Population-level analysis of microspike dynamics over 15 minutes for cells treated with or without PMA/ionomycin.
In conclusion, we describe a simple method for programming the adhesive properties of mammalian cells independent of proteins, glycans, or their endogenous adhesion machinery. Our approach involves the solid-phase modification of synthetic oligonucleotides with a lipidic phosphoramidite that is synthesized in one step from commercially available starting materials. The modified oligonucleotides passively incorporate into cell membranes from dilute solutions in five minutes or less and, when incorporating suitable chemically defined linkers, can rapidly and specifically direct physical interactions between cells and complementary-labeled cells or surfaces. The resulting chemically adherent cells maintain high viability, proliferative capacity, and behave identically to unmodified cells when analyzed by light microscopy. Because cells are unaltered by the immobilization process, changes to their behaviors are easily imaged upon perturbation by addition of small molecule drugs. We anticipate these chemically homogeneous lipid-modified oligonucleotides will also facilitate numerous other applications, including the study of cell-cell interactions, membrane mechanics, the bottom-up assembly of modular tissues25, or the study of biological processes occurring within 50 nm of the cell surface.
Supplementary Material
Acknowledgments
We thank Professors Wendell Lim and Mark Labarge for materials. We thank Professors Kevan Shokat, Pam England, and Jack Taunton for helpful comments and for sharing instruments and facilities, and Bioforce Nanosciences for help with the Nano eNabler. The Bio-Organic Biomedical Mass Spectrometry Resource at UCSF (A.L. Burlingame, Director) is supported by NIH NCRR P41RR001614. This work is partially supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE-0648991 to J. Liu, by a UCSF CTSI-SOS pilot grant, by the Program for Breakthrough Biological Research, by Grant IRG 97-150-10 from the American Cancer Society, by the Kimmel Family Foundation, and by DOD BCRP grant W81XWH-10-1-1023.
ABBREVIATIONS
- FACS
Fluorescence Activated Cell Sorting
- DNA
Deoxyribonucleic Acids
- DIFO
Difluorocyclooctyne
- NHS
N-hydroxysuccinimide
- PEG
polyethylene glycol
- MFI
median fold fluorescence increase
- dT
deoxythymidine
- MEF
mouse embryonic fibroblast
- PBS
phosphate buffered saline
- TEAA
triethylammonium acetate
- HPLC
high pressure liquid chromatography
- P/I
phorbol-12-myristate-13-acetate (PMA) and ionomycin
- FITC
fluoroscein isothiocyanate
Footnotes
Author Contributions
The manuscript was written by ZJG and NSS. ZJG, NSS, and MET designed the experiments. NSS, MET, NYJ, JSL, and KEB performed the experiments and interpreted the data. All authors have given approval to the final version of the manuscript.
Supporting Information. Schemes, figures, time-lapse movies, and experimental methods. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Ferguson GJ, Milne L, Kulkarni S, Sasaki T, Walker S, Andrews S, Crabbe T, Finan P, Jones G, Jackson S, Camps M, Rommel C, Wymann M, Hirsch E, Hawkins P, Stephens L. Nat Cell Biol. 2007;9:86–91. doi: 10.1038/ncb1517. [DOI] [PubMed] [Google Scholar]
- 2.Aldaye FA, Senapedis WT, Silver PA, Way JC. J Am Chem Soc. 2010;132:14727–9. doi: 10.1021/ja105431h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Science. 1997;276:1425–8. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 4.Desai RA, Gao L, Raghavan S, Liu WF, Chen CS. J Cell Sci. 2009;122:905–11. doi: 10.1242/jcs.028183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato M, Mrksich M. J Am Chem Soc. 2004;126:6504–5. doi: 10.1021/ja039058e. [DOI] [PubMed] [Google Scholar]
- 6.Dutta D, Pulsipher A, Luo W, Yousaf MN. J Am Chem Soc. 2011;133:8704–13. doi: 10.1021/ja2022569. [DOI] [PubMed] [Google Scholar]
- 7.Liu X, Yan H, Liu Y, Chang Y. Small. 2011;7:1673–82. doi: 10.1002/smll.201002292. [DOI] [PubMed] [Google Scholar]
- 8.Bailey RC, Kwong GA, Radu CG, Witte ON, Heath JR. J Am Chem Soc. 2007;129:1959–67. doi: 10.1021/ja065930i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandra RA, Douglas ES, Mathies RA, Bertozzi CR, Francis MB. Angew Chem Int Ed Engl. 2006;45:896–901. doi: 10.1002/anie.200502421. [DOI] [PubMed] [Google Scholar]
- 10.Hsiao SC, Shum BJ, Onoe H, Douglas ES, Gartner ZJ, Mathies RA, Bertozzi CR, Francis MB. Langmuir. 2009;25:6985–91. doi: 10.1021/la900150n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwata H, Teramura Y, Minh LN, Kawamoto T. Bioconjugate Chemistry. 2010;21:792–796. doi: 10.1021/bc900494x. [DOI] [PubMed] [Google Scholar]
- 12.Gartner ZJ, Bertozzi CR. Proc Natl Acad Sci U S A. 2009;106:4606–10. doi: 10.1073/pnas.0900717106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao W, Loh W, Droujinine IA, Teo W, Kumar N, Schafer S, Cui CH, Zhang L, Sarkar D, Karnik R, Karp JM. Faseb J. 2011 doi: 10.1096/fj.10-178384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfeiffer I, Hook F. J Am Chem Soc. 2004;126:10224–5. doi: 10.1021/ja048514b. [DOI] [PubMed] [Google Scholar]
- 15.Teramura Y, Chen H, Kawamoto T, Iwata H. Biomaterials. 2010;31:2229–35. doi: 10.1016/j.biomaterials.2009.11.098. [DOI] [PubMed] [Google Scholar]
- 16.Borisenko GG, Zaitseva MA, Chuvilin AN, Pozmogova GE. Nucleic Acids Res. 2009;37:e28. doi: 10.1093/nar/gkn1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan YH, van Lengerich B, Boxer SG. Proc Natl Acad Sci U S A. 2009;106:979–84. doi: 10.1073/pnas.0812356106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H, Kwong B, Irvine DJ. Angew Chem Int Ed Engl. 2011 doi: 10.1002/anie.201101266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshina-Ishii C, Miller GP, Kraft ML, Kool ET, Boxer SG. J Am Chem Soc. 2005;127:1356–7. doi: 10.1021/ja043299k. [DOI] [PubMed] [Google Scholar]
- 20.Borjesson K, Tumpane J, Ljungdahl T, Wilhelmsson LM, Norden B, Brown T, Martensson J, Albinsson B. J Am Chem Soc. 2009;131:2831–9. doi: 10.1021/ja8038294. [DOI] [PubMed] [Google Scholar]
- 21.Silvius JR, Leventis R. Biochemistry. 1993;32:13318–26. doi: 10.1021/bi00211a045. [DOI] [PubMed] [Google Scholar]
- 22.Rabuka D, Forstner MB, Groves JT, Bertozzi CR. J Am Chem Soc. 2008;130:5947–53. doi: 10.1021/ja710644g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato K, Itoh C, Yasukouchi T, Nagamune T. Biotechnology Progress. 2004;20:897–904. doi: 10.1021/bp0342093. [DOI] [PubMed] [Google Scholar]
- 24.Mills JB, Vacano E, Hagerman PJ. J Mol Biol. 1999;285:245–57. doi: 10.1006/jmbi.1998.2287. [DOI] [PubMed] [Google Scholar]
- 25.Kachouie NN, Du Y, Bae H, Khabiry M, Ahari AF, Zamanian B, Fukuda J, Khademhosseini A. Organogenesis. 2010;6:234–44. doi: 10.4161/org.6.4.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.