Abstract
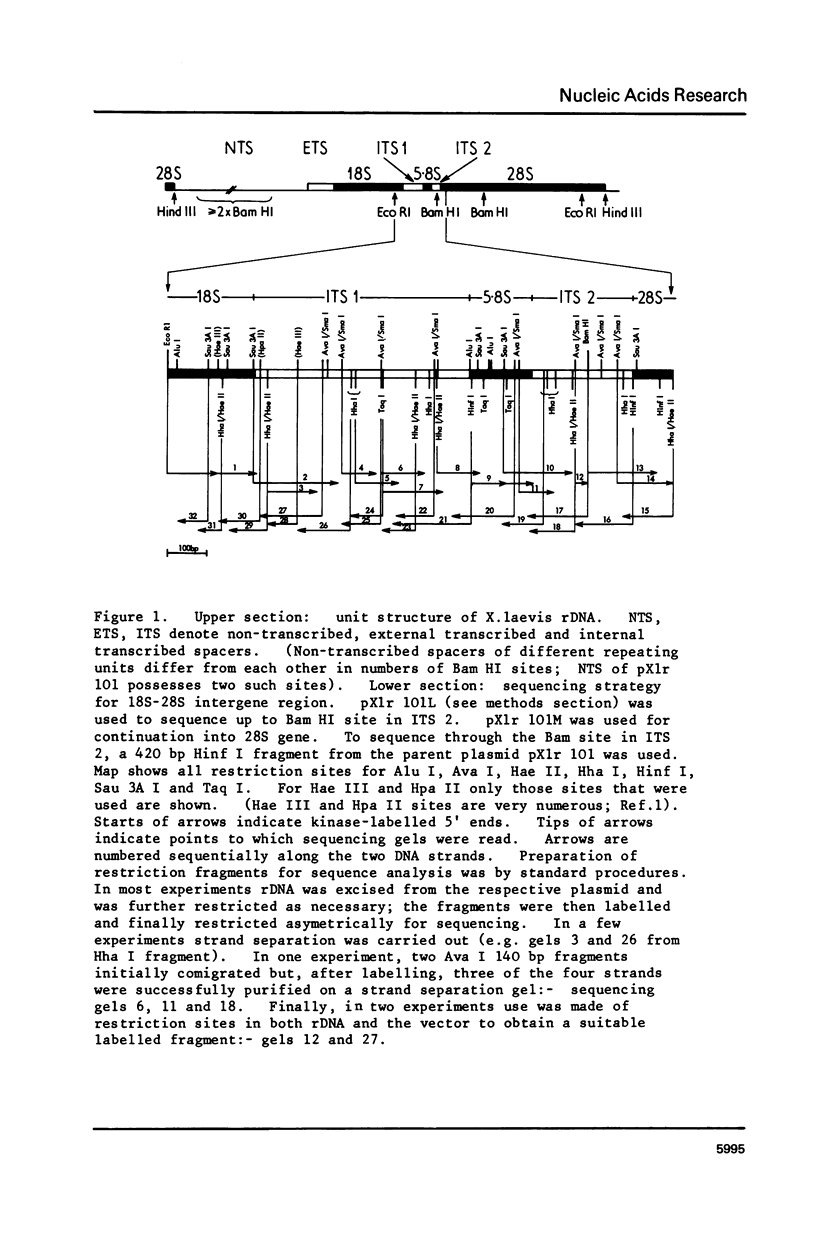
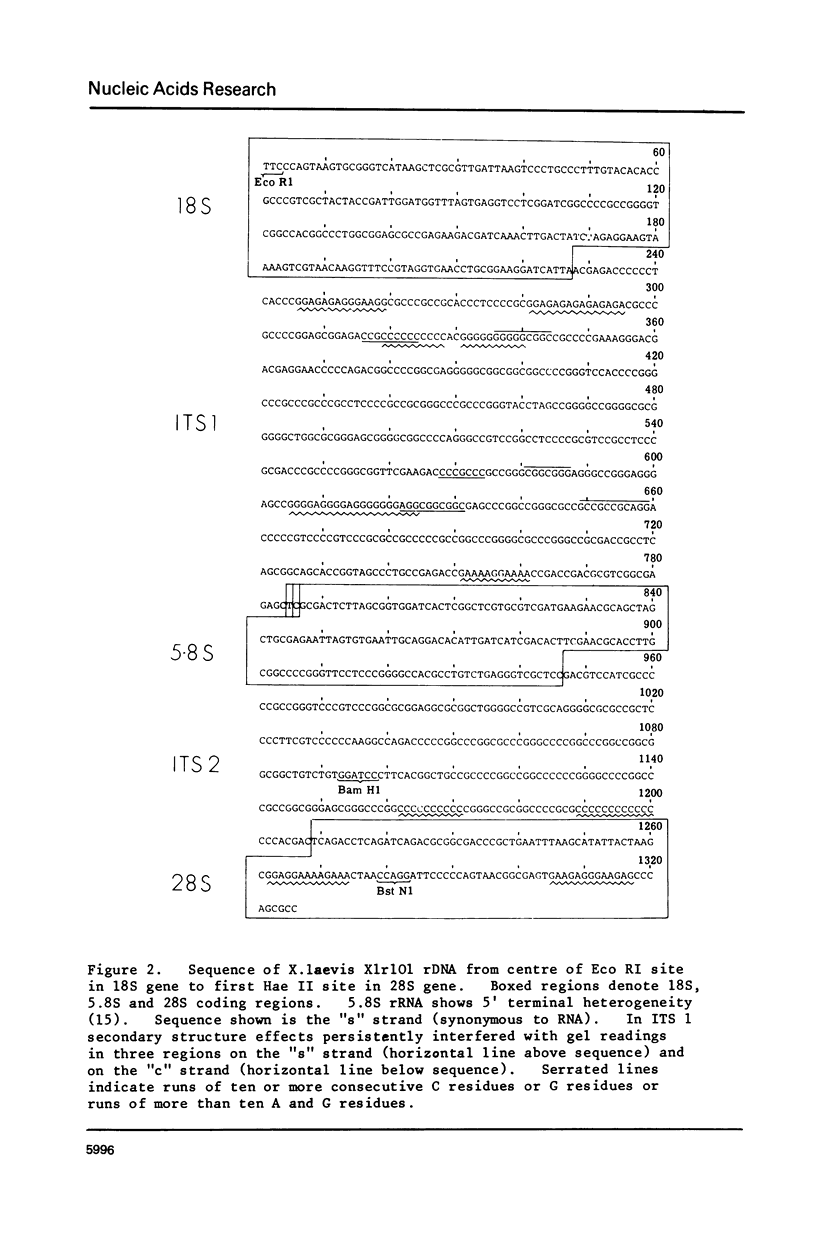
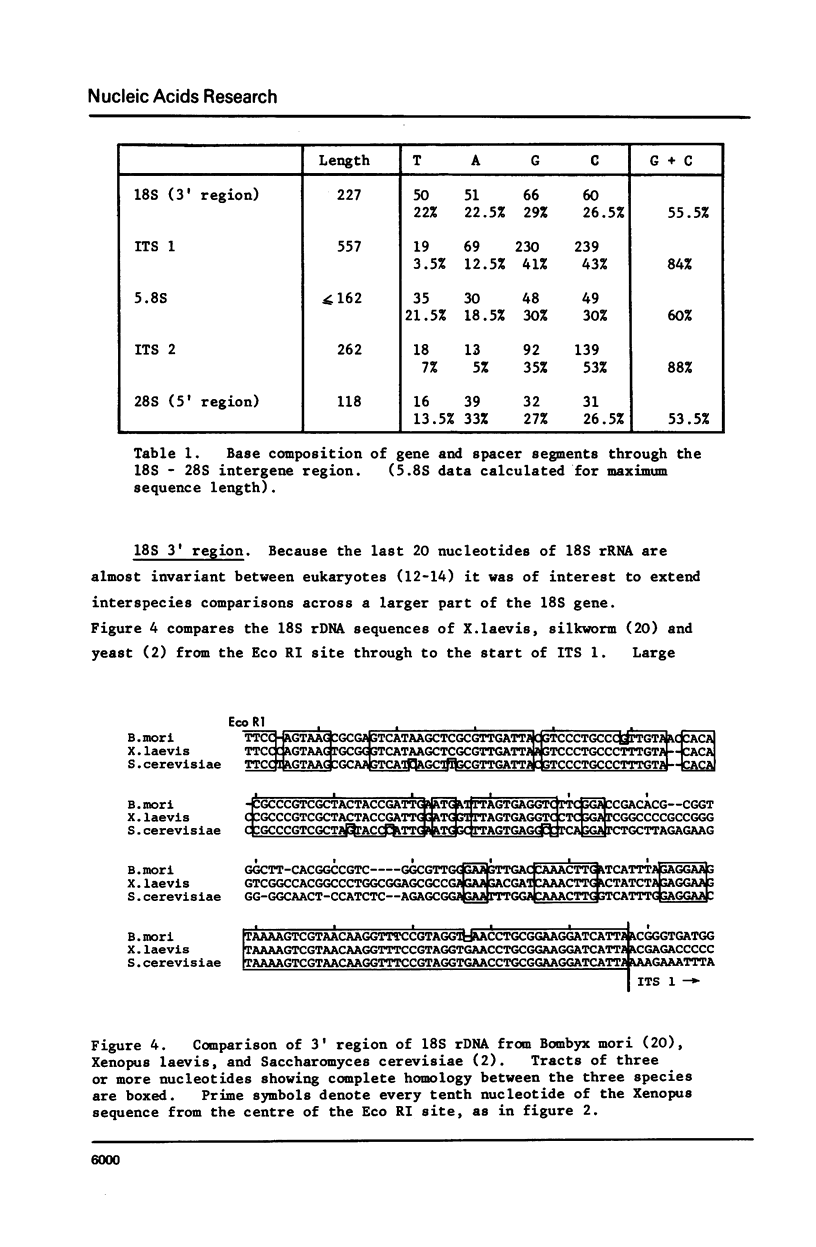
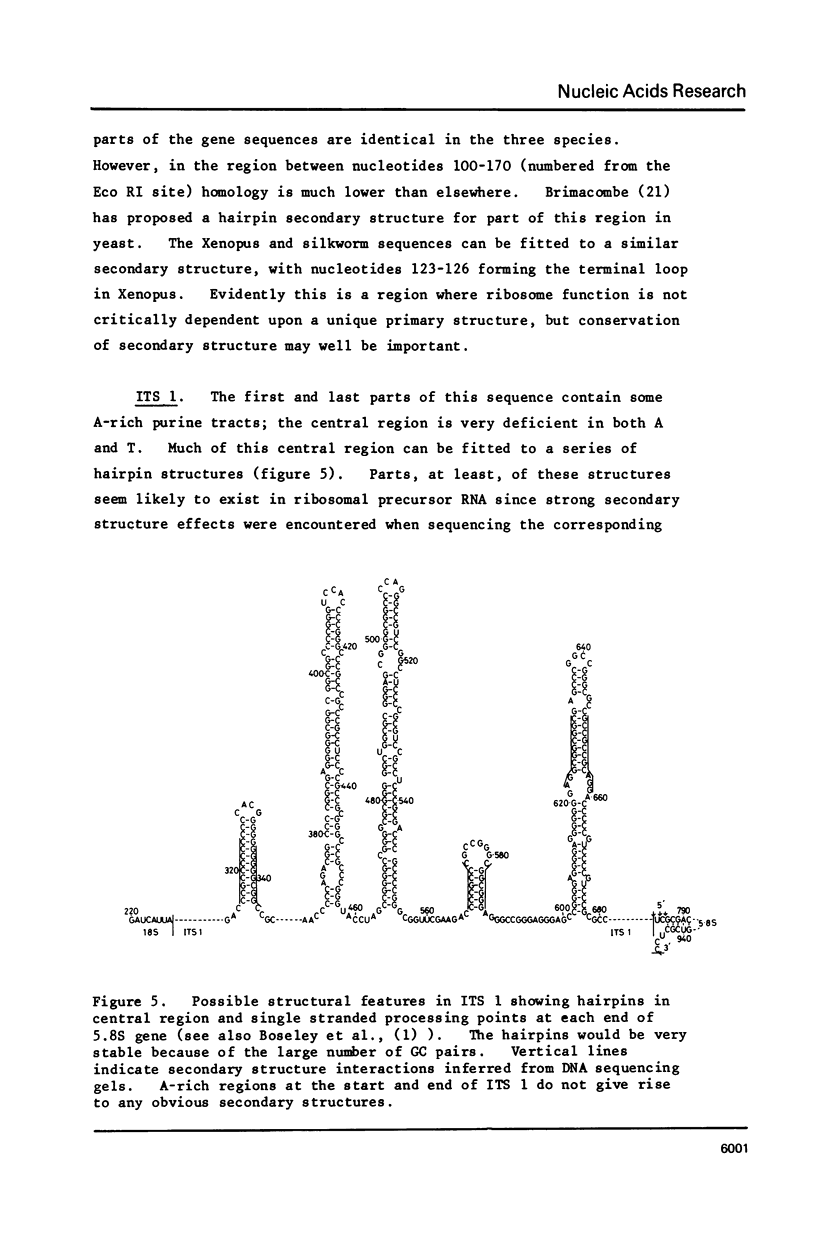
We have determined the nucleotide sequence of part of a cloned ribosomal transcription unit from Xenopus laevis extending from the 3' region of the 18S gene through the 18S-28S intergene region into the start of the 28S gene. The 18S 3' region possess two tracts of high homology with the corresponding segments of other eukaryotic 18S genes (yeast and Bombyx mori) separated by a tract of low homology which in X. laevis is rich in G plus C. The first internal transcribed spacer, between the 18S and 5.8S genes, is 557 nucleotides long, very rich in G plus C (84%) and shows no sequence homology with the corresponding yeast sequence. The 5.8S rRNA sequence is revised slightly in the light of the DNA sequence. The second internal transcribed spacer, between the 5.8S and 28S genes, is 262 nucleotides long and is even richer in G plus C (88%) than the first internal spacer. 28S rRNA starts with the sequence pUCAG. This is encoded at the first of three closely linked TCAG sites in rDNA.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberty H., Raba M., Gross H. J. Isolation from rat liver and sequence of a RNA fragment containing 32 nucleotides from position 5 to 36 from the 3' end of ribosomal 18S RNA. Nucleic Acids Res. 1978 Feb;5(2):425–434. doi: 10.1093/nar/5.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boseley P. G., Tuyns A., Birnstiel M. L. Mapping of the Xenopus laevis 5.8S rDNA by restriction and DNA sequencing. Nucleic Acids Res. 1978 Apr;5(4):1121–1137. doi: 10.1093/nar/5.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Young R. A., Steitz J. A. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980 Feb;19(2):393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- Brand R. C., Gerbi S. A. Fine structure of ribosomal RNA. II. Distribution of methylated sequences within Xenopus laevis rRNA. Nucleic Acids Res. 1979 Nov 24;7(6):1497–1511. doi: 10.1093/nar/7.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jonge P., Klootwijk J., Planta R. J. Sequence of the 3'-terminal 21 nucleotides of yeast 17S ribosomal RNA. Nucleic Acids Res. 1977 Oct;4(10):3655–3663. doi: 10.1093/nar/4.10.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon I. C., Anderson S., Nierlich D. P. Distinctive sequence of human mitochondrial ribosomal RNA genes. Nature. 1980 Jul 31;286(5772):460–467. doi: 10.1038/286460a0. [DOI] [PubMed] [Google Scholar]
- Ford P. J., Mathieson T. The nucleotide sequences of 5.8-S ribosomal RNA from Xenopus laevis and Xenopus borealis. Eur J Biochem. 1978 Jun 1;87(1):199–214. doi: 10.1111/j.1432-1033.1978.tb12367.x. [DOI] [PubMed] [Google Scholar]
- Hagenbüchle O., Santer M., Steitz J. A., Mans R. J. Conservation of the primary structure at the 3' end of 18S rRNA from eucaryotic cells. Cell. 1978 Mar;13(3):551–563. doi: 10.1016/0092-8674(78)90328-8. [DOI] [PubMed] [Google Scholar]
- Hampe A., Eladari M. E., Galibert F. Nucleotide sequence study of mouse 5.8S ribosomal RNA. Biochimie. 1976;58(8):943–951. doi: 10.1016/s0300-9084(76)80282-9. [DOI] [PubMed] [Google Scholar]
- Khan M. S., Maden B. E. Nucleotide sequence relationships between vertebrate 5.8 S ribosomal RNAs. Nucleic Acids Res. 1977 Jul;4(7):2495–2505. doi: 10.1093/nar/4.7.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. S., Maden B. E. Nucleotide sequences within the ribosomal ribonucleic acids of HeLa cells, Xenopus laevis and chick embryo fibroblasts. J Mol Biol. 1976 Feb 25;101(2):235–254. doi: 10.1016/0022-2836(76)90375-2. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Nazar R. N., Sitz T. O., Busch H. Structural analyses of mammalian ribosomal ribonucleic acid and its precursors. Nucleotide sequence of ribosomal 5.8 S ribonucleic acid. J Biol Chem. 1975 Nov 25;250(22):8591–8597. [PubMed] [Google Scholar]
- Rubin G. M. The nucleotide sequence of Saccharomyces cerevisiae 5.8 S ribosomal ribonucleic acid. J Biol Chem. 1973 Jun 10;248(11):3860–3875. [PubMed] [Google Scholar]
- Salim M., Maden B. E. Nucleotide sequence encoding the 5' end of Xenopus laevis 18S rRNA. Nucleic Acids Res. 1980 Jul 11;8(13):2871–2884. doi: 10.1093/nar/8.13.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samols D. R., Hagenbuchle O., Gage L. P. Homology of the 3' terminal sequences of the 18S rRNA of Bombyx mori and the 16S rRNA of Escherchia coli. Nucleic Acids Res. 1979 Nov 10;7(5):1109–1119. doi: 10.1093/nar/7.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Skriabin K. G., Kraev A. S., Rubtsov P. M., Baev A. A. Polnaia posledovatel'nost' nukleotidov speisernoi oblasti, raspolozhennoi mezhdu genami 18S i 5.8S RNK drozhzhei. Dokl Akad Nauk SSSR. 1979;247(3):761–765. [PubMed] [Google Scholar]
- Slack J. M., Loening U. E. 5'-Ends of ribosomal and ribosomal precursor RNAs form Xenopus laevis. Eur J Biochem. 1974 Mar 15;43(1):59–67. doi: 10.1111/j.1432-1033.1974.tb03384.x. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Trendelenburg M. F., Gurdon J. B. Transcription of cloned Xenopus ribosomal genes visualised after injection into oocyte nuclei. Nature. 1978 Nov 16;276(5685):292–294. doi: 10.1038/276292a0. [DOI] [PubMed] [Google Scholar]
- Vass J. K., Maden B. E. Studies on the conformation of the 3' terminus of 18-S rRNA. Eur J Biochem. 1978 Apr;85(1):241–247. doi: 10.1111/j.1432-1033.1978.tb12232.x. [DOI] [PubMed] [Google Scholar]
- Veldman G. M., Brand R. C., Klootwijk J., Planta R. Some characteristics of processing sites in ribosomal precursor RNA of yeast. Nucleic Acids Res. 1980 Jul 11;8(13):2907–2920. doi: 10.1093/nar/8.13.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. Secondary structure maps of ribosomal RNA and DNA. I. Processing of Xenopus laevis ribosomal RNA and structure of single-stranded ribosomal DNA. J Mol Biol. 1974 Oct 25;89(2):379–395. doi: 10.1016/0022-2836(74)90526-9. [DOI] [PubMed] [Google Scholar]
- Young R. A., Macklis R., Steitz J. A. Sequence of the 16 S-23 s spacer region in two ribosomal RNA operons of Escherichia coli. J Biol Chem. 1979 May 10;254(9):3264–3271. [PubMed] [Google Scholar]
- Young R. A., Steitz J. A. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3593–3597. doi: 10.1073/pnas.75.8.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]




