Abstract
Several lines of evidence indicate that the diacylglycerol kinase eta (DGKH) gene is implicated in the etiology of bipolar disorder (BD). However, the functional neural mechanisms of DGKH's risk association remain unknown. Therefore, we examined the effects of three haplotype-tagging risk variants in DGKH (single nucleotide polymorphisms rs9315885, rs1012053, and rs1170191) on brain activation using a verbal fluency functional magnetic resonance imaging task. The subject groups consisted of young individuals at high familial risk of BD (n=81) and a comparison group of healthy controls (n=75). Individuals were grouped based on risk haplotypes described in previous studies. There was a significant risk haplotype*group interaction in the left medial frontal gyrus (BA10, involving anterior cingulate BA32), left precuneus, and right parahippocampal gyrus. All regions demonstrated greater activation during the baseline condition than sentence completion. Individuals at high familial risk for BD homozygous for the DGKH risk haplotype demonstrated relatively greater activation (poor suppression) of these regions during the task vs the low-risk haplotype subjects. The reverse pattern was seen for the control subjects. These findings suggest that there are differential effects of the DGKH gene in healthy controls vs the bipolar high-risk group, which manifests as a failure to disengage default-mode regions in those at familial risk carrying the risk haplotype.
Keywords: bipolar disorder, fMRI, DGKH, default-mode, high risk, lithium
INTRODUCTION
Bipolar disorder (BD) is a highly debilitating condition marked by recurrent episodes of depression and mania. Epidemiological research has shown that BD has a strong genetic component, with heritability estimates ranging from 59 to 93% (Bertelsen et al, 1977; McGuffin et al, 2003). Genetic linkage and candidate gene association studies indicate that BD is likely to be a complex polygenetic disorder with individual genes conferring small influences to overall risk of disorder (Barnett and Smoller, 2009).
One of the first susceptibility genes for BD to be identified by the genome-wide association study (GWAS) approach in two independent samples of European origin was diacylglycerol kinase eta (DGKH) (Baum et al, 2008). Association was found for three single nucleotide polymorphisms (SNPs) located in the first intron of the DGKH gene (rs9315885, rs1012053, and rs1170191). Although some studies have not confirmed this association (Sklar et al, 2008; Tesli et al, 2009; Yosifova et al, 2009), numerous lines of evidence indicate that DGKH might be involved in the etiology of the disorder. First of all, DGKH is located in the region 13q14 that has been linked to BD in previous studies (Badner and Gershon, 2002; Detera-Wadleigh and McMahon, 2006). Further, an association of DGKH with BD has been replicated in a Sardinian as well as a Chinese sample at the haplotype level (Zeng et al, 2011; Squassina et al, 2009). A recent study also reported an association of DGKH with BD, as well as unipolar depression and adult attention deficits/hyperactivity disorder (Weber et al, 2011). In addition, SNP rs9315885 has been associated with BD in a Finnish family cohort (Ollila et al, 2009), and the GWAS carried out by the Wellcome Trust Case Control Consortium (The Wellcome Trust Case Control Consortium, 2007) indicated association for several SNPs near and within DGKH (Manchia et al, 2009; Robbins and Arnsten, 2009). Finally, gene expression level of DGKH in the prefrontal cortex has been reported to be significantly increased in BD patients (Moya et al, 2010). The DGKH gene is also of particular interest as it encodes the η isoform of diacylglycerol kinase that is involved in the phosphoinositol pathway through which lithium, one of the most effective pharmacological treatments for BD, is thought to exert its therapeutic effects (Manji and Lenox, 1999).
Together, these findings suggest the involvement of the DGKH gene in the pathophysiology of BD. To the best of our knowledge, however, no study has examined association between genetic variation in the DGKH gene on brain activation. Therefore, we investigated the effects of three SNPs of the DGKH gene that have been associated with BD (Baum et al, 2008) on brain activation during a functional magnetic resonance imaging (fMRI) paradigm in healthy controls and a group of individuals at high familial risk of the disorder. The advantage of studying brain activation patterns in individuals who are unaffected by BD themselves but have a close relative suffering from the disorder is that findings are likely to reflect the influence of shared risk genes for the disorder, excluding confounding effects of chronic illness or medication.
We examined the effects of the DGKH haplotype in those at familial risk for the disorder in relation to healthy controls. We hypothesized that as DGKH has been associated with risk for BD, the effects of the low-risk vs high-risk haplotype may differ in those who have an already present genetic loading for the disorder vs those without. As there is limited literature regarding this particular susceptibility gene in terms of regional brain activation, we hypothesized that these differences would occur in regions previously implicated in the disorder, specifically the prefrontal cortex. We employed the verbal initiation section of the Hayling Sentence Completion Test (Burgess and Shallice, 1996) involving verbal fluency and word production to test this hypothesis. The task was chosen as it has been shown to differentiate BD patients and those at increased familial risk for the disorder from healthy controls with respect to differing brain activation patterns (McIntosh et al, 2008b; Whalley et al, 2011). It has also been found to activate left superior and middle prefrontal regions (Allen et al, 2010), anterior cingulate areas (Nathaniel-James et al, 1997), as well as the striatum (McIntosh et al, 2008b), which are considered to be involved in the pathophysiology of BD (Strakowski et al, 2005; Cerullo et al, 2009). Moreover, it probes neuropsychological deficits in verbal initiation and verbal fluency commonly observed in BD (Arts et al, 2008).
MATERIALS AND METHODS
Subjects
Individuals at high genetic risk of BD I (BDI) because of a close family history of the disorder and control subjects were recruited as part of the Scottish Bipolar Family Study that has been described in detail elsewhere (Sprooten et al, 2011; Whalley et al, 2011). To identify high-risk participants, caseloads of psychiatrists across Scotland were searched for individuals diagnosed with BDI. Diagnoses were confirmed with the Structural Clinical Interview for DSM-IV-TR Axis I Disorders (SCID-I) (First et al, 2002) or the symptom checklist of the Operational Criteria (McGuffin et al, 1991). Subsequently, subjects affected by BDI were asked to identify a first- or second-degree relative aged 16–25 years not suffering from the disorder. These unaffected individuals were invited to participate in this study providing that they had at least one first-degree, or two second-degree relatives suffering from BDI. It should be noted, owing to the age of the subjects, the high-risk cohort will still be within the risk period for development of the disorder, hence this group will likely contain those at high risk who will remain well and those who will subsequently become unwell. The results should be considered in this light. Control subjects with no personal history of BD or family history of a mood disorder in first-degree relatives were identified from the social networks of the bipolar high-risk subjects. Only unrelated individuals were included in the current analysis. Exclusion criteria for all groups included a personal history of major depression, mania or hypomania, psychosis, or any major neurological or psychiatric disorder, a history of substance dependence, a history of learning disability or any history of head injury that included loss of consciousness, and any contraindications to fMRI. A total of 81 bipolar high-risk subjects and 75 controls provided suitable fMRI data along with genetic information. All participants provided written informed consent, and the study was approved by the multicentre research ethics committee for Scotland.
Genotyping
Genomic DNA was extracted from venous blood samples. The genotyping was conducted by the Wellcome Trust Clinical Research Facility, Edinburgh, UK (www.wtcrf.ed.ac.uk), and used standard TaqMan assays, by the TaqMan PCR-based method (TaqMan, AssayByDesign, Applied Biosystems, Foster City, California). Subjects were typed at SNPs rs9315885, rs1012053, and rs1170191 that have been previously shown to be associated with BDI (Baum et al, 2008). As there was evidence of linkage disequilibrium (LD) between the SNPs (rs9315885 and rs1012053 D'=0.999, p<0.001; rs9315885 and rs1170191 D'=0.642, p<0.001), we studied the effects of genetic variation of the SNPs combined as a three-marker haplotype in line with a previous study (Squassina et al, 2009). In this previous study, the most common haplotype was T-A-C (for rs9315885, rs1012053, and rs1170191, respectively). This haplotype was the only one that showed significant association with BD, remaining significant after permutation testing (Squassina et al, 2009). Subjects in the current study were therefore grouped according to the presence of this presumed risk haplotype (T-A-C). If they were homozygous for this risk haplotype they are referred to as ‘RISK+', if they were not carriers of the risk haplotype or heterozygous they are referred to as ‘RISK−'. Differences from the Hardy–Weinberg equilibrium (HWE) and assessments of linkage disequilibrium were performed using the genetics package in ‘R' (version 2.12.1, the R Foundation for Statistical Computing).
Clinical Assessments
All participants were interviewed by one of the two experienced psychiatrists (AMM and JES) using the SCID (First et al, 2002) to confirm the absence of any lifetime axis I disorders. Current manic and depressive symptoms were rated using the Young Mania Rating Scale (Young et al, 1978) and Hamilton Rating Scale for Depression (HAM-D) (Hamilton 1960). Estimates of temperamental variations in minor affective symptoms were assessed using the Temperament Evaluation of Memphis, Pisa, Paris and San Diego Autoquestionnaire (TEMPS-A) (Akiskal et al, 2005), a validated self-rated questionnaire that provides measures of cyclothymic, depressive, hyperthymic, irritable, and anxious temperament. Statistical analysis of demographic characteristics was conducted using independent t-tests or χ2 tests. For the clinical assessments and measures of temperament, comparison of groups was conducted using Mann–Whitney U-tests.
Experimental Paradigm
Subjects performed the verbal initiation section of the Hayling Sentence Completion Test (Burgess and Shallice, 1997) in the scanner (Whalley et al, 2004). This task is considered an extension of the verbal fluency task, where constraint is based on sentence context rather than letter or semantic category. Briefly, subjects were shown sentences with the last word missing and asked to silently think of an appropriate word to complete the sentence and press a button when they had done so. The task has four levels of difficulty, according to the range of suitable completion words suggested by the sentence context. This design allows both a standard subtraction analysis (sentence completion vs baseline) and the more constrained parametric analysis (examining areas of increasing activation with increasing task difficulty). Sentences were presented in blocks of fixed difficulty. Each block lasted 40 s and included eight sentences. Sentences were presented for a period of 3 s followed by a fixation cross for 2 s. The baseline condition consisted of viewing a screen of white circles on a black background for 40 s. The order of the blocks was pseudo-random, and each block was repeated four times using different sentences. Standardized verbal instructions were given before scanning.
Immediately after scanning, subjects were given the same sequence of sentences on paper and requested to complete each sentence with the word they first thought of in the scanner. ‘Word appropriateness' scores were determined from the word frequency list of sentence completion norms (Bloom and Fischler, 1980). A score of one was given to the most frequently produced word in the word frequency list, a score of two for the next most frequently produced word, etc.
Scanning Procedure
Imaging was carried out at the Brain Imaging Research Centre (BIRC) for Scotland on a GE 1.5. T Signa scanner (GE Medical, Milwaukee, WI, USA). The functional imaging protocol consisted of axial gradient-echo planar images (EPI) (TR/TE=2000/40 ms; matrix=64 × 64; field of view (fov)=24 cm) acquired continually during the experimental paradigm. Twenty-seven contiguous 5 mm slices were acquired within each TR. Each EPI acquisition was run for 404 volumes, the first four of which were discarded. The T1 sequence yielded 180 contiguous 1.2 mm coronal slices (matrix=192 × 192; fov=24 cm; flip angle=8°). Visual stimuli were presented using a screen (IFIS, MRI Devices, Waukesha, WI, USA) placed in the bore of the magnet.
Image Processing and Analysis
The EPI and T1 images were reconstructed into nifti format (Mayo Foundation, Rochester, MN, USA) using DICOM convert functions available in SPM5 (Statistical Parametric Mapping: The Wellcome Department of Cognitive Neurology and collaborators, Institute of Neurology, London) running in Matlab (The MathWorks, Natick, MA, USA). Images were preprocessed using standard protocols available in SPM5. All EPI images were realigned to the mean volume in the series. The functional images were then normalized according to the standard co-registration procedures using the individual's structural scan. Finally, all realigned and normalized images were smoothed with an 8 × 8 × 8 mm full-width half-maximum Gaussian filter.
First-level statistical analysis was performed using the general linear model approach. At the individual subject level, the data were modeled with four conditions corresponding to the four difficulty levels each modeled by a boxcar convolved with a synthetic hemodynamic response function. Estimates of the subject's movement during the scan were entered as ‘covariates of no interest'. Before fitting the model, the participants data were filtered in the time domain using high pass filter (128 s cutoff), and serial correlations were accounted for by using the autoregressive (AR(1)) model. Contrasts were constructed to examine all four sentence completion conditions vs baseline, and areas of increasing activation with increasing task difficulty (the parametric contrast).
Second-Level Analysis
All second-level statistical analyses were conducted in SPM5. In order to directly test our hypothesis of differential effects of the risk haplotype between the controls and bipolar high-risk groups, our approach was to examine regions where there was a significant genotype*group interaction. This was followed by analyzing pair-wise comparisons of the genotype groups within the controls and high risk separately to explore the origin of these interaction effects. For each contrast of interest (sentence completion vs baseline and parametric effects), one contrast image per subject was entered into a second-level random effects analysis. Haplotype*group interactions were examined using a full factorial ANOVA model comparing the bipolar high-risk group against the healthy controls. Haplotype and group were entered as two factors in the design matrix with two levels of genotype (risk haplotype homozygotes vs the remainder) and two diagnostic groups (healthy controls vs individuals at high risk of BD). Where significant interactions were found, pair-wise group comparisons were explored and the effect of haplotype within each diagnostic group was examined.
Statistical maps were thresholded at a level of p<0.005 (uncorrected), and regions were considered significant at a cluster level of p<0.05, corrected for multiple comparisons. All coordinates are quoted in Montreal Neurological Institute (MNI) convention (http://www.mni.mcgill.ca), and images are overlaid onto standard brain in MNI space using Mango software package (http://ric.uthscsa.edu/mango). Based on our previous hypothesis, small volume corrections were applied for the prefrontal cortex, created using the WFU PickAtlas (Tzourio-Mazoyer et al, 2002; Maldjian et al, 2003).
RESULTS
Demographic, Clinical, and Behavioral Measures
Controls
Thirty control subjects were homozygous for the risk haplotype (T-A-C), referred to as RISK+. Of the remainder (n=45), referred to as RISK−, 38 individuals carried one copy of the (T-A-C) haplotype.
Bipolar high risk
For the bipolar high-risk group, 35 subjects were homozygous for the risk haplotype. Of the remainder (n=46), 39 individuals carried one copy of the (T-A-C) haplotype.
For each of the SNPs individually, none of the allele frequencies differed from HWE, and the haplotype frequencies did not differ between the groups (χ2=0.16, p=0.69). Measures of linkage disequilibrium are provided in the Materials and Methods. Further details describing the individual haplotype frequencies are presented in Supplementary Table. These frequencies were consistent with those reported previously (Squassina et al, 2009).
Demographic details are presented in Table 1. There were no significant differences between the haplotype groups or between high-risk subjects and controls in terms of age, gender, handedness, or verbal intelligence as estimated with the National Adult Reading Test (Nelson 1982).
Table 1. Demographic, Clinical, and Behavioral Characteristics.
|
Controls (n=75) |
Bipolar high-risk subjects (n=81) |
Between-group comparison | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RISK− (n=45) | RISK+ (n=30) | Within-group comparison | RISK− (n=46) | RISK+ (n=35) | Within-group comparison | |||||
| Demographics | ||||||||||
| |
Mean/n (SD) |
Mean/n (SD) |
T/χ2 |
p |
Mean/n (SD) |
Mean/n (SD) |
T/χ2 |
p |
T/χ2 |
p |
| Age (years) | 20.99 (2.35) | 20.53 (2.50) | 0.81 | 0.42 | 21.00 (2.52) | 21.75 (3.08) | 1.21 | 0.23 | 1.23 | 0.22 |
| Gender (male:female) | 20 : 25 | 13 : 17 | 0.01 | 0.92 | 21 : 25 | 19 : 16 | 0.59 | 0.44 | 0.45 | 0.50 |
| Handedness (right:other) | 41 : 1 | 27 : 3 | 2.16 | 0.14 | 42 : 4 | 31 : 4 | 1.33 | 0.51 | 1.62 | 0.45 |
| Verbal IQ (NART) | 110.49 (5.91) | 109.67 (9.41) | 0.46 | 0.64 | 109.24 (7.25) | 111.60 (6.17) | 1.55 | 0.13 | 0.09 | 0.93 |
| Clinical characteristics | ||||||||||
| |
Median (IQR) |
Median (IQR) |
U |
p |
Median (IQR) |
Median (IQR) |
U |
p |
U |
P |
| YMRS sum score | 0 (0) | 0 (0) | 0.12 | 0.90 | 0 (0) | 0 (0) | 0.19 | 0.19 | 1.67 | 0.10 |
| HAM-D sum score | 0 (1) | 0 (1) | 1.33 | 0.18 | 0 (2) | 1 (2) | 1.54 | 0.12 | 3.72 | <0.01 |
| Temperament characteristics (TEMPS-A) | ||||||||||
| |
Median (IQR) |
Median (IQR) |
U |
p |
Median (IQR) |
Median (IQR) |
U |
p |
U |
p |
| Cyclothymia | 1.00 (2.75) | 1.00 (2.75) | 0.02 | 0.99 | 2.00 (4.00) | 2.00 (3.00) | 0.01 | 0.99 | 2.91 | <0.01 |
| Depressive | 0.00 (2.00) | 0.00 (1.00) | 0.16 | 0.87 | 0.00 (2.00) | 0.00 (1.00) | 0.20 | 0.85 | 0.95 | 0.34 |
| Irritability | 1.00(2.00) | 1.00 (2.00) | 0.39 | 0.69 | 1.00 (1.00) | 1.00 (2.00) | 1.54 | 0.12 | 1.70 | 0.09 |
| Hyperthymia | 2.00 (2.00) | 1.50 (4.00) | 0.48 | 0.64 | 2.00 (2.00) | 1.00 (3.50) | 0.72 | 0.47 | 0.55 | 0.58 |
| Anxious | 0.50 (1.00) | 0.50 (2.00) | 0.09 | 0.93 | 0.00 (1.00) | 1.00 (2.00) | 0.99 | 0.32 | 0.17 | 0.86 |
| Behavioral characteristics (Hayling sentence completion test) | ||||||||||
| |
Mean (SD) |
Mean (SD) |
T |
p |
Mean (SD) |
Mean (SD) |
T |
p |
T |
p |
| Reaction time (s) | 2466 (584) | 2440 (543) | 0.71 | 0.81 | 2505 (679) | 2471 (543) | 0.25 | 0.81 | 0.33 | 0.74 |
| Word Appro-priateness | 3.01 (0.51) | 3.02 (0.64) | 0.17 | 0.96 | 2.86 (0.561) | 2.91 (0.56) | 0.40 | 0.69 | 1.25 | 0.22 |
Abbreviations: HAM-D, Hamilton Rating Scale for Depression; IQR, interquartile range; NART, National Adult Reading Test; RISK−, non-risk haplotype carriers; RISK+, risk haplotype homozygotes; TEMPS-A, Temperament Evaluation of Memphis, Pisa, Paris and San Diego Autoquestionnaire; YMRS, Young Mania Rating Scale.
For the clinical measures, there were significant differences between controls and high-risk subjects on measures of depression and cyclothymia as measured with the HAM-D (Hamilton 1960) and a short version of the TEMPS-A (Akiskal et al, 2005), respectively with the high-risk subjects having higher scores on both measures. These findings have been described previously in this sample (Sprooten et al, 2011; Whalley et al, 2011). There were, however, no significant differences within the groups based on haplotype.
Statistical analyses revealed no significant differences in Hayling Sentence Completion Test reaction time or word appropriateness scores between the two haplotype groups or between the controls and high-risk subjects.
Task-Related Brain Activation Patterns
All subjects demonstrated the expected patterns of brain activation and behavioral responses indicating subjects were performing the tasks appropriately in the scanner (see Supplementary Figure) (Whalley et al, 2004; McIntosh et al, 2008a; Whalley et al, 2011). Regions activated across the groups for the sentence completion vs baseline contrast included the left medial and lateral prefrontal regions, left lateral temporal cortex, subcortical structures, left lateral parietal cortex, occipital lobes bilaterally, and right cerebellum. For the parametric contrast, all groups demonstrated activation in similar areas including left lateral and medial prefrontal cortex, left lateral temporal cortex, and right cerebellum. Regions of reduced activation during the task (greater activation during baseline) involved anterior medial prefrontal regions, precuneus, posterior cingulate cortex, and parahippocampal regions.
Haplotype-Dependent Differences in Brain Activation
Sentence completion vs baseline
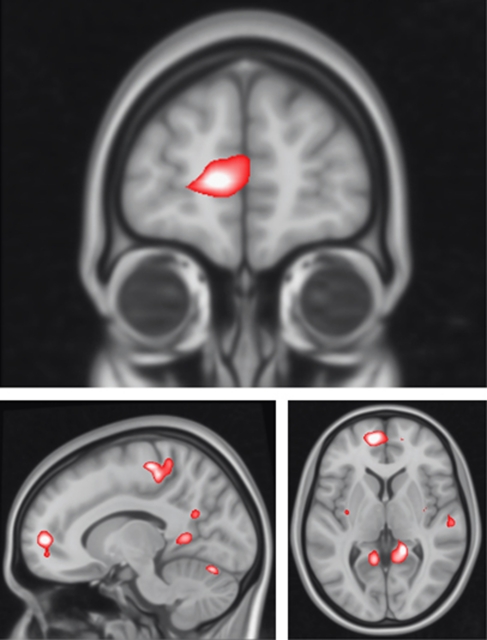
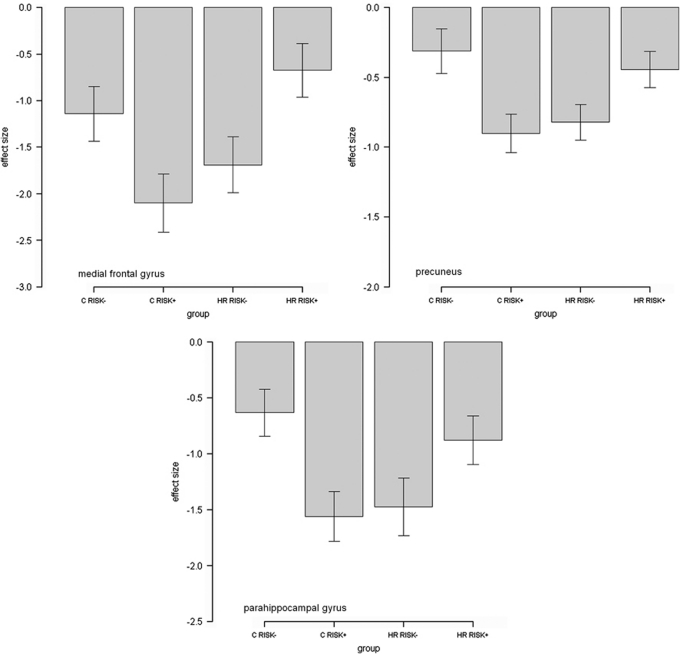
For sentence completion vs baseline, there were significant haplotype*group interactions in several clusters in the left medial frontal gyrus extending to the anterior cingulate (p=0.04, KE=470, Z=3.76, x=−10, y=54, and z=4, with a small volume correction for the prefrontal cortex), in the left precuneus extending to posterior cingulate (p<0.01, KE=1149, Z=3.80, x=−6, y=−32, and z=50, at the whole brain level), and in the right parahippocampal region (p=0.04, KE=7 07, Z=3.86, x=16, y=−42, and z=2, at the whole brain level), see Figure 1, Table 2. For all regions, there was relatively greater activation during task conditions in the high-risk RISK+ group vs the high-risk RISK− group, with the reverse seen in the control groups, see Figure 2.
Figure 1.
Haplotype*group interactions for DGKH for sentence completion vs baseline. Images are overlaid onto standard brain in MNI space using Mango software package (http://ric.uthscsa.edu/mango). Map represents F-statistic images thresholded at F⩾8, equivalent to p uncorrected=0.005, see Materials and Methods for further details.
Table 2. Significant Haplotype*Group Interactions.
| Contrast | Region | MNI coordinates x, y, z (mm) | Cluster size (k) | Z | p |
|---|---|---|---|---|---|
| Sentence completion vs baseline | Left medial frontal gyrus | −10, 54, 4 | 470 | 3.76 | <0.05 |
| Left precuneus | −6, −32, 50 | 1149 | 3.80 | <0.01 | |
| Right parahippocampus | 16, −42, 2 | 707 | 3.86 | <0.05 | |
| Parametric | Right posterior cingulate | 4, −66, 20 | 1317 | 4.07 | <0.01 |
Abbreviations: MNI, Montreal Neurological Institute; k, number of voxels.
Figure 2.
Graph of effect size for clusters of significant interaction. For sentence completion vs baseline, RISK− represents non-risk haplotype carriers and RISK+ represents risk haplotype homozygotes. The Y axis denotes BOLD effect size.
Post-hoc pair-wise significance tests between RISK− and RISK+ within the control and high-risk groups separately indicated these clusters were also significantly different between the haplotype groups (controls RISK− vs RISK+: p<0.05, p<0.01, p<0.01; high-risk RISK− group vs RISK+ group: p<0.01, p<0.05, p<0.05 for prefrontal, precuneus, and parahippocampal clusters, respectively).
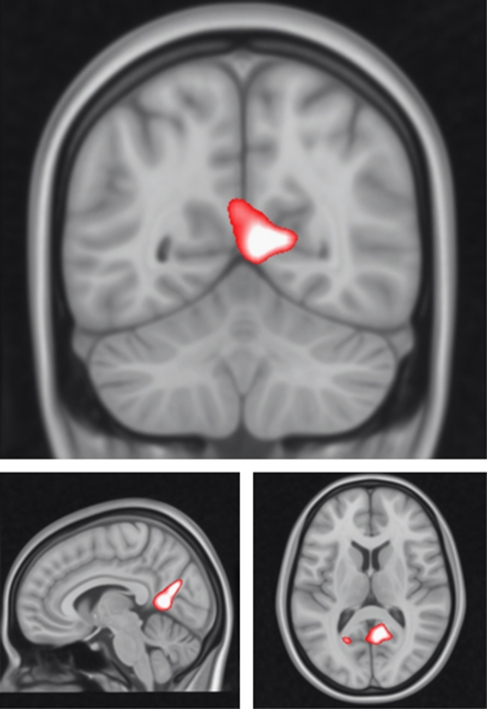
Parametric contrast
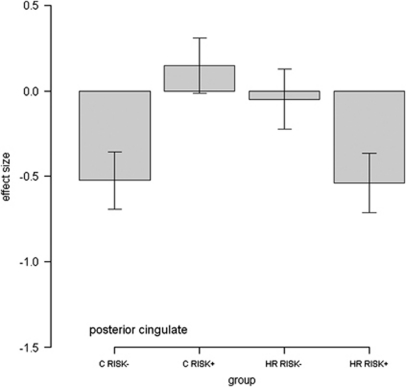
For the parametric contrast, there was a significant haplotype*group interaction in the posterior cingulate cortex (p<0.01, KE=1317, Z=4.07, x=4, y=−66, and z=20, at the whole brain level, see Figure 3, Table 2). This interaction was in the direction of a decreased gradient of activation with increasing sentence difficulty in RISK+ vs RISK− high-risk subjects. The reverse pattern was seen in the control group.
Figure 3.
Haplotype*group interactions for DGKH for the parametric contrast. Images are overlaid onto standard brain in Montreal Neurological Institute space using Mango software package (http://ric.uthscsa.edu/mango). Map represents F-statistic images thresholded at F⩾8, equivalent to p uncorrected=0.005.
Post-hoc pair-wise significance tests between RISK− and RISK+ within the control and high-risk groups separately indicated this cluster was also significantly different between haplotype groups (controls RISK− vs RISK+: p<0.01; high-risk RISK− vs RISK+ subjects: p<0.05, for posterior cingulate cluster).
Correlation Analyses
We examined correlations across all subjects between data extracted from the clusters of difference reported above (medial prefrontal cortex, precuneus, parahippocampal gyrus, and posterior cingulate) and the clinical data (HAM-D score, Young Mania Rating Scale score, and individual components of the TEMPS-A). None remained significant after controlling for multiple comparisons.
DISCUSSION
In the current study, we have demonstrated differential effects of DGKH haplotypes on brain activation in the medial prefrontal cortex, precuneus, parahippocampus, and posterior cingulate between healthy controls and a cohort at familial risk of BD during a verbal initiation task. The task was found to engage typical language-related regions, as well as demonstrating reduced activation in regions typically disengaged during cognitive paradigms as previously described (Raichle et al, 2001; Whalley et al, 2004; Whalley et al, 2011), and see Supplementary Figure 1. The advantage of studying such effects in a high genetic risk group allows the examination of the impact of the gene on brain function in the absence of confounding disease and medication effects associated with studies on patient groups.
From Figure 2, the origin of the haplotype*group interactions for the clusters originating from the sentence completion vs baseline contrast appeared to be a reduced disengagement, or failure to suppress activation in the high-risk RISK+ haplotype group vs high-risk RISK− during task conditions. The reverse was demonstrated in the control group, Figure 2 (ie., there was greater suppression in the control RISK+ group vs the control RISK− group). The clusters demonstrating interaction effects for this contrast, the medial prefrontal cortex, parahippocampal gyrus, precuneus, and posterior cingulate are indeed typically associated with default-mode patterns of activation. The default-mode network refers to a set of regions that consistently demonstrate synchronous task-independent deactivation across a range of cognitive tasks compared with baseline conditions (Raichle et al, 2001; Greicius et al, 2003; Raichle and Snyder, 2007). The posterior cingulate cortex was also found to demonstrate significant interaction effects for the parametric contrast, Figure 4. This contrast focusses on a gradation of neural activity in relation to task difficulty, rather than the sentence completion contrast, which relates activation across all levels of task condition to a simple visual baseline. In the posterior cingulate for the parametric contrast, the RISK− high-risk subjects and RISK+ controls demonstrate small increases in activation with increasing task difficulty. The RISK+ high-risk subjects, however, demonstrated decreases in activation with increasing task difficulty. In summary therefore, in the posterior cingulate, the RISK+ high-risk subjects have poor suppression when looking across all task conditions; however, when looking at the relationship between activation and task load, the RISK+ high-risk subjects demonstrate a greater decrease in activation with increasing task difficulty.
Figure 4.
Graph of effect size for cluster of significant interaction. For parametric contrast, RISK− represents non-risk haplotype carriers, RISK+ represents risk haplotype homozygotes. The Y axis denotes BOLD effect size.
Default-mode regions are considered to subserve self-referential mental activity, autobiographical memory, and aspects of internal emotion processing (Gusnard and Raichle, 2001; Cabeza et al, 2002). Failure to deactivate these regions has been linked to performance deficits in healthy subjects (Weissman et al, 2006; Mason et al, 2007; Eichele et al, 2008). It has also been suggested that abnormalities within this network may underlie deficits in cognitive and affective processing seen in psychiatric disorders (Ongur et al, 2010). Indeed, abnormalities in the default-mode network have been reported in BD patients (Calhoun et al, 2008; Ongur et al, 2010; Costafreda et al, 2011). Further, the default-mode network is also considered to be strongly influenced by genetic factors (Glahn et al, 2010).
As the specific molecular role of DGKH in not yet known, the precise mechanisms of action of DGKH variation in conferring risk for the disorder is as yet unclear. All SNPs contributing to this haplotype are indeed located within intronic regions and are therefore noncoding. We would argue, however, that this does not necessarily mean that they are nonfunctional, rather that any function is currently unknown. We could speculate that, given previous evidence for the involvement of the gene in BD, they may be involved in gene regulation, or may be in LD with another as yet unknown functional variant of the gene, which may have downstream influence on BOLD signal. In the context of the current findings, they suggest that there is an influence of the DGKH gene on function in default-mode brain regions, and this differs between those at risk for the disorder and healthy controls. In other words the risk haplotype appears to alter the ability to disengage regions involved in self-referential and internal emotional processing during cognitive tasks in the bipolar high-risk group, as compared to healthy controls. One possible explanation for this difference is that additional genetic or environmental risk factors interacting with the effects of DGKH in the high-risk group. Presumably, controls carrying the risk haplotype are able to compensate and allocate cognitive resources more efficiently during task conditions than the bipolar high-risk subjects carrying the risk haplotype, who also importantly carry additional background risk factors. We interpret these results to suggest therefore that the relationship between DGKH genotype and functional architecture in the high-risk group may differ from controls based on an already present genetic loading for the disorder.
In terms of its biological relevance, DGKH is an upstream regulator of protein kinase C (PKC). It catalyzes the conversion of diacylglycerol to phosphatidic acid, thereby leading to reductions in diacylglycerol. The latter is essential for activating several isoforms of PKC; a family of enzymes that is heterogeneously distributed in the brain and has a pivotal role in regulating pre- and postsynaptic neurotransmission (Casabona 1997; Catapano and Manji, 2008). Accordingly, it has been proposed that variations in the DGKH gene might result in reductions of diacylglycerol kinase activity, thereby leading to increased PKC signaling (Robbins and Arnsten, 2009). Numerous lines of evidence suggest the importance of PKC in the pathophysiology and treatment of BD. Investigations of peripheral blood and postmortem studies indicate altered PKC levels in BD, and lithium interacts with the PKC pathway resulting in decreasing downstream PKC levels and activity (Catapano and Manji, 2008). Similarly, the mood stabilizer valproate inhibits PKC, suggesting a commonality in the pathway of therapeutic effect (Catapano and Manji, 2008). Moreover, the PKC antagonist tamoxifen has been shown to reduce manic symptoms in BD patients (Yildiz et al, 2008). Hence, DGKH is predicted to have a role in lithium response and in mood control and therefore in the etiology of the disorder.
Although there is limited literature relating specifically to the effects of DGKH on brain functioning, there is evidence of increased gene expression of DGKH in the prefrontal cortex of patients with BD vs controls (Moya et al, 2010). There are also several lines of evidence suggesting that PKC impacts on cognitive functioning. Research has focussed on its role in mediating the effects of chronic stress on the prefrontal cortex (Hains et al, 2009), in regulating optimal prefrontal activity during working memory performance (Birnbaum et al, 2004), and in association with age-related cognitive decline (Brennan et al, 2009). Although speculative, these may fit with neurobiological models of BD where there is altered prefrontal modulation of other cortical and subcortical structures involved in emotional processing leading to impaired emotional regulation and increased mood fluctuation in the disorder (Phillips et al, 2003).
In summary, these findings suggest that variation in the DGKH gene is associated with differential modulation of default network suppression during a sentence completion paradigm in bipolar high-risk individuals when compared with healthy controls. Differential effects of DGKH haplotypes in those at high familial risk of BD vs controls supports previous evidence that the DGKH gene may be involved in the pathogenesis of BD. Further replication in patient groups and/or during resting state scanning may help confirm the nature of these effects.
Acknowledgments
We would like to thank all of the participants who took part in the study and the radiographers who acquired the MRI scans. This study was conducted at the Scottish Brain Imaging Research Centre, which is supported by SINAPSE (Scottish Imaging Network, a Platform for Scientific Excellence, www.sinapse.ac.uk). The investigators also acknowledge the financial support of National Health Service (NHS) Research Scotland, through the Scottish Mental Health Research Network (www.smhrn.org.uk) who provided assistance with subject recruitment and cognitive assessments.
The author HCW is supported by a Dorothy Hodgkin Fellowship from the Royal Society of Edinburgh (DH080018). MP is supported by a studentship from the Medical Research Council. JH is supported by a Scottish Senior Clinical Fellowship from the Chief Scientists Office in Scotland. JES is supported by a Clinical Research Training Fellowship from the Wellcome Trust. AMM is currently supported by the Health Foundation through a Clinician Scientist Fellowship (Ref: 2268/4295) and by the National Alliance for Research on Schizophrenia and Depression through an Independent Investigator Award. The investigators also acknowledge the financial support of National Health Service (NHS) Research Scotland, through the Scottish Mental Health Research Network (www.smhrn.org.uk) who provided assistance with subject recruitment and cognitive assessments. HCW, JH, SML, and AMM have received financial support from Pfizer (formerly Wyeth) in relation to imaging studies of people with schizophrenia and BD. The authors MP, LR, ECJ, and JES have no competing interests to declare.
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- Akiskal HS, Mendlowicz MV, Jean-Louis G, Rapaport MH, Kelsoe JR, Gillin JC, et al. TEMPS-A: validation of a short version of a self-rated instrument designed to measure variations in temperament. J Affect Disord. 2005;85:45–52. doi: 10.1016/j.jad.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Allen P, Stephan KE, Mechelli A, Day F, Ward N, Dalton J, et al. Cingulate activity and fronto-temporal connectivity in people with prodromal signs of psychosis. Neuroimage. 2010;49:947–955. doi: 10.1016/j.neuroimage.2009.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts B, Jabben N, Krabbendam L, van Os J. Meta-analyses of cognitive functioning in euthymic bipolar patients and their first-degree relatives. Psychol Med. 2008;38:771–785. doi: 10.1017/S0033291707001675. [DOI] [PubMed] [Google Scholar]
- Badner JA, Gershon ES. Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry. 2002;7:405–411. doi: 10.1038/sj.mp.4001012. [DOI] [PubMed] [Google Scholar]
- Barnett JH, Smoller JW. The genetics of bipolar disorder. Neuroscience. 2009;164:331–343. doi: 10.1016/j.neuroscience.2009.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum AE, Akula N, Cabanero M, Cardona I, Corona W, Klemens B, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2008;13:197–207. doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelsen A, Harvald B, Hauge M. A Danish twin study of manic-depressive disorders. Br J Psychiatry. 1977;130:330–351. doi: 10.1192/bjp.130.4.330. [DOI] [PubMed] [Google Scholar]
- Birnbaum SG, Yuan PX, Wang M, Vijayraghavan S, Bloom AK, Davis DJ, et al. Protein kinase C overactivity impairs prefrontal cortical regulation of working memory. Science. 2004;306:882–884. doi: 10.1126/science.1100021. [DOI] [PubMed] [Google Scholar]
- Bloom PA, Fischler I. Completion norms for 329 sentence contexts. Mem Cogn. 1980;8:631–642. doi: 10.3758/bf03213783. [DOI] [PubMed] [Google Scholar]
- Brennan AR, Yuan P, Dickstein DL, Rocher AB, Hof PR, Manji H, et al. Protein kinase C activity is associated with prefrontal cortical decline in aging. Neurobiol Aging. 2009;30:782–792. doi: 10.1016/j.neurobiolaging.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess P, Shallice T. The Hayling and Brixton Tests. Thames Valley Test Company Limited: Bury St. Edmunds, UK; 1997. [Google Scholar]
- Burgess PW, Shallice T. Response suppression, initiation and strategy use following frontal lobe lesions. Neuropsychologia. 1996;34:263–272. doi: 10.1016/0028-3932(95)00104-2. [DOI] [PubMed] [Google Scholar]
- Cabeza R, Dolcos F, Graham R, Nyberg L. Similarities and differences in the neural correlates of episodic memory retrieval and working memory. Neuroimage. 2002;16:317–330. doi: 10.1006/nimg.2002.1063. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Maciejewski PK, Pearlson GD, Kiehl KA. Temporal lobe and ‘default' hemodynamic brain modes discriminate between schizophrenia and bipolar disorder. Hum Brain Mapp. 2008;29:1265–1275. doi: 10.1002/hbm.20463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casabona G. Intracellular signal modulation: a pivotal role for protein kinase C. Prog Neuropsychopharmacol Biol Psychiatry. 1997;21:407–425. doi: 10.1016/s0278-5846(97)00011-0. [DOI] [PubMed] [Google Scholar]
- Catapano LA, Manji HK. Kinases as drug targets in the treatment of bipolar disorder. Drug Discov Today. 2008;13:295–302. doi: 10.1016/j.drudis.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Cerullo MA, Adler CM, Delbello MP, Strakowski SM. The functional neuroanatomy of bipolar disorder. Int Rev Psychiatry. 2009;21:314–322. doi: 10.1080/09540260902962107. [DOI] [PubMed] [Google Scholar]
- Costafreda SG, Fu CH, Picchioni M, Toulopoulou T, McDonald C, Kravariti E, et al. Pattern of neural responses to verbal fluency shows diagnostic specificity for schizophrenia and bipolar disorder. BMC Psychiatry. 2011;11:18. doi: 10.1186/1471-244X-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detera-Wadleigh SD, McMahon FJ. G72/G30 in schizophrenia and bipolar disorder: review and meta-analysis. Biol Psychiatry. 2006;60:106–114. doi: 10.1016/j.biopsych.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Eichele T, Debener S, Calhoun VD, Specht K, Engel AK, Hugdahl K, von Cramon DY, et al. Prediction of human errors by maladaptive changes in event-related brain networks. Proc Natl Acad Sci USA. 2008;105:6173–6178. doi: 10.1073/pnas.0708965105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Gibbon M, Williams JBW. Structured Clinical Interview for DSM-IV-TR Axis I Disorders, Research Version, Patient Edition with Psychotic Screen. Biometrics Research, New York State Psychiatric Institute: New York; 2002. [Google Scholar]
- Glahn DC, Winkler AM, Kochunov P, Almasy L, Duggirala R, Carless MA, et al. Genetic control over the resting brain. Proc Natl Acad Sci USA. 2010;107:1223–1228. doi: 10.1073/pnas.0909969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc Natl Acad Sci USA. 2003;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusnard DA, Raichle ME. Searching for a baseline: functional imaging and the resting human brain. Nat Rev Neurosci. 2001;2:685–694. doi: 10.1038/35094500. [DOI] [PubMed] [Google Scholar]
- Hains AB, Vu MA, Maciejewski PK, van Dyck CH, Gottron M, Arnsten AF. Inhibition of protein kinase C signaling protects prefrontal cortex dendritic spines and cognition from the effects of chronic stress. Proc Natl Acad Sci USA. 2009;106:17957–17962. doi: 10.1073/pnas.0908563106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton M. A rating scale for depression. J Neurol Neurosurg Psychiatry. 1960;23:56–62. doi: 10.1136/jnnp.23.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldjian JA, Laurienti PJ, Kraft RA, Burdette JH. An automated method for neuroanatomic and cytoarchitectonic atlas-based interrogation of fMRI data sets. Neuroimage. 2003;19:1233–1239. doi: 10.1016/s1053-8119(03)00169-1. [DOI] [PubMed] [Google Scholar]
- Manchia M, Squassina A, Congiu D, Chillotti C, Ardau R, Severino G, et al. Interacting genes in lithium prophylaxis: preliminary results of an exploratory analysis on the role of DGKH and NR1D1 gene polymorphisms in 199 Sardinian bipolar patients. Neurosci Lett. 2009;467:67–71. doi: 10.1016/j.neulet.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Manji HK, Lenox RH. Ziskind-Somerfeld Research Award. Protein kinase C signaling in the brain: molecular transduction of mood stabilization in the treatment of manic-depressive illness. Biol Psychiatry. 1999;46:1328–1351. doi: 10.1016/s0006-3223(99)00235-8. [DOI] [PubMed] [Google Scholar]
- Mason MF, Norton MI, Van Horn JD, Wegner DM, Grafton ST, Macrae CN. Wandering minds: the default network and stimulus-independent thought. Science. 2007;315:393–395. doi: 10.1126/science.1131295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin P, Farmer A, Harvey I. A polydiagnostic application of operational criteria in studies of psychotic illness. Development and reliability of the OPCRIT system. Arch Gen Psychiatry. 1991;48:764–770. doi: 10.1001/archpsyc.1991.01810320088015. [DOI] [PubMed] [Google Scholar]
- McGuffin P, Rijsdijk F, Andrew M, Sham P, Katz R, Cardno A. The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch Gen Psychiatry. 2003;60:497–502. doi: 10.1001/archpsyc.60.5.497. [DOI] [PubMed] [Google Scholar]
- McIntosh A, Whalley HC, McKirdy J, Hall J, Sussmann J, Shankar P, et al. Differences in dorsal and ventral prefrontal function separate bipolar disorder from schizophrenia. Schizopr Res. 2008a;98:40. [Google Scholar]
- McIntosh AM, Whalley HC, McKirdy J, Hall J, Sussmann JE, Shankar P, et al. Prefrontal function and activation in bipolar disorder and schizophrenia. Am J Psychiatry. 2008b;165:378–384. doi: 10.1176/appi.ajp.2007.07020365. [DOI] [PubMed] [Google Scholar]
- Moya PR, Murphy DL, McMahon FJ, Wendland JR. Increased gene expression of diacylglycerol kinase eta in bipolar disorder. Int J Neuropsychopharmacology. 2010;13:1127–1128. doi: 10.1017/S1461145710000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathaniel-James DA, Fletcher P, Frith CD. The functional anatomy of verbal initiation and suppression using the Hayling test. Neuropsychologia. 1997;35:559–566. doi: 10.1016/s0028-3932(96)00104-2. [DOI] [PubMed] [Google Scholar]
- Nelson H. The National Adult Reading Test Manual. NFER-Nelson: Windsor; 1982. [Google Scholar]
- Ollila HM, Soronen P, Silander K, Palo OM, Kieseppa T, Kaunisto MA, et al. Findings from bipolar disorder genome-wide association studies replicate in a Finnish bipolar family-cohort. Mol Psychiatry. 2009;14:351–353. doi: 10.1038/mp.2008.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongur D, Lundy M, Greenhouse I, Shinn AK, Menon V, Cohen BM, et al. Default mode network abnormalities in bipolar disorder and schizophrenia. Psychiatry Res. 2010;183:59–68. doi: 10.1016/j.pscychresns.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips ML, Drevets WC, Rauch SL, Lane R. Neurobiology of emotion perception II: implications for major psychiatric disorders. Biol Psychiatry. 2003;54:515–528. doi: 10.1016/s0006-3223(03)00171-9. [DOI] [PubMed] [Google Scholar]
- Raichle ME, Snyder AZ.2007A default mode of brain function: a brief history of an evolving idea Neuroimage 371083–1090.discussion 1097–9. [DOI] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proc Natl Acad Sci USA. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins TW, Arnsten AF. The neuropsychopharmacology of fronto-executive function: monoaminergic modulation. Annu Rev Neurosci. 2009;32:267–287. doi: 10.1146/annurev.neuro.051508.135535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P, Smoller JW, Fan J, Ferreira MA, Perlis RH, Chambert K, et al. Whole-genome association study of bipolar disorder. Mol Psychiatry. 2008;13:558–569. doi: 10.1038/sj.mp.4002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprooten E, Sussmann JE, Clugston A, Peel A, McKirdy J, Moorhead TWJ, et al. White matter integrity in individuals at high genetic risk of bipolar disorder. Biol Psychiatry. 2011;70:350–356. doi: 10.1016/j.biopsych.2011.01.021. [DOI] [PubMed] [Google Scholar]
- Squassina A, Manchia M, Congiu D, Severino G, Chillotti C, Ardau R, et al. The diacylglycerol kinase eta gene and bipolar disorder: a replication study in a Sardinian sample. Mol Psychiatry. 2009;14:350–351. doi: 10.1038/mp.2008.93. [DOI] [PubMed] [Google Scholar]
- Strakowski SM, Delbello MP, Adler CM. The functional neuroanatomy of bipolar disorder: a review of neuroimaging findings. Mol Psychiatry. 2005;10:105–116. doi: 10.1038/sj.mp.4001585. [DOI] [PubMed] [Google Scholar]
- Tesli M, Kahler AK, Andreassen BK, Werge T, Mors O, Mellerup E, et al. No association between DGKH and bipolar disorder in a Scandinavian case-control sample. Psychiatr Genet. 2009;19:269–272. doi: 10.1097/YPG.0b013e32832d302f. [DOI] [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium Genome-wide association study of 14 000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Weber H, Kittel-Schneider S, Gessner A, Domschke K, Neuner M, Jacob CP, et al. Cross-disorder analysis of bipolar risk genes: further evidence of DGKH as a risk gene for bipolar disorder, but also unipolar depression and adult ADHD. Neuropsychopharmacology. 2011;36:2076–2085. doi: 10.1038/npp.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman DG, Roberts KC, Visscher KM, Woldorff MG. The neural basis of momentary lapses of attention. Nat Neurosci. 2006;9:971–978. doi: 10.1038/nn1727. [DOI] [PubMed] [Google Scholar]
- Whalley HC, Simonotto E, Flett S, Marshall I, Ebmeier KP, Owens DG, et al. fMRI correlates of state and trait effects in subjects at genetically enhanced risk of schizophrenia. [see comment] Brain. 2004;127:478–490. doi: 10.1093/brain/awh070. [DOI] [PubMed] [Google Scholar]
- Whalley HC, Sussmann JE, Chakirova G, Mukerjee P, Peel A, McKirdy J, et al. The neural basis of familial risk and temperamental variation in individuals at high risk of bipolar disorder. Biol Psychiatry. 2011;70:343–349. doi: 10.1016/j.biopsych.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Yildiz A, Guleryuz S, Ankerst DP, Ongur D, Renshaw PF. Protein kinase C inhibition in the treatment of mania: a double-blind, placebo-controlled trial of tamoxifen. Arch Gen Psychiatry. 2008;65:255–263. doi: 10.1001/archgenpsychiatry.2007.43. [DOI] [PubMed] [Google Scholar]
- Yosifova A, Mushiroda T, Stoianov D, Vazharova R, Dimova I, Karachanak S, et al. Case-control association study of 65 candidate genes revealed a possible association of a SNP of HTR5A to be a factor susceptible to bipolar disease in Bulgarian population. J Affect Disord. 2009;117:87–97. doi: 10.1016/j.jad.2008.12.021. [DOI] [PubMed] [Google Scholar]
- Young RC, Biggs JT, Ziegler VE, Meyer DA. A rating scale for mania: reliability, validity and sensitivity. Br J Psychiatry. 1978;133:429–435. doi: 10.1192/bjp.133.5.429. [DOI] [PubMed] [Google Scholar]
- Zeng Z, Wang T, Li T, Li Y, Chen P, Zhao Q, et al. Common SNPs and haplotypes in DGKH are associated with bipolar disorder and schizophrenia in the Chinese Han population. Mol Psychiatry. 2011;16:473–475. doi: 10.1038/mp.2010.86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.