Abstract
The adaptor protein R7 family binding protein (R7BP) modulates G protein coupled receptor (GPCR) signaling and desensitization by controlling the function of regulator of G protein signaling (RGS) proteins. R7BP is expressed throughout the brain and appears to modulate the membrane localization and stability of three proteins that belong to R7 RGS family: RGS6, RGS7, and RGS9-2. RGS9-2 is a potent negative modulator of opiate and psychostimulant addiction and promotes the development of analgesic tolerance to morphine, whereas the role of RGS6 and RGS7 in addiction remains unknown. Recent studies revealed that functional deletion of R7BP reduces R7 protein activity by preventing their anchoring to the cell membrane and enhances GPCR responsiveness in the basal ganglia. Here, we take advantage of R7BP knockout mice in order to examine the way interventions in R7 proteins function throughout the brain affect opiate actions. Our results suggest that R7BP is a negative modulator of the analgesic and locomotor activating actions of morphine. We also report that R7BP contributes to the development of morphine tolerance. Finally, our data suggest that although prevention of R7BP actions enhances the analgesic responses to morphine, it does not affect the severity of somatic withdrawal signs. Our data suggest that interventions in R7BP actions enhance the analgesic effect of morphine and prevent tolerance, without affecting withdrawal, pointing to R7BP complexes as potential new targets for analgesic drugs.
Keywords: hot plate, neuropathic pain, RGS9-2, RGS7, morphine, fentanyl
INTRODUCTION
Regulators of G protein signaling (RGS) are a large family of multifunctional proteins that control fundamental physiological functions as well as pharmacological responses mediated by the G protein coupled receptors (GPCRs). RGS proteins control signal transduction by associating with activated Gα subunits to promote their termination, as well as by several other mechanisms that affect activation of effector enzymes and receptor desensitization (Berman and Gilman, 1998; Dohlman and Thorner, 1997). Several RGS proteins contain additional domains that modify their catalytic activity or provide additional functions. RGS9-2 as well as highly homologous RGS6 and RGS7 (RGS6/7) are among the most abundant RGSs in the CNS (Gold et al, 1997; Terzi et al, 2009). They belong to the R7 subfamily that additionally contains RGS9-1 and RGS11 proteins prevalent in the retina (Hooks et al, 2003; Anderson et al, 2009). Members of the R7 family share several domains and form constitutive complexes with the type 5 G protein β subunit (Gβ5) (Sondek and Siderovski, 2001; Cheever et al, 2008). The R7–Gβ5 association is essential for the protein's stability and function (He et al, 2000; Chen et al, 2003). R7 proteins also share a DEP domain (Martemyanov et al, 2003; Ballon et al, 2006), which is required for their interaction with the adaptor molecules R9AP in the retina (Hu and Wensel, 2002) and R7 family binding protein (R7BP) in the brain (Martemyanov et al, 2005). The R7–R7BP interaction is essential for the cellular localization and function of R7 proteins, as it determines the localization of the protein at the cell membrane (Drenan et al, 2005; Anderson et al, 2009; Jayaraman et al, 2009).
A number of studies have shown the functional significance of R7 interactions with their binding partners (Jayaraman et al, 2009; Martemyanov et al, 2003; Psifogeorgou et al, 2011). These studies revealed that Gβ5 and R7BP knockout mice have dramatically diminished levels of RGS9-2 protein (Chen et al, 2003; Anderson et al, 2009), that loss of R7BP affects RGS9-2 localization and striatal signaling (Anderson et al, 2009), and that pharmacological manipulations, or changes in neuronal excitability affect the composition of R7 complexes in the striatum (Anderson et al, 2009; Psifogeorgou et al, 2011). Recent evidence from genetic mouse models suggests that R7BP has a key role in regulation of GPCR responses in the basal ganglia, via interactions with RGS9-2 and RGS7. RGS9-2 is expressed in very high amounts in the striatum, where it has been shown to modulate responsiveness of μ-opioid and D2 dopamine GPCRs (Rahman et al, 2003; Zachariou et al, 2003; Cabrera-Vera et al, 2004; Kovoor et al, 2005; Gold et al, 2007; Traynor et al, 2009). Specifically, our earlier work revealed a potent role of RGS9-2 in opiate actions (Zachariou et al, 2003; Psifogeorgou et al, 2007; Psifogeorgou et al, 2011). Opiates, such as morphine, have several applications as analgesics for some forms of pain, but their prolonged use is associated with severe side effects such as the development of dependence and analgesic tolerance (Kreek, 2001; Inturrisi, 2002). A number of recent reports implicate a number of signal transduction molecules, such as β-arrestin-2, several protein kinases or scaffolding molecules in the mechanisms underlying analgesic tolerance (Bohn et al, 1999; Chen et al, 2008; Charlton et al, 2008; Melief et al, 2010) but the exact brain network specific events underlying this phenomenon are not fully understood. RGS9-2 actions in the striatum modulate opiate reward, dependence and analgesia, and contribute to the development of analgesic tolerance (Traynor et al, 2009). RGS6 and RGS7 show a broader pattern of distribution, and, in addition to striatum, they are present at significant levels in the hippocampus as well as in the locus coeruleus (LC) (a noradrenergic nucleus involved in the expression of opiate withdrawal) (Gold et al, 1997). This expression pattern suggests that R7 RGS protein complexes may potently modulate opiate addiction. Since R7BP is an essential component of such complexes, we hypothesized that blockade of R7BP/R7 interactions affects opiate actions. To examine this hypothesis, we took advantage of genetically modified mice, lacking the R7BP gene (Anderson et al, 2007) and performed a series of behavioral and biochemical studies. Our findings suggest that R7BP acts as a negative modulator of the analgesic and locomotor activating actions of morphine, has a major role in morphine tolerance but does not affect morphine withdrawal. These data point to R7BP as a potent new pharmacological target, as blockade of its function may enhance the analgesic actions of morphine and prevent the development of analgesic tolerance without affecting the severity of withdrawal.
MATERIALS AND METHODS
Animals
For all behavioral assays, we used naive 2–4 month R7BP knockout mice (R7BPKO) and their wild-type littermates (R7BPWT), as described before (Anderson et al, 2010). All assays were performed in male mice except for morphine withdrawal assays, which were performed in female mice. For immunoprecipitation assays, striata were extracted 30 min following saline, fentanyl or morphine injections as described earlier (Charlton et al, 2008). For immunoblotting analysis, striata from 2-month-old C57Bl/6 mice were extracted 30 min, 2 h, or 4 days after saline or drug treatment as described (Zachariou et al, 2003). Animals were housed in a 12-h dark/light cycle room. All experiments were performed in accordance with the Declaration of Helsinki and with the Guide for the Care and Use of Laboratory Animals by the National Institutes of Health and were approved by the animal care and use committees of the University of Crete and the Scripps Research Institute.
Co-immunoprecipitation Assays and Western Blotting
Striatal tissue of mice treated with saline, morphine, or fentanyl for 30 min were rapidly dissected as described earlier (Psifogeorgou et al, 2011). Lysates were precleared with 20 μl of G agarose beads (Roche, Mannheim, Germany) by 1–2 h incubation at 4 °C. Supernatants were incubated overnight with primary antibody on a rotating platform, followed by incubation with 20 μl protein G agarose beads for 2 h at 4 °C. Beads were washed three times in lysis buffer, precipitated, and resuspended in equal volume of loading buffer. Immunoprecipitated proteins were analyzed by 10% SDS–polyacrylamide gel electrophoresis followed by electrotransfer to 0.45 mm nitrocellulose membrane (Bio-Rad Labs, Hercules, CA). For western blot analysis studies, Gβ5 (a protein shown not to be regulated by opiate administration) was used as a loading control. The following antibodies were used for IP and western blot assays: rabbit protein A-purified anti-RGS9-antibody (1 : 10 000; Psifogeorgou et al, 2007) and a rabbit anti-Gβ5 (C-terminus) antibody (1 : 20 000; W Simonds, National Institute of Diabetes, Digestive and Kidney Diseases, Bethesda, MD), a rabbit anti-RGS6/7 antibody (Wyeth–Ayerst Pharmaceuticals, Madison, NJ, USA), and a rabbit anti-R7BP antibody (Martemyanov et al, 2005). For synaptosomal preparations, striatum was dissected and homogenized in 1 ml 0.32 M sucrose 0.1 mM CaCl2 (containing phosphatase-NaF, Na3VO4-, and protease inhibitors) using a glass homogenizing tube and a Teflon pestle. After centrifugation at 4000 r.p.m. for 5 min at 4 °C, the pellet was discarded and the supernatant was centrifuged at 13 200 r.p.m. for 20 min. The pellet was resuspended in 1% SDS plus inhibitors in a final volume of 150 μl and placed on ice (Moron et al, 2003). For all western blotting and co-immunoprecipitation assays, levels were quantified by measuring the optical density of specific bands using NIH image. Data are expressed as percentage of saline-treated group.
Hot Plate Analgesia and Tolerance Paradigms
For acute analgesia studies, baseline latencies (time to leak paw or jump) were monitored using the 52 °C hot plate paradigm (IITC, Life Sciences, CA) as described before (Psifogeorgou et al, 2011). Following that mice were injected with morphine (or fentanyl) and hot plate responses were monitored 30 min later. For analgesic tolerance assays, mice were monitored at baseline and 30 min after drug administration for 5 consecutive days. All hot plate data are expressed as percentage maximal possible effect (MPE=[latency–baseline]/[cutoff–latency]). A cutoff time of 40 s was used for all hot plate experiments.
Locomotor Activity Assays
For locomotor activity assays, on test day, littermate mice were habituated to the locomotor cages for 1 h before testing. Mice were injected subcutaneously with saline daily for 3 days for habituation. Morphine (5, 10, 20 mg/kg) freshly dissolved in saline was injected with 3-day interval between the different doses. After each injection, the mouse was placed in the center of locomotor activity chambers (ENV515; 170 W × −170 L × 120 H; Med Associates, St Albans, VT). Ambulatory activity was monitored for 180 min by an automated tracking system consisting of three 16 beam infrared arrays as described previously (Anderson et al, 2010).
Spared Nerve Injury Model
The surgical procedure for spare nerve injury was performed under avertine (Sigma-Aldrich) general anesthesia. Using stereomicroscope, an incision of the skin and muscle of the left hind-paw at mid-thigh level was performed to reach the sciatic nerve and its three branches. The common peroneal and the sural nerves were carefully ligated with 6.0 silk suture (Ethicon, Johnson & Johnson Intl.), transected and a 1- to 2-mm section of these nerves was removed, while the tibia nerve was left intact. Skin incisions were then closed with silk 4.0 sutures.
Von Frey Assay for Tactile Allodynia
Mechanical threshold for hind-paw withdrawal was determined using von Frey hairs with ascending forces expressed in grams (0.1–3.6 g) (Electronic von Frey Anesthesiometer, IITC, CA). Mice were single-placed in Plexiglas boxes on an elevated mesh screen. Animals were habituated to the apparatus every other day for 2 weeks before surgery. For assessment of mechanical allodynia, each von Frey hair was applied on the plantar surface of the left hind-paw, five times per paw, and the mechanical threshold was defined as three or more positive responses.
Morphine Withdrawal Paradigm
Mice were injected with increasing doses of morphine every 12 h for 4 consecutive days (day 1: 20 mg/kg, day 2: 40 mg/kg, day 3: 60 mg/kg, day 4: 80 mg/kg) and on the fifth day they received a morphine injection (80 mg/kg) and 3 h later, withdrawal was precipitated with a s.c. injection of naloxone (1 mg/kg; Sigma). Withdrawal signs (jumps, wet dog shakes, tremor, ptosis, diarrhea, weight loss) were monitored for 30 min immediately after naloxone injection, and the experimenter was blind to the animal genotype. We used female mice from the R7BP line as no sex differences are observed in the high intensity opiate withdrawal paradigm we use.
Data Analysis
For hot plate analgesic tolerance, von Frey mechanical allodynia and opiate withdrawal behavioral assays, we used two-way ANOVA and Bonferroni post hoc tests for within groups comparisons whenever analysis revealed a genotype effect. For co-immunoprecipitation assays, we used one-way ANOVA followed by Dunnett's post hoc test. For western blot analysis, and acute hot plate assays we used Student's t-test.
RESULTS
Based on earlier findings demonstrating that RGS9-2 is a potent modulator of opiate actions (Zachariou et al, 2003) and on more recent evidence showing a modulatory role of R7BP on stability and cellular localization of R7 RGS proteins (Anderson et al, 2009), we hypothesized that R7BP affects multiple aspects of opiate actions in the brain.
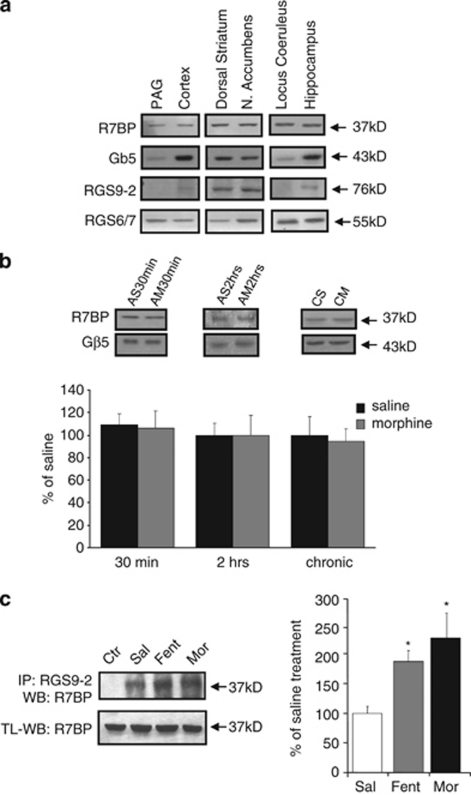
Figure 1a shows the expression of R7BP, Gβ5, RGS6/7, and RGS9-2 in various brain regions known to express μ-opioid receptors (MOPR). As shown in Figure 1a, R7BP is abundant in the dorsal striatum, nucleus accumbens and in brain regions such as the LC and the hippocampus, where RGS6/7 is enriched compared with Gβ5 and RGS9-2.
Figure 1.
Expression of R7 family members and interacting proteins in the mouse brain. (a) The expression of R7BP, Gβ5, RGS9-2, and RGS6/7 and in different brain regions of the mouse. Western blot analysis was also used to determine possible regulation of R7BP levels in the mouse striatum by acute (30 min or 2 h, n=4–5 per group) or repeated morphine application (once a day for 4 days, n=4 per group). As shown in (b), acute or repeated morphine treatments do not affect R7BP levels. Co-immunoprecipitation assays in the striatum demonstrate an increase in RGS9-2/R7BP complexes following acute fentanyl or morphine administration (c; n=3 per group). Striata from C57/Bl/6 mice treated with saline (sal) fentanyl (fent) or morphine (mor) for 30 min were immunoprecipitated with anti-RGS9-2 antibody and assayed with anti-R7BP antibody. Total lysates (TL) were assayed for R7BP levels. Data are expressed as mean±SEM, n=3 per group *p<0.05, one-way ANOVA followed by Dunnett's post hoc test.
We also used western blot analysis in order to investigate the way morphine treatment modulates R7BP levels in the striatum. We have previously reported that in this brain region, RGS9-2 levels are increased 2 h following morphine application (Psifogeorgou et al, 2007). Interestingly, the other components of this complex, R7BP and Gβ5 are not regulated by morphine (Figure 1b). R7BP and Gβ5 levels remain unchanged 30 min or 2 h post-morphine injections or following repeated morphine administration (once a day for 4 consecutive days). In this set of studies, we also used synaptosomal preparations, in order to examine if redistribution of R7BP after morphine application results in changes in the levels of the protein in synaptosomes. As shown in Supplementary Figure 1, morphine does not promote any changes in R7BP levels in striatal synaptosomes. Based on recent findings demonstrating robust changes in RGS9-2 protein interactions in the striatum following exposure to opiate agonists (Psifogeorgou et al, 2011), we hypothesized that morphine treatment affects the composition of R7BP complexes in this brain region. Our immunoprecipitation assays demonstrate that fentanyl and morphine enhance the association of RGS9-2 with R7BP in the striatum by 86±19% and 127±44.5%, respectively (Figure 1c; n=3 per group *p<0.05, one-way ANOVA followed by Dunnett's post hoc test and Supplementary Figure 2). Thus, although opiates do not affect R7BP levels, they promote R7BP/RGS9-2 protein interactions.
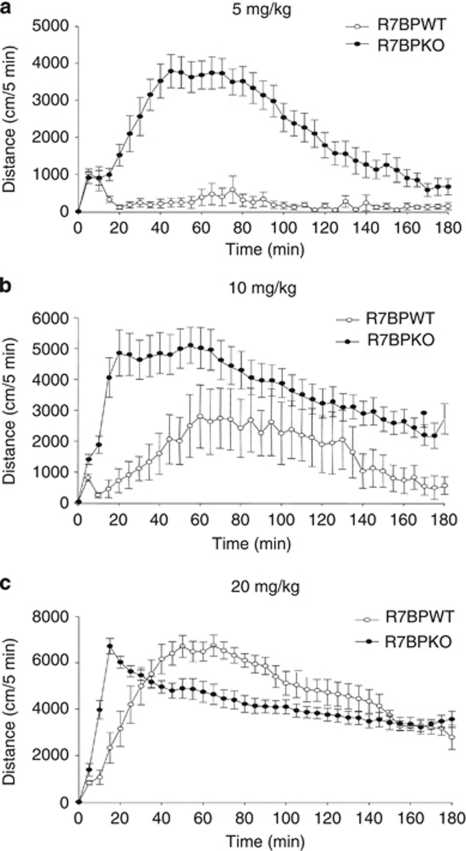
In the next set of studies, we examined the role of R7BP in behavioral responses to morphine using R7BPKO mice. Consistent with earlier findings, knockout of R7BP increases sensitivity to the locomotor activating actions of morphine, as mutant animals show locomotor activation at much lower doses than those required for their wild-type controls (Figure 2). A two-way ANOVA (time by genotype) revealed a significant effect of genotype (F(1,444)=549.718, p<0.001), a significant effect of time (F(36,444)=6.340, p<0.001) and a significant interaction between time and genotype (F(36,444)=5.635, p<0.001). Furthermore, at doses causing locomotor activation in both genotypes, the activation started significantly earlier in R7BPKO mice compared with their wild-type littermates (maximal response times to 10 mg/kg morphine were 20 min for R7BPKO and 60 min for R7BPWT, whereas responses to 20 mg/kg were 15 min for R7BPKO vs 45 min for R7BPWT mice). A two-way ANOVA (time by genotype) revealed a significant effect of genotype (10 mg/kg, F(1,444)=181.918, p<0.001; 20 mg/kg, F(1,407)=25.459, p<0.001) and a significant effect of time (10 mg/kg, F(36,444)=4.350, p<0.001; 20 mg/kg, F(36,407)=21.711, p<0.001). There was a significant interaction between time and genotype at 20 mg/kg dose (F(36,407)=7.309, p<0.001) but not at 10 mg/kg (F(36,444)=1.064, p=0.373). These findings suggest that genetic elimination of R7BP increases sensitivity to the locomotor activating actions of morphine.
Figure 2.
A role of R7BP in the locomotor activating actions of morphine. Knockout of R7BP increases sensitivity to the locomotor activating actions of morphine. In particular, R7BPKO mice show a significant locomotor activation at a dose of 5 mg/kg (a). Wild-type animals do not show activation at this dose (p<0.001 two-way ANOVA followed by Bonferroni test). At higher doses of morphine (10 and 20 mg/kg), locomotor activation starts much earlier in R7BPKO mice compared with their wild-type controls (b, c, p<0.001, followed by Bonferroni post hoc test). Data are expressed as mean±SEM, n=5–9 per group.
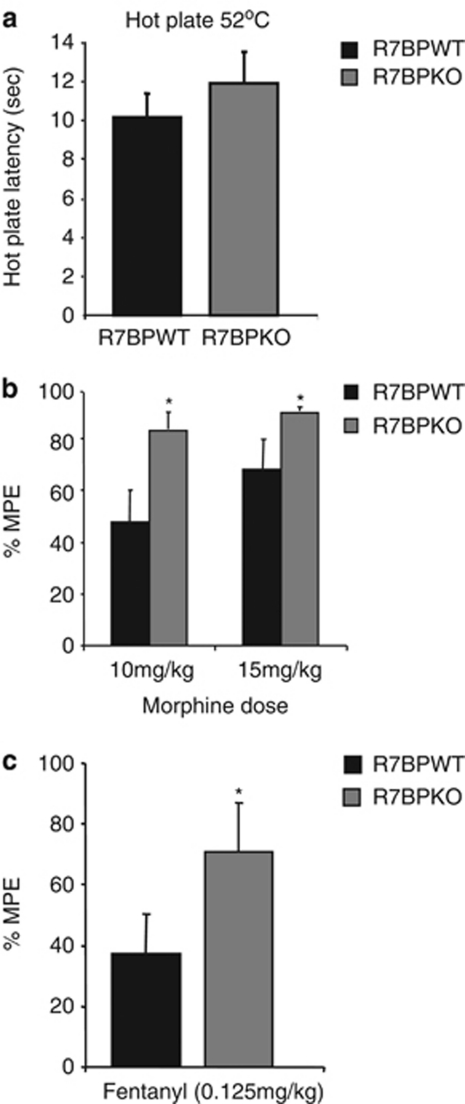
We next examined the role of R7BP in the analgesic actions of morphine in the hot plate assay. As shown in Figure 3, R7BPKO mice have normal thermal nociceptive thresholds at 52 °C (Figure 3a). As shown in the next graph, R7BPKO mice show a greater analgesic response to low morphine doses (10 and 15 mg/kg) than their wild-type controls (Figure 3b; *p<0.05, t-test). The onset or duration of the analgesic response was not affected by the deletion of the R7BP gene (not shown). A similar phenotype is observed following fentanyl administration (Figure 3c; *p<0.05, t-test).
Figure 3.
A role of R7BP in opiate analgesia. Knockout of R7BP does not affect paw withdrawal latencies in the hot plate test (a). R7BPKO mice show greater responses to morphine (10 and 15 mg/kg, n=10–11 per group) than their wild-type controls (b). R7BPKO mice show greater response than their wild-type controls to the analgesic actions of fentanyl (0.125 mg/kg, n=9–10 per group) in the hot plate assay (c). *p<0.05, t-test. Data are expressed as mean±SEM.
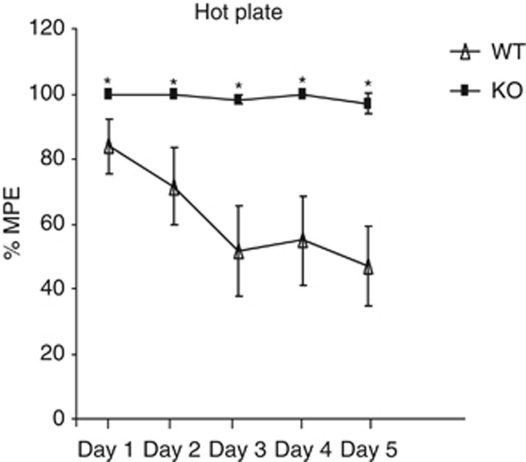
We also investigated the impact of R7BP on the chronic actions of morphine. Using the 52 °C hot plate paradigm, we monitored analgesic responses to morphine (20 mg/kg) once a day for 5 consecutive days. As shown in Figure 4, while wild-type animals show a reduced analgesic response to morphine by day 3 and complete tolerance by day 5, R7BPKO mice show maximal analgesic response until day 5. A two-way ANOVA analysis followed by the Bonferroni post hoc test revealed a significant genotype (F(1,68)=17.05, p<0.001), time (F(4,68)=9.149, p<0.0001), and interaction effect (F(4,68)=7.064, p<0.0001), n=8–11 per group.
Figure 4.
Blockade of R7BP actions prevents the development of morphine tolerance. Mice received 20 mg/kg morphine once a day for 5 consecutive days and 52 °C hot plate responses were monitored before and 30 min after morphine administration. R7BPKO mice show the same analgesic response throughout the study, whereas wild-type animals develop tolerance by the third day of morphine administration. Data are expressed as mean±SEM, *p<0.001, two-way ANOVA, followed by the Bonferroni post hoc test (n=8–11 per group).
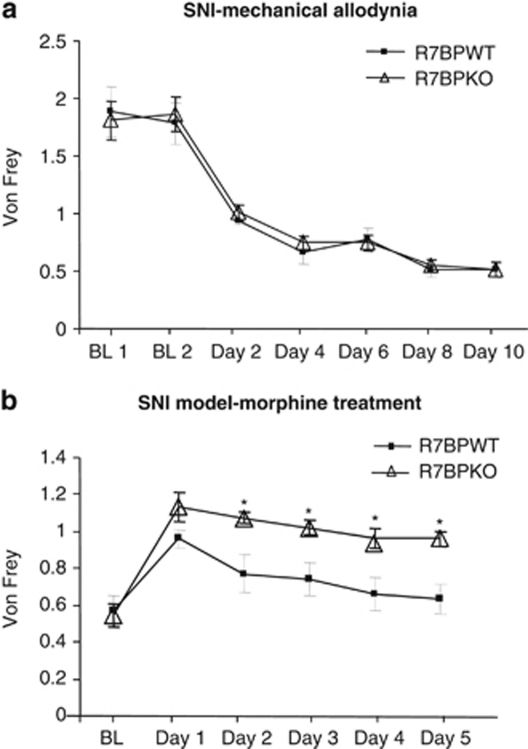
Since our studies so far revealed a modulatory role of R7BP in the antinociceptive actions of morphine in response to acute noxious stimuli, our next set of studies investigated the role of R7BP on morphine actions using chronic pain models. In particular, we used a neuropathic pain paradigm, the spared nerve injury model, and evaluated the antiallodynic effect of morphine in R7BPWT and mutant mice. First, we examined the influence of R7BP in neuropathic pain symptoms such as mechanical allodynia. Figure 5a demonstrates mechanical allodynia responses of the ipsilateral (left) hind-paw in the von Frey assay. As suggested by these data, mechanical nociceptive thresholds are not different between genotypes. The next set of studies evaluated the effect of morphine in mechanical allodynia responses in SNI groups of R7BPWT and R7BPKO mice. Morphine was administered for 5 consecutive days twice a day, in order to examine the effect of genotype on the rate of tolerance development. R7BPKO mice showed a higher response to the antiallodynic actions of morphine (3 mg/kg) during the first day of morphine application, but this effect was not statistically significant. R7BPWT mice showed a reduced response to morphine starting from the second day of the drug administration as a result of tolerance to the actions of morphine. R7BPKO appeared to develop tolerance to morphine at a much slower rate (Figure 5b). A two-way ANOVA followed by Bonferroni post hoc test revealed significant genotype (F(1,112)=18.78, p<0.001), time (F(7,112)=11.15, p<0.0001), and interaction effect (F(7,112)=2.563, p<0.05).
Figure 5.
Knockout of R7BP delays tolerance to the antiallodynic effects of morphine in neuropathic pain-suffering animals. We examined the influence of R7BP in the mechanical allodynia in the SNI model of neuropathic pain. (a) The von Frey responses of R7BPWT and R7BPKO mice following 1–10 days of spared nerve injury. Both wild-type and mutant animals develop the same degree of mechanical allodynia. Morphine (3 mg/kg s.c.) produces an antiallodynic effect on both genotypes, but responses of R7BPKOs are greater than those of their WT controls (n=7–11 per group). A delay in the development of morphine tolerance is also observed in R7BP mutants (b). Data are expressed as mean±SEM, *p<0.001 two-way ANOVA, followed by the Bonferroni post hoc test.
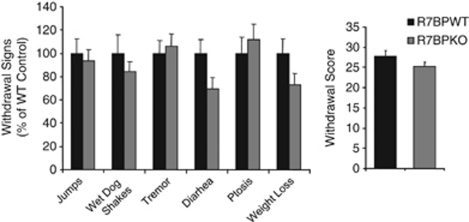
The last part of the study examined the influence of R7BP on morphine withdrawal. R7BP is present in several brain regions modulating opiate reward and dependence, including the LC, the hippocampus, and the striatum. Earlier work, revealed a preventive role of the R7 member RGS9-2 in morphine dependence (Zachariou et al, 2003). Based on this information, we hypothesized that regulation of R7 protein family members by R7BP in any of these brain regions could affect the expression of morphine withdrawal. We used a paradigm in which increasing doses of morphine were administered for 5 days, followed by an acute injection of the MOPR antagonist naloxone (1 mg/kg) 3 h after the last morphine injection to precipitate withdrawal. Several opiate withdrawal signs (jumps, wet dog shakes, tremor, diarrhea ptosis, and weight loss) were monitored for 30 min following naloxone administration. As shown in Figure 6, there were no significant differences between genotypes in opiate withdrawal behavior, although there was a trend for milder diarrhea and weight loss in R7BPKO mice. The overall withdrawal score for R7BPWT mice=27.7±1.5 and for R7BP mutants=25.2±1.2.
Figure 6.
R7BP does not affect morphine withdrawal. Mice received increasing morphine doses as described in Methods and withdrawal was precipitated using naloxone hydrochloride (1 mg/kg). Several withdrawal signs (jumps, wet dog shakes, tremor, diarrhea, and weight loss) were monitored for 30 min after naloxone administration. No significant difference was observed between genotypes, although there was a trend for decreased diarrhea and weigh loss in mutant mice. Data are expressed as mean±SEM, n=12–14 per group.
DISCUSSION
Our findings support a unique role of R7BP in morphine actions, as a negative modulator of morphine analgesia and a key component for the development of morphine tolerance. Our study provides novel information on the role of R7BP in opiate actions and suggests that R7 protein complexes have an essential role in modulation of opiate actions in the CNS. A number of recent studies reveal a dynamic role of several RGS family members in neuronal responses. Regarding opiate actions, RGS4 appears to have a potent role in modulation of opiate dependence in the LC (Han et al, 2010). Several recent studies also suggest that RGS9-2 complexes in the striatum modulate MOPR signaling and affect opiate analgesia, reward, and dependence (Zachariou et al, 2003; Psifogeorgou et al, 2011). To date there is no information about the role of other RGS proteins in opiate addiction. As shown by our western blot analysis studies, R7BP is distributed in several brain regions, which are also expressing other components of the R7 complexes, namely Gβ5, RGS9-2, RGS6, and RGS7. Our first set of studies explored the regulation of R7BP and R7BP complexes by opiates. We first monitored R7BP levels following acute or chronic morphine application. R7BP levels are not regulated by morphine in the striatum a brain region that we have observed an upregulation of RGS9-2 2 h after acute morphine application, and a downregulation of RGS9-2 following chronic morphine exposure (Zachariou et al, 2003; Psifogeorgou et al, 2007). Notably, Gβ5 levels also remain unchanged by acute or chronic opiate treatment (Psifogeorgou et al, 2007). This finding suggests that opiates produce specific alterations in the levels of RGS proteins participating in R7 complexes, but not in their interacting partners. Our immunoprecipitation studies provide evidence about the influence of opiate agonists on R7BP protein interactions, showing that activation of the MOPR by morphine or fentanyl promotes the formation of complexes between RGS9-2 and R7BP. These results suggest that association of RGS9-2 with R7BP following MOPR activation is essential for the regulation of MOPR regulation by RGS9-2. Recent studies from our group revealed that RGS9-2 acts as a negative modulator of morphine actions but a positive modulator of fentanyl effects (Psifogeorgou et al, 2011). Interestingly, R7BP is required for both positive and negative modulation of MOPR by RGS9-2, as R7BP/RGS9-2 complexes are formed by both fentanyl and morphine.
We predicted that R7BP has a prominent role in opiate dependence and analgesia based on its effects on the function of RGS9-2 and RGS6/7 in the brain. Earlier work revealed that genetic ablation of the R7BP gene modulates motor activity and psychostimulant responses as well as motor learning (Anderson et al, 2010). Our work extends these findings and shows that blockade of R7BP actions enhances opiate analgesia and prevents the development of morphine tolerance. Interestingly, the R7BP phenotype is not identical to the one observed in mice lacking RGS9, as blockade of RGS9-2 actions enhances morphine analgesia but also exacerbates morphine withdrawal, whereas genetic deletion of R7BP affects analgesia but not withdrawal. As R7BP interacts with RGS6/7 and RGS9-2 in the brain (Martemyanov et al, 2005), and it is present in the LC, we speculate that actions of R7 complexes in the LC oppose the actions of R7 complexes in the striatum and for this reason, the overall withdrawal expression does not change in R7BP mutants. Our study provides solid evidence on a role of R7BP protein in morphine tolerance, using several paradigms of morphine administration in pain-free as well as in chronic pain-suffering animals. The enhanced response to the analgesic actions of morphine and the prevention of analgesic tolerance are not a result of altered sensory processing in the mutant mice, as R7BPKO mice show normal hot plate latency and normal responses to the von Frey assay for mechanical allodynia. Naive, pain-free and neuropathic pain-suffering R7BP mutants show enhanced responses to opiate analgesics, compared with their wild-type controls. Notably, R7BP appears to have a more prominent role in the modulation of morphine antinociceptive actions in noxious thermal stimuli, as shown by hot plate assay data, compared with the antiallodynic actions of the drug. However, in both cases, R7BP complexes appear to have an essential role in morphine actions. Given the limited pharmacological tools available for the study of R7BP complexes, constitutive knockout mice provide an important tool for an initial evaluation of R7BP function in the brain. So far, our studies confirm that ablation of the factors contributing to R7 RGS proteins stability has major consequences for their stability and function, and provide the first evidence on the role of R7BP in morphine tolerance. Being aware of the limitations of constitutive knockout mice, we avoided using this line in reward paradigms involving learning and memory mechanisms: as R7BP is expressed in the hippocampus, and earlier data suggest that its interacting partner Gβ5 has a prominent role in hippocampal function (Xie et al, 2010), we avoided using the conditioned place preference or self-administration paradigms. Our future studies aim to the use of more elaborate genetic tools, which will permit brain region-specific manipulations of R7BP levels in adult mice, as it is obvious that R7BP complexes modulate MOPR (and other GPCR) function in several brain regions. It is also important to identify the composition of the exact R7BP/R7 protein complexes associated with morphine actions in different brain regions.
Several studies in the past have demonstrated a functional role of signal transduction molecules modulating MOPR signaling and desensitization. Molecules like RGS9-2 and spinophilin affect opiate reward, dependence, and analgesia (Zachariou et al, 2003; Charlton et al, 2008), whereas other signal transduction proteins appear to selectively influence some of the drug's actions: as mentioned earlier, RGS4 knockout mice show severe morphine dependence but no tolerance phenotypes (Han et al, 2010) and deletion of the β-arrestin-2 gene prevents analgesic tolerance to morphine but does not affect the development of morphine dependence (Bohn et al, 2004). These differences may be a result of tissue- or cell type-specific actions of signal transduction molecules or the fact that some of these proteins affect several GPCRs besides MOPR. Clearly, there are several factors that contribute to the development of morphine tolerance and brain region-specific cellular adaptations associated with MOPR desensitization. The presence of R7BP in all brain regions expressing MOPR suggests that this protein modulates MOPR function in various networks, perhaps via interactions with RGS9-2 and RGS6/7. We speculate that R7BP actions in the brain reward center, in the hippocampus and possibly in the spinal cord modulate the different actions of opiates.
Our findings provide a better understanding on the mechanisms underlying the acute and chronic actions of opiates. Elucidating the cellular mechanism of opiate actions is essential for the more efficient use of the currently available opiate analgesics, but also for the development of novel drug targets. In the case of R7BP, a potent modulator of MOPR actions has a prominent role in the development of analgesic tolerance but does not affect opiate withdrawal. Moreover, R7BP may be a more attractive drug target than the individual R7 RGS proteins because of its wide distribution: thus, prevention of R7BP actions does not only affect RGS9-2 function in the striatum, but also the function of R6/7 proteins expressed in many other brain regions implicated in addiction and analgesia. These properties of R7BP make it an attractive pharmacological target and suggest that the development of agents preventing specific protein interactions may offer a powerful new tool for the management of certain forms of pain.
Acknowledgments
This study was supported by the Greek Secretariat for research and technology (PENED03), the Manassaki Fellowship Foundation (DT), and National Institutes of Health DA021743 and DA026405 to KAM.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- Anderson GR, Cao Y, Davidson S, Truong HV, Pravetoni M, Thomas MJ, et al. R7BP complexes with RGS9-2 and RGS7 in the striatum differentially control motor learning and locomotor responses to cocaine. Neuropsychopharmacology. 2010;35:1040–1050. doi: 10.1038/npp.2009.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GR, Lujan R, Martemyanov KA. Changes in striatal signaling induce remodeling of RGS complexes containing Gbeta5 and R7BP subunits. Mol Cell Biol. 2009;29:3033–3044. doi: 10.1128/MCB.01449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GR, Lujan R, Semenov A, Pravetoni M, Posokhova EN, Song JH, et al. Expression and localization of RGS9-2/G 5/R7BP complex in vivo is set by dynamic control of its constitutive degradation by cellular cysteine proteases. J Neurosci. 2007;27:14117–14127. doi: 10.1523/JNEUROSCI.3884-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballon DR, Flanary PL, Gladue DP, Konopka JB, Dohlman HG, Thorner J. DEP-domain-mediated regulation of GPCR signaling responses. Cell. 2006;126:1079–1093. doi: 10.1016/j.cell.2006.07.030. [DOI] [PubMed] [Google Scholar]
- Berman DM, Gilman AG. Mammalian RGS proteins: Barbarians at the gates. J Biol Chem. 1998;273:1269–1272. doi: 10.1074/jbc.273.3.1269. [DOI] [PubMed] [Google Scholar]
- Bohn LM, Dykstra LA, Lefkowitz RJ, Caron MG, Barak LS. Relative efficacy is determined by the complements of the G protein-coupled receptor desensitization machinery. Mol Pharmacol. 2004;66:106–112. doi: 10.1124/mol.66.1.106. [DOI] [PubMed] [Google Scholar]
- Bohn LM, Lefkowitz RJ, Gainetdinov RR, Peppel K, Caron MG, Lin FT. Enhanced morphine analgesia in mice lacking β-arrestin 2. Science. 1999;286:2495–2498. doi: 10.1126/science.286.5449.2495. [DOI] [PubMed] [Google Scholar]
- Cabrera-Vera TM, Hernandez S, Earls LR, Medkova M, Sundgren-Andersson AK, Surmeier JD, et al. RGS9-2 modulates D2 dopamine receptor-mediated Ca2_ channel inhibition in rat striatal cholinergic interneurons. Proc Natl Acad Sci. 2004;101:16339–16344. doi: 10.1073/pnas.0407416101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton JJ, Allen BP, Psifogeorgou K, Chakravarty S, Gomes I, Neve RL, et al. Multiple actions of spinophilin modulate mu opioid receptor function. Neuron. 2008;58:238–247. doi: 10.1016/j.neuron.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever ML, Snyder JT, Gershnurg S, Siderovski DP, Harden KT, Sondek J. Crystal structure of the multifunctional RGS9-2-Gb5 complex. Nat Strct Mol Biol. 2008;15:155–162. doi: 10.1038/nsmb.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Eversole-Cire P, Zhang H, Mancino V, Chen YJ, He W, et al. Instability of GGL domain containing RGS proteins in mice lacking the G protein beta subunit Gβ5. Proc Natl Acad Sci. 2003;100:6604–6609. doi: 10.1073/pnas.0631825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Geis C, Sommer C. Activation of TRPV1 contributes to morphine tolerance: involvement of the mitogen-activated protein kinase signaling pathway. J Neurosci. 2008;28:5836–5845. doi: 10.1523/JNEUROSCI.4170-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman HG, Thorner J. RGS proteins and signaling by heterotrimeric G proteins. J Biol Chem. 1997;272:3871–3874. doi: 10.1074/jbc.272.7.3871. [DOI] [PubMed] [Google Scholar]
- Drenan RM, Doupnik CA, Boyle MP, Muglia LJ, Huettner JE, Linder ME, et al. Palmitoylation regulates plasma membrane-nuclear shuttling of R7BP, a novel membrane anchor for the RGS7 family. J Cell Biol. 2005;169:623–633. doi: 10.1083/jcb.200502007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold SJ, Hoang CV, Potts BW, Porras G, Pioli E, Kim KW, et al. RGS9-2 negatively modulates L-3,4-dihydroxyphenylalanine-induced dyskinesia in experimental Parkinson's disease. J Neurosci. 2007;27:14338–14348. doi: 10.1523/JNEUROSCI.4223-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold SJ, Ni YG, Dohlman HG, Nestler EJ. Regulators of G-protein signaling (RGS) proteins: region-specific expression of nine subtypes in rat brain. J Neurosci. 1997;17:8024–8037. doi: 10.1523/JNEUROSCI.17-20-08024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MH, Renthal W, Ring RH, Rahman Z, Psifogeorgou K, Howland D, et al. Brain region specific actions of regulator of G protein signaling 4 oppose morphine reward and dependence but promote analgesia. Biol Psychiatry. 2010;67:761–769. doi: 10.1016/j.biopsych.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Lu L, Zhang X, El-Hodiri HM, Chen CK, Slep KC, et al. Modules in the photoreceptor RGS9-1.Gbeta 5L GTPase-accelerating protein complex control effector coupling, GTPase acceleration, protein folding, and stability. J Biol Chem. 2000;275:37093–37100. doi: 10.1074/jbc.M006982200. [DOI] [PubMed] [Google Scholar]
- Hooks SB, Waldo GL, Corbitt ET, Krumins AM, Harden TK. RGS6, RGS7, RGS9 and RGS11 stimulate GTPase activity of Gi family of G proteins with differential selectivity and maximal activity. J Biol Chem. 2003;278:10087–10093. doi: 10.1074/jbc.M211382200. [DOI] [PubMed] [Google Scholar]
- Hu G, Wensel TG. R9AP, a membrane anchor for the photoreceptor GTPase accelerating protein, RGS9-1. Proc Natl Acad Sci. 2002;99:9755–9760. doi: 10.1073/pnas.152094799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inturrisi CE. Clinical pharmacology of opioids for pain. Clin J Pain. 2002;18 (4 Suppl:S3–13. doi: 10.1097/00002508-200207001-00002. [DOI] [PubMed] [Google Scholar]
- Jayaraman M, Zhou H, Jia L, Cain MD, Blumer KJ. R9AP and R7BP: traffic cops for the RGS7 family in phototransduction and neuronal GPCR signaling. Trends Pharmacol Sci. 2009;30:17–24. doi: 10.1016/j.tips.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovoor A, Seyffarth P, Ebert J, Barghshoon S, Chen CK, Schwarz S, et al. D2 dopamine receptors colocalize regulator of G protein signalling 9-2 via the RGS9 DEP domain, and RGS9 knockout mice develop dyskinesias associated with dopamine pathways. J Neurosci. 2005;280:5133–5136. doi: 10.1523/JNEUROSCI.2840-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreek MJ. Drug addictions. Molecular and cellular endpoints. Ann NY Acad Sci. 2001;937:27–49. [PubMed] [Google Scholar]
- Martemyanov KA, Yoo PJ, Skiba NP, Arshavsky VY. R7BP, a novel neuronal protein interacting with RGS proteins and the R7 subfamily. J Biol Chem. 2005;280:5133–5136. doi: 10.1074/jbc.C400596200. [DOI] [PubMed] [Google Scholar]
- Martemyanov KA, Lishko PV, Calero N, Keresztes G, Solokow M, Strissel KJ, et al. The DEP domain determines subcellular targeting of the GTPase activating protein RGS9 in vivo. J Neurosci. 2003;23:10175–10181. doi: 10.1523/JNEUROSCI.23-32-10175.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melief EJ, Miyatake M, Bruchas MR, Chavkin C. Ligand-directed c-Jun N-terminal kinase activation disrupts opioid receptor signaling. Proc Natl Acad Sci USA. 2010;107:11608–11613. doi: 10.1073/pnas.1000751107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moron JA, Zakharova M, Ferrer JE, Merrill GA, Hope B, Lafer EM, et al. Mitogen-activated protein kinase regulates dopamine transporter surface expression and dopamine transport capacity. J Neurosci. 2003;23:8480–8488. doi: 10.1523/JNEUROSCI.23-24-08480.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psifogeorgou K, Papakosta V, Gold SJ, Kardassis D, Zachariou V. RGS9-2 is a negative modulator of mu opioid receptor function. J Neurochem. 2007;103:617–625. doi: 10.1111/j.1471-4159.2007.04812.x. [DOI] [PubMed] [Google Scholar]
- Psifogeorgou K, Terzi D, Papachatzaki MM, Varidaki A, Ferguson D, Gold SJ, et al. A unique role of RGS9-2 in the striatum as a positive or negative modulator of opiate actions. J Neurosci. 2011;31:5617–5624. doi: 10.1523/JNEUROSCI.4146-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman Z, Schwarz J, Gold SJ, Zachariou V, Wein MN, Choi KH, et al. RGS9 modulates dopamine signaling in the basal ganglia. Neuron. 2003;38:941–952. doi: 10.1016/s0896-6273(03)00321-0. [DOI] [PubMed] [Google Scholar]
- Sondek J, Siderovski JP. G-γ like domains: new frontiers in G protein signalling and β-propeller scaffolding. Biochem Pharmacol. 2001;61:1329–1337. doi: 10.1016/s0006-2952(01)00633-5. [DOI] [PubMed] [Google Scholar]
- Terzi D, Stergiou E, King SL, Zachariou V. RGS proteins in neuropsychiatric disorders. Progress Mol Biol Trans Sci. 2009;86C:299–333. doi: 10.1016/S1877-1173(09)86010-9. [DOI] [PubMed] [Google Scholar]
- Traynor JR, Terzi D, Calbarone BJ, Zachariou V. RGS9-2: probing an intracellular modulator of behaviour as a drug target. Trends Pharmacol Sci. 2009;30:105–111. doi: 10.1016/j.tips.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Allen KL, Kourrich S, Colón-Saez J, Thomas MJ, Wickman K, et al. Gbeta5 recruits R7 RGS proteins to GIRK channels to regulate the timing of neuronal inhibitory signaling. Nat Neurosci. 2010;13:661–663. doi: 10.1038/nn.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariou V, Georgescu D, Sanchez N, Rahman Z, DiLeone R, Berton O, et al. Essential role for RGS9 in opiate action. Proc Natl Acad Sci. 2003;100:13656–13661. doi: 10.1073/pnas.2232594100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.