Abstract
Glial tumors are the main primary adult brain tumor. Even with the most advanced treatments, which include stereotactic microscope aided surgical resection, internal and external radiation therapy and local and systemic chemotherapy, median survival time for patients diagnosed with these malignancies is about 12 months. We explore here the possibility that the endoplasmic reticulum stress response (ERSR) could be a possible target to develop chemotherapeutic agents to induce toxicity in glioma cells. ERSR has the dual capacity of activating repair and/or cytotoxic mechanisms. ERSR is triggered by the accumulation of unfolded proteins in the ER. The presence of unfolded proteins in the ER regulates, via a complex biochemical cascade, the upregulation of molecular chaperones, inhibition of protein synthesis, and an increase of proteasome mediated unfolded protein degradation. ERSR in particular conditions can also contribute to cell death via activation of programmed cell death. Apoptosis activation during ERSR is usually caused by the activation of one or a combination of three biochemical cascades. Induction of these pathways ultimately leads to caspase 3 activation culminating in apoptosis. Glioma cells are in a condition of constant low grade ERSR, which possibly contributes to their resistance to treatment protocols. It is conceivable that small molecules that interact with this phenomenon ultimately could be used to modulate the system to activate apoptosis and cause gliotoxicity. We will discuss here ERSR biochemically relevant features to death mechanisms and already identified small molecules that by modulating ERSR are able to activate glioma cell death.
Keywords: Endoplasmic reticulum stress response, glioma, calcium, apoptosis, caspase 4/12, small molecules, gliotoxicity
1. INTRODUCTION
Malignant gliomas are the most common adult primary brain tumor. Gliomas, which include astrocytomas, oligodendrocytomas, ependymomas, and glioblastomas, although relatively rare (1.3% incidence, 2.2% total cancer deaths) typically represent 40% of all adult brain tumors and 80% of all adult primary brain malignant tumors (www.cancer.org accessed December 13, 2010). They are characterized by local proliferation, insidious infiltration throughout the brain parenchyma, and robust angiogenesis. Current conventional treatment protocols include microscope aided surgical resection, internal and external radiation therapy and local and systemic chemotherapy. Several chemotherapeutic agents, such as temozolomide (TMZ), cisplatin, carmustine, and lomustine, have been used to slow the progression of these incurable cancers. However, glioma cells often acquire resistance to these agents, typically through epigenetic inactivation of the DNA repairing enzyme methylguanine methyltransferase (MGMT) [1]. The median survival time for patients diagnosed with glioblastomas is about 12 months [2]. These dismal outcomes warrant an expansion of the research efforts to search for molecular pathways to develop novel antiglioma agents and therapies.
The endoplasmic reticulum stress response (ERSR) has recently received much attention as a molecular pathway that can be modulated to cause cytotoxicity in glioma cells, mostly via apoptosis. This review will examine our current understanding of ERSR induced apoptosis and address its role in inhibiting oncogenic activity and promoting selective cytotoxicity in glioma cells. We will also review agents used to induce ERSR and their possible role as antiglioma therapies.
2. ERSR
The ER is central to many cellular functions related to protein synthesis, storage, sorting, and targeting and is also heavily involved in Ca2+ signaling. The high Ca2+ concentration in the ER environment is essential for the rapid Ca2+ mobilization following the elevation of the second messenger inositol (1, 4, 5) trisphosphate triggered by agonist-induced phospholipase C activation on the plasma membrane. Additionally, the high ion concentration in the ER is also intended for the purpose of aiding protein folding by promoting the binding of molecular chaperones (e.g. glucose regulated proteins GRP78 and 94) to growing polypeptide chains. The presence of Ca2+ binding proteins in the ER, in turn allows Ca2+ storage to occur at high concentrations. The ER also contains proteins that participate in posttranslational modifications of nascent proteins, such as disulfide bond formation and N-linked glycosylation (NLG) [3]. The highly oxidizing ER lumen, favors disulfide bond formation used to stabilize protein folding. Disturbances in any of these vital functions results in an increase of unfolded proteins in the ER, which initiate the phenomenon known as ERSR [3]. The goal of the ERSR is to enhance protein folding capabilities, reduce new protein synthesis, and clear malformed proteins to allow the return of normal cellular function. However, when it fails to do so, ERSR can trigger apoptotic cell death to eliminate compromised cells.
The ability of the ERSR to reduce the presence of unfolded/nonfunctional proteins is a complex task. The reduction of unfolded proteins are accomplished in four ways: (a) increased protein folding capabilities via upregulation of molecular chaperones, mainly GRP78 but also GRP94; (b) reduction of general protein translation, (c) enhanced degradation of unfolded or damaged proteins, and (d) activation of the antiapoptotic transcription factor nuclear factor kappa B (NF-κB) [4]. These tasks can be modulated through the disinhibition of three factors, activating transcription factor 6 (ATF6), inositol requiring enzyme (IRE1), and PKR like endoplasmic reticulum kinase (PERK).
In normal conditions (with low amounts of unfolded proteins in the ER), GRP78 binds the three ER transmembrane proteins, ATF6, IRE1, and PERK and inhibits their activity. GRP78 is therefore the main sensor for protein folding and the main actuator of the ERSR. GRP78 is formed by two functional domains. The larger 44 kDa domain possesses ATPase activity and the smaller domain of 20 kDa constitutes the protein binding domain [5]. A third domain largely composed of helical structure of 10 kDa has unknown functions [5]. GRP78 is constitutively present, although at relatively low levels, in all cells [6]. In the absence of unfolded proteins, this chaperone exists in its inactive state and bound to ATP. Following binding of unfolded proteins to the 20 kDa moiety, conformational changes trigger ATP hydrolysis in the 44 kDa moiety. The presence of ADP in the 44 kDa subunit increases the affinity for unfolded protein and in parallel reduces the ability of the chaperone to bind AFT6, IRE1, PERK, and other GRP78 bound proteins [5]. The latter proteins are now free and available to exert their roles to transduce ERSR signals [7–9].
3. DEPLOYMENT OF ERSR
3.1. ATF6
Once ATF6 is freed from GRP78, ATF6 translocates from the ER to the Golgi where it is cleaved into two fragments by the Golgi enzymes, site 1 protease (S1P) and site 2 protease (S2P) [10, 11]. This proteolytic processing yields an inactive smaller C terminal fragment and an active 50kDa cytosolic N-terminal fragment that encodes a basic region leucine zipper (bZIP) transcription factor [12]. The 50kDa active ATF6, that now exposes the nuclear targeting sequence, translocates to the nucleus, where it binds in association with NF-Y to promoters that contain ERSR elements (ERSE) resulting in their transcriptional initiation [13]. GRP78 and the transcription factor, CCAAT/enhancer binding homologous protein (CHOP), are the classical ERSR genes, whose promoters contain ERSE domains and, therefore, are highly induced during ERSR by the aforementioned mechanism [14]. ATF6 and X-box binding protein 1 (XBP1) [15] can also heterodimerize and upregulate the expression of genes that participate in protein removal e.g. ER degradation-enhancing α mannosidase like protein 1 (EDEM1) [16], which processes unfolded proteins so that they can be promptly ubiquitined and destroyed through the energy dependent proteasomal degradation activated by the ER associated degradation system (ERAD) [3].
3.2. IRE1
IRE1 is a 100kDa ER transmembrane protein with kinase and endonuclease activities [17]. Following dissociation from GRP78, IRE1 undergoes oligomerization and activation via autophosphorylation [18]. Active IRE1 facilitates the removal of a 26 nucleotide 3′ intron [19] from XBP1, thus activating the 41kDa bZIP transcription factor (XBP1), and hence causing the upregulation of genes responsible for protein folding (GRP78) and degradation (e.g. EDEM1) [20].
3.3. PERK
Like IRE1, PERK undergoes oligomerization and activation via autophosphorylation [21], following dissociation from GRP78 [22]. Activated PERK phosphorylates the eukaryotic initiating factor 2α (eif2α), which results in its activation [23]. Activated eif2α causes the inhibition of mRNA translation, which selectively inhibits protein synthesis and thereby causes a reduction of the overall ER protein load [3]. Certain mRNA, such as GRP78, CHOP, activating transcription factor 4 (ATF4), and NF-κB, can selectively opt out of such an inhibition of translation so that ERSR driven positive gene expression can be sustained. Phosphorylation of eif2α also results in ATF4 activation and its nuclear translocation [24]. Although with lower efficiency, ATF4 can activate ERSE sequence containing promoters in a manner analogous to ATF6, however the main trust of ATF4 is its cooperation with cAMP response element-binding (CREB) system [25], requiring simultaneous adenylate cyclase activation. Redundantly, ATF4 results in the enhancement of the transcription of genes involved in protein folding and degradation [25, 26]. Additionally, activated eif2α can also induce phosphorylation of the inhibitor of NF-κB leading to NF-κB activation and translocation [27]. Among many other effects, NF-κB activation promotes upregulation of antiapoptotic Bcl2 [28].
4. ACTIVATION OF APOPTOSIS DURING ERSR
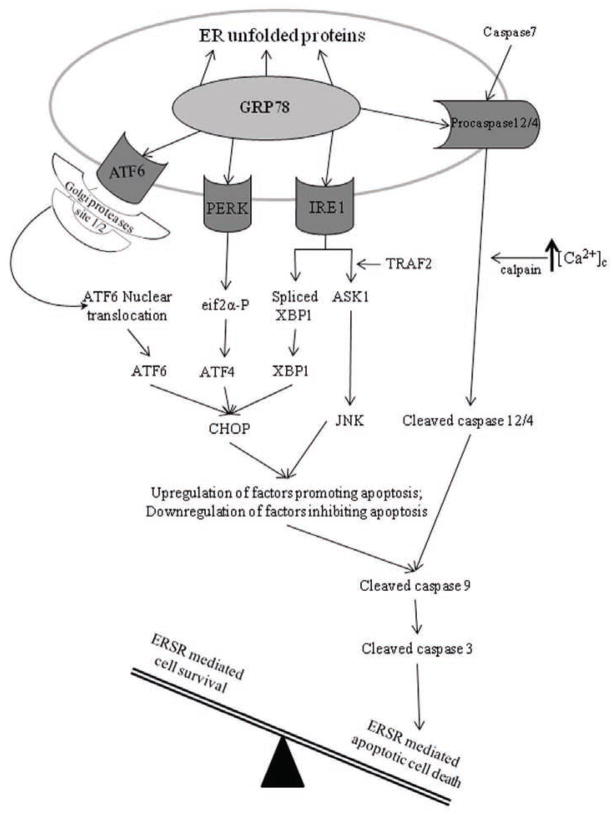
In addition, to its protective role, ERSR has been shown to initiate cellular death in various cell types. ERSR can trigger apoptosis when high levels of unfolded proteins are persistent [17, 29, 30]. However, the mode of such a switch is still obscure. To date, three ERSR mediated apoptotic pathways have been characterized: (1) activation of CHOP, (2) activation of ER associated caspases, and (3) c-Jun N-terminal kinase (JNK)-mediated apoptosis activation (Fig. 1).
Fig. 1. Schematic representation of the ERSR proapoptotic pathways.
During ERSR, ATF6, PERK, IRE1 and ER-associated caspases 4/12 pathways are used to activate apoptosis. ATF6, PERK, and IRE1 pathways can all converge and increase the expression of CHOP, which promotes the transcriptional upregulation of proapoptotic genes and the inhibition of antiapoptotic genes. During ERSR deployment, IRE1 and TRAF2 form a complex, which results in activation of ASK1. Following ASK1 activation, JNK can transcriptionally modulate genes that suppress cell survival and promote apoptosis. During ERSR, caspase 7 and calpain can cleave/activate procaspase 4/12. CHOP and JNK activated signaling results in mitochondrial apoptosis activation, which can synergize with active caspase 4/12 to cause the activation of caspase 9, which in turn activates caspase 3 ultimately culminating in apoptosis.
4.1. Role of CHOP in ERSR Mediated Apoptosis
CHOP activation has been the target of intense study and is the most characterized apoptotic pathway activated by ERSR. CHOP is normally expressed at low levels in unchallenged conditions [36–38]. The CHOP promoter, like GRP78, ATF4, and NF-κB, contains binding sites known as ERSE and their mRNA have the “opt out” sequence that is used to circumvent the eif2α mediated inhibition of global protein synthesis. Therefore these factors can be significantly unregulated during ERSR [31]. Activated ATF6, IRE1, and PERK signals converge on the CHOP promoter and result in a significant enhancement of transcription of this factor [24, 32–34] (Fig. 1). The role of CHOP (also known as growth arrest and DNA damage-inducible gene 153 (GADD153)) in ERSR and apoptosis has been extensively studied. CHOP−/− cells are resistant to ERSR induced apoptosis [35]. Furthermore, overexpression of CHOP in cells results in the enhancement of apoptotic cell death [36]. While it is unlikely that CHOP directly induces apoptotic death, CHOP transcriptionally regulates genes that participate in the apoptotic pathway.
CHOP level increase is associated with indirect inhibition of Bcl2 expression which, in turns, unleashes the apoptotic triggering effect of Bax/Bad systems in the mitochondria, resulting in caspase 9 and then caspase 3 activation [37] (Fig. 1). Of interest is that the effect of CHOP on Bcl2 can be antagonized by a concomitant activation of NF-κB, which results in upregulation of Bcl2 expression [36, 37].
CHOP also promotes ERSR mediated apoptosis by activating the proapoptotic factor Bim [38]. Another possible CHOP target that could result in apoptosis induction is the death receptor 5 (DR5), which is upregulated following the induction of CHOP as it shown in human carcinoma cells [39]. Following siRNA knockdown of CHOP, reduced DR5 upregulation and apoptosis were observed in the presence of ERSR caused by thapsigargin (THAP), an ERSR inducer that depletes ER Ca2+ by preventing its ER uptake [39, 40].
4.2. Role of ER Associated Caspase Activation in ERSR Mediated Apoptosis
A second ERSR directed apoptosis activation system is constituted by the species specific ER associated caspases 4 and 12. These particular caspases are located in the ER membrane and are bound, like ATF6, IRE1 and PERK, to GRP78 in resting conditions, and hence their activation is inhibited. Unfolded proteins appearance, similar to the other GRP78 inhibited factors, also decreases GRP78’s affinity for caspase 4/12 and results in their dissociation, rendering them ready for further activation [41, 42]. Free procaspases 4/12 are cleaved by calpain in the presence of elevated cytoplasmic Ca2+ concentrations ([Ca2+]c) into the active forms that can lead to the initiation of apoptosis [43] (Fig. 1).
Caspase 12 is a rodent specific [44, 45] cysteine aspartate dependent selective protease that is localized on the cytosolic side of the ER [43, 46]. As in the case of most caspases, caspase 12 exist as a zymogen whose cleavage results in an active caspase [47]. Caspase 12 −/− cells have been shown to be resistant to cell death instigated by ER stress suggesting caspase 12 has a major role in ERSR mediated apoptosis [46].
ERSR results in cytosolic translocation of cleaved caspase 12 which can now activate caspase 9, and initiate apoptosis [45] (Fig. 1). A causal relationship between caspase 9 and 12’s ability to induce cell death was suggested by experiments. Caspase 9 carrying an inactive mutated catalytic domain could not interact with caspase 12 and ERSR mediated apoptosis was reduced [45].
Caspase 4 is a human specific caspase, which has 57% homology to rodent caspase 12, and is considered to be an orthologue [48]. Caspase 4, like caspase 12, is specifically readied for activation when released by GRP78 during ERSR, and its calpain dependent cleavage in a high Ca2+ environment generates the active fragment that is involved in apoptosis initiated by ERSR [49, 50]. Cancer cell lines, such as HeLa and SK-N-SH, treated with siRNA against caspase 4 exhibited a reduction of ERSR mediated apoptosis. However, caspase 4 knockdown cells could still activate apoptosis through other mechanisms, independently of ERSR induction [49]. It has also been shown that caspase 4 is activated during ERSR induction in malignant human glioma [51–53].
Studies associated with the Cancer Genome Atlas project indicate that caspase 4 is upregulated in patient derived glioma specimens (http://tcga.cancer.gov accessed November 1, 2010), thus suggesting that ERSR activation in glioma cells could be associated with enhancement of apoptosis given the presence of higher levels of caspase 4.
4.2.1. ER associated Caspase Activators - Ca2+ calpain System
Cytoplasmic Ca2+ concentration ([Ca2+]c) plays a major role in the activation of ER associated caspases that ultimately leads to ERSR induced apoptosis. Caspase 12 activation has been shown to occur when the ER Ca2+ stores are depleted by THAP, a condition that results also in a significant and prolonged elevation of [Ca2+]c. The prolonged reduction of [Ca2+]ER leads to the unfolding of ER proteins because of the chaperone dependence on ER Ca2+ and the lack of the ion that serves as nucleation factor for many folded proteins. Increased levels of unfolded proteins in the ER cause the concurrent release of ER associated caspases from GRP78. In the presence of the simultaneous elevation of [Ca2+]c, freed procaspase 12 can be cleaved by calpain into its active form (Fig. 1). In mouse embryonic fibroblasts (MEF) deficient in calpain, both caspase 12 proteolytic cleavage and ERSR mediated apoptosis were abolished [54]. Similarly, caspase 4 cleavage and the subsequent apoptosis were attenuated in cells where [Ca2+]c in the cytoplasm was kept low by high extracellular EGTA concentration treatment [50]. The one-two punch, consisting of disinhibition of ER associated procaspases and elevation of [Ca2+]c, is responsible for the activation of the caspases and is an essential part of ERSR induced apoptosis (Fig. 1).
4.2.2. ER Associated Caspase Activators – Caspase 7
ER associated caspases, such as caspase 4/12, can also be activated using another signaling system. In resting conditions, caspase 12 is bound to GRP78 and is shielded from proteolytic cleavage and activation. In the presence of unfolded proteins, caspase 12 dissociates from GRP78 and the procaspase can be cleaved by activated caspase 7, which translocates from the cytosol to the ER surface during ERSR [45] (Fig. 1). Caspase 7 proteolytic action on caspase 12 takes place on the residues Asp-94 and Asp-341, which interestingly, are different from the activation sites utilized by calpain [43]. The implication for this differential processing are not completely understood. Dominant negatives for the catalytic domain of caspase 7 attenuate caspase 12 dependent apoptosis supporting the role of this pathway on ERSR induced cell death [45].
4.3. Role of JNK Activation in ERSR Mediated Apoptosis
IRE1 activation has also been implicated in ERSR mediated apoptosis involving JNK activation. Activated IRE1 recruits cytosolic TNF receptor-associated factor 2 (TRAF2) to the ER membrane. The IRE1-TRAF2 complex, established during ERSR induction, activates TNF-dependent apoptosis-signaling kinase 1 (ASK1) [55], which causes the activation of mitogen activated protein kinase (MAPK), JNK [55, 56] (Fig. 1). The resultant JNK activation results in both Bcl2 downregulation, which in turns removes suppression of the mitochondrial apoptosis cascade, and proapoptotic Bim activation, [57–59] resulting in a strong induction of apoptotic cell death. The importance of the IRE1-TRAF2-ASK-JNK pathway in ERSR mediated apoptosis has been demonstrated in several studies. Primary neurons from ASK−/− mice displayed reduced JNK activation and were resistant to apoptosis following ERSR induction [60]. ASK1 ablation via siRNA in HeLa cells attenuated ERSR mediated apoptosis following THAP treatment [61]. In addition, IRE1 inhibition resulted in increased glioma cell survival and angiogenesis in glioma implanted mice [62].
4.4. Cross Talk of Apoptosis Inducing Pathways in ERSR
Cross talk of the ERSR apoptotic pathways seems likely due to their shared molecular components. The CHOP induced events promote cell death by suppressing Bcl2 expression and thereby eliminating a checkpoint on mitochondrial determined apoptotic events by also promoting proapoptotic gene expression to directly cause apoptosis. During ERSR, increases in [Ca2+]c promote calpain mediated activation of ER associated caspases. The mitochondria- and the ER-dependent activated caspases converge on caspase 3, the final activator of apoptosis, and could result in a potentiation of the apoptotic program execution to trigger cell death.
5. ROLE OF ERSR IN TUMOR CELLS
Exaggerated growth and competition for nutritional resources in the tumor determines the hostile environment typical of the cancer mass, which results in increased levels of hypoxia and decreased glucose levels. Tumor cells have adapted to these harsh living conditions with aerobic glycolysis, the Warburg effect, to convert glucose, glutamine and serine to lactate for energy [63]. The lactic acid, which is a major catabolic product in these conditions, decreases the pH and aggravates local distress. The resulting limited amount of energy and oxygen affects ATP availability and therefore renders ATP-dependent pump function difficult. During high rates of proliferation, many processes are affected and could suffer from higher levels of misfolded proteins, DNA damage, and insufficient supply of nutrients in the ER to meet the demands of the rapidly dividing cells [64, 65]. For these reasons, an adaptive defense strategy is employed by the cell to counteract the continued exposure to stress. Chronically elevated levels of ERSR assist in protecting the cancer cell from the unfavorable environment in which they exist by causing a higher chaperone availability opposed to mildly increased unfolded protein levels [66]. However, additional ERSR, beyond a certain critical point, results in activation of a series of events that culminate in apoptosis due to the lack of compensating resources.
High levels of GRP78 have been reported in many cancer cell lines and patient samples, including glioma cells [67], and has been inversely correlated with disease outcome and tumor proliferation rates [53]. Constitutively, activated ERSR has been shown to be a protective mechanism in cancer cells and even participate in tolerance to chemotherapeutic agents. Protection could occur through suppression of proapoptotic pathways in mild ERSR conditions. Elevated GRP78 levels would increase the direct binding and inhibition of caspase 4/12 and other ERSR effectors, which are known to form inactive complexes with GRP78 [42]. Interestingly, GRP78 molecular overexpression, in the absence of unfolded proteins, leads to CHOP downregulation possibly contributing to tumor cells’ resistance to apoptosis [68]. Bax inhibition, and subsequently inhibition of the release of cytochrome c from the mitochondria, has also been associated with isolated elevated GRP78 levels [69].
ATF6, IRE1, or PERK cascades have been previously shown to be activated in tumor cells probably due to the conditions present within the hostile tumor environment, such as hypoxia and ischemia. ATF6 has been shown to be activated and translocated to the nucleus during ischemia [70]. When ATF6 expression was inhibited by microRNA (miRNA), cardiac myocyte death increased during ischemic conditions [70]. These findings support a protective role for ATF6 during ischemia. The IRE1-XBP1 arm of the ERSR cascade has been suggested to be essential for tumor growth under oncogenic stress. Fibroblast tumor cells lacking XBP1 were unable to grow as xenografts [71], showed increased levels of apoptosis, and decreased angiogenesis [72]. Indeed, IRE1 has been shown to directly increase angiogenesis through upregulation of vascular endothelial growth factor-A (VEGF-A) in response to hypoxia and hypoglycemia. Dominant negative IRE1 tumor cells displayed less vascularization and more invasive tumors in an orthotopic glioma model [73]. Hypoxia, which can also trigger ERSR, has been shown to activate PERK, leading to the phosphorylation of eif2α [74]. Tumors derived from PERK deficient animals were unable to cause sufficient angiogenesis to self sustain [75]. While many of these studies have not been performed in glioma, it is safe to hypothesize that the mildly elevated GRP78 levels present in glioma cells, likely associated with mildly elevated levels of unfolded proteins in the ER, are associated with low level activation of these three factors contributing to cell resilience.
While the basally elevated level of ERSR in tumor cells may actually be protective against chemotherapies, evidence also shows that further stimulation of ERSR in these cells is accompanied with enhanced cell death. Several studies have shown that downregulating GRP78, which leads to the activation of ERSR, augments the effectiveness of antineoplastic agents [53, 69], while increasing levels of GRP78, (which decreases ERSR), correlate to drug resistance [76, 77]. Knockdown of GRP78 augmented the effectiveness of temozolomide (TMZ), the chemotherapeutic current standard of care in malignant glioma [53]. Overexpression of GRP78 in glioma cells rendered cells resistant to apoptosis, decreased caspase 7 activation, and prevented sensitivity to cisplatin and etoposide [67]. Conversely, downregulation of GRP78 rendered the cells susceptible to cisplatin and etoposide and inhibited growth [67]. In conclusion, while low levels of ERSR are undoubtedly protective, higher levels of induction can push the cells coping mechanisms toward recruiting death mechanisms, a situation that can be exploited for antineoplastic treatments.
6. TARGETING ERSR WITH SMALL MOLECULES TO CAUSE CELL DEATH
Gliomas have been shown to be sensitive to agents that interfere with the ERSR. Agents affecting ER Ca2+ homeostasis, such as THAP, flavonoids (FLAV), curcumin (CUR), and non steroidal anti-inflammatory drugs (NSAID), such as celecoxib (CELE), cause protein unfolding and highly activate ERSR and cell death in glioma cells [40, 78–82]. Agents directly interfering with protein folding or maturation, such as tunicamycin (TUN) or brefeldin A (BFA), cause induction of ERSR and cell death. Agents able to affect misfolded protein removal, inhibitors of proteasome activity, such as specific agents like bortezomib (BOR) [51] and human immunodeficiency virus protease inhibitors (HIV-PIs) [51, 52], are associated with high levels of ERSR induction in glioma cells and cause gliotoxicity. Ideal candidate molecules for chemotherapeutic potential would increase ERSR in cancer cells enough to activate the prodeath arms of ERSR, yet only induce the prosurvival arms in normal cells, resulting in tumor specific effects. Such a strategy could result from the decreased ability of tumor cells to cope with additional ER stress compared to normal cells. This provides two options for targeting ERSR as an antineoplastic therapy: (1) the ERSR cascade itself could be targeted to induce cell death directly, or (2) ERSR could be exacerbated to make cells susceptible to other chemotherapies [66]. The following part of this review will focus on specific small molecules that have been found to elicit ERSR and cause apoptotic cell death in tumor cells and more specifically in glioma cells.
6.1. Agents Interfering with Ca2+ Regulated ERSR Phenomenon
Ca2+ has long been known to initiate and participate in multiple cell signaling pathways. This ubiquitous ion is stored in the ER, where its high concentrations aid in protein folding and serve as a fast acting reservoir for Ca2+ signaling. Sarco/endoplasmic reticulum ATPase (SERCA) 2 is the sole pump responsible for concentrating Ca2+ into the ER [83]. Ca2+ release from the ER is attained actively through inositol trisphosphate receptors (IP3R), ryanodine receptors (RynR) modulation, and passively through chemical diffusion and leakage associated with translocon activity during protein synthesis [84–86]. Temporary ER Ca2+ release is an event that occurs often during normal Ca2+ signaling [87]. However, prolonged ER Ca2+ store depletion, an unusual condition, has been shown to trigger protein unfolding. Protein unfolding in low [Ca2+]ER is due to failure in chaperone function and direct interference with protein nucleation [87]. Small molecules, interfering with this complex Ca2+ homeostatic process, have been shown to elicit ERSR mediated apoptosis in several cancer cell types.
6.1.1. Flavonoids
Flavonoids (FLAV) are plant-derived, naturally occurring polyphenolic compounds that possess the ability to elevate [Ca2+]c in U87MG and T98G human glioma cells [79]. FLAVs have therapeutic potential in cancer treatments due to their ability to induce apoptosis. GRP78 expression is decreased by (−)-epigallocatechin-3-gallate (EGCG), a flavonoid derived from green tea (Fig. 2). This effect could be associated with activation of the prodeath arm of ERSR. EGCG has been shown to directly interact with the ATP binding domain of GRP78 and render GRP78 unable to bind to the caspase 7 complex, ultimately culminating in increased activation of caspase 7 and apoptosis in etoposide treated cancer cells [80]. Interestingly, the FLAVs apigenin, (−)-epigallocatechin, genistein, and EGCG have been shown to have proapoptotic effects in U87MG and T98G human glioma cells while normal astrocytes are largely resistant [79]. FLAV triggered an increase in [Ca2+]c and caspase 4 activation in glioma cells [79] (Fig. 2). Additionally, ERSR mediated apoptosis following FLAV exposure was also accompanied by JNK activation, Bax upregulation, and cytochrome c release [79]. Activation of the mitochondrial dependent apoptotic pathway resulted in caspase 9 and 3 activation following exposure to FLAV [79]. Although it has not been definitively proven that ERSR induction was directly related to apoptosis of glioma cells induced by FLAV exposure, these data strongly suggest that ERSR could be a key participant in FLAV induced cell death.
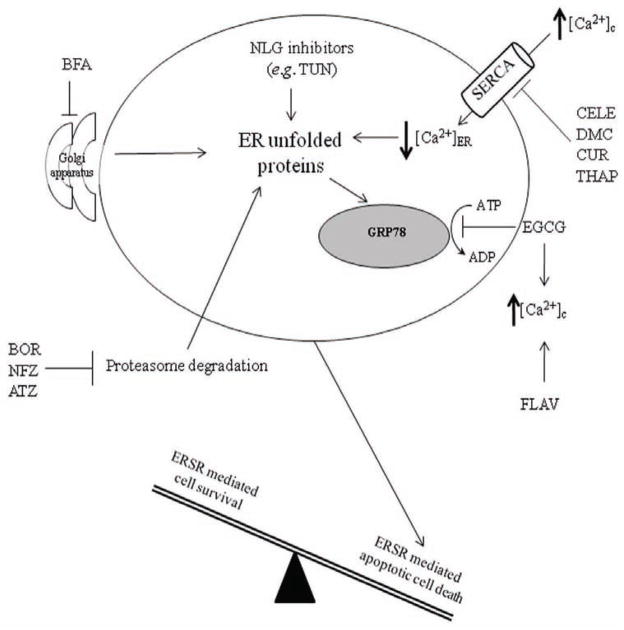
Fig. 2. Known interactions between small molecules and the ERSR in glioma.
Several compounds have been investigated for their ERSR inducing apoptotic properties in glioma cells. FLAV treatment results in increased [Ca2+]c. EGCG binds to GRP78 and prevents the formation of antiapoptotic GRP78-caspase 7 complex while also promoting elevated [Ca2+]c. THAP, CELE, DMC, and CUR are potent SERCA inhibitors that lead to reduced [Ca2+]ER. NLG inhibitors (e.g. TUN) prevent N-linked glycosylation of proteins leading to ER retention of unfolded proteins. BFA inhibits protein export from the ER to the Golgi thus promoting ER protein accumulation. BOR, NFZ, ATZ inhibit the proteasome and cause aged and unfolded protein accumulation in the ER.
6.1.2. SERCA inhibitors
THAP, a plant derived sesquiterpene lactone, deprives the ER of Ca2+ by irreversibly inhibiting SERCA, leading to ERSR activation culminating in cell death [88]. THAP has been widely regarded as a highly effective cytotoxic agent (Fig. 2). However, THAP is not generally considered a superior anticancer choice because (1) THAP is considered a tumor promoter [89], (2) THAP is not well tolerated experimentally in in vivo studies [66], and (3) irreversible, generalized SERCA inhibition, in the absence of selective tumor targeting, results in global cytotoxic effects extending to normal tissues [90–92]. Researchers have averted some of these undesirable effects by utilizing an inactive THAP prodrug that is selectively activated by a tumor specific antigen found only in prostate cancer cells and has shown favorable anticancer properties in animal studies [90].
SERCA inhibiting compounds may still be a viable option for tumor cytotoxicity despite some of the therapeutic setbacks of THAP. Recently, diaryl substituted pyrazoles have been identified to inhibit SERCA and have antitumor activity [81, 82]. A prominent member of this family is the well characterized NSAID, CELE. This NSAID, a COX2 inhibitor, has been shown to be a reversible SERCA inhibitor [81], with potent cytotoxic potential in glioma cells [82]. It has been suggested that CELE induced SERCA inhibition elicits ERSR independently of its COX2 inhibitory properties. In fact, the CELE structural analog, 2,5–dimethyl – celecoxib (DMC), which is devoid of COX 2 inhibitory activity, completely retains SERCA inhibitory activity and induces ERSR and apoptosis [82]. CELE and DMC effects correlated with the inhibition of SERCA, ER Ca2+ depletion, and upregulation in GRP78, CHOP, and caspase 4 typically observed during ERSR [82] (Fig. 2). Furthermore, CELE and DMC elicited ERSR and apoptosis in in vivo studies using U87MG tumor xenografts [82]. However, both molecules are not good drug candidates because of their unfavorable CNS bioavailability and side effects at the dosages required to elicit these effects.
Curcumin (CUR) is an anticancer polyphenolic compound found in the culinary spice, turmeric. CUR is a potent reversible inhibitor of all three known SERCA isoforms [78], and like THAP has been shown to induce ERSR and apoptosis in tumor cells (Fig. 2). Recent reports have confirmed activation of ERSR following CUR treatment, as demonstrated by upregulation of GRP78, CHOP, and induction of apoptosis in human non-small cell lung cancer cells [93]. CUR treated glioma cells exhibit increased calpain activation, in addition, to an increase in Bax:Bcl2 ratio and caspase 3 cleavage [94]. Finally, CUR treatment inhibited brain tumor formation in orthotopic mouse models [95].
6.2. Small Molecule that Affect Protein Maturation and Sorting
6.2.1. N-linked Glycosylation Inhibitors
Tunicamycin (TUN) inhibits N-linked glycosylation (NLG) of proteins and represents a classic ERSR inducer (Fig. 2). The role of inhibiting NLG and its effects on glioma receptor tyrosine kinases (RTK) expression have recently been investigated [96, 97]. In U251 human glioma cells, expression of epidermal growth factor receptor (EGFR), an RTK that is overexpressed in 40–90% of glioblastomas [98], was shown to be particularly sensitive to NLG inhibition following TUN treatment [96]. NLG disruption resulted in decreased RTK signaling, possibly due to the inability of receptor maturation culminating in ER retention [96]. Glioma cells were highly sensitive to radiation following exposure to NLG inhibitors [96]. Furthermore, NLG inhibition had no effect on normal tissue, indicating a favorable therapeutic ratio between normal and malignant tissues [96]. This novel potential antiglioma cancer therapy approach was further validated in vivo using D54 and U87MG glioma xenograft tumor models [97]. NLG inhibition yielded significant reductions in tumor growth following concomitant treatment with TUN and radiation therapy [97]. These results suggest that ERSR induction, via NLG inhibition, could prove to be a selective inducer of apoptosis in glioma cells (Fig. 2).
6.2.2. Golgi Disruptors: Brefeldin A
Brefeldin A (BFA), a macrocyclic lactone, destroys Golgi maturation and therefore disrupts the flow of proteins from the ER, through the Golgi and then to the plasma membrane, effectively halting the normal sorting of membrane proteins [99]. The accumulation of these unshuttled proteins in the ER activates ERSR [100, 101]. Apoptosis has been shown to occur in glioma cells following BFA treatment [102]. BFA has also been investigated in glioma cells and shown to downregulate membrane type 1 matrix metalloproteinase (MT1-MMP), an enzyme involved in glioma invasion and metastasis [103]. Downregulation of MT1-MMP expression might prove to be advantageous in targeting glioma cells toward apoptotic cell death. Prouix Bonneau et al. sought to determine whether MT1-MMP expression in glioma cells could be attenuated following ERSR induction [104]. BFA treated cells failed to express MT1-MMP on the cell surface and showed increased GRP78 expression, suggesting that abrogated protein trafficking leads to ERSR activation. Pommepuy et al. observed increased apoptotic glioma cell death after exposure to BFA [102] (Fig. 2). Further studies are needed to determine whether BFA induced downregulation of MT1-MMP results in apoptotic cell death.
6.2.3. Proteasome Inhibitors
Proteasome degradation is a major component of relieving ERSR through the clearance of unfolded proteins. ERSR induced EDEM activity prepares unfolded proteins for the action of the ERAD system, which promotes export of unfolded proteins to the cytosol, where they are subsequently tagged with ubiquitin and degraded by the proteasome [3]. Following IRE1 activation, XBP1 increases the expression of proteins (e.g. EDEM) [105–108], whose action prepare unfolded proteins to be processed by ubiquitin ligases, the first step in proteasome mediated protein degradation [109]. Failure of the proteasome to degrade proteins results in an accumulation of unfolded proteins in the ER and aggravates ERSR. Several agents can induce ERSR by inhibiting the proteasome. Small molecules, such as BOR and HIV-PIs, have been reported to inhibit the proteasome and cause a backlog of unfolded proteins. BOR, a classic selective inhibitor of 26S proteasome, has been FDA approved for treatment of multiple myeloma. Its anticancer effects have also been investigated in the treatment of glioma. Single agent treatment with BOR activated ERSR as indicated by increased GRP78 and CHOP expression and caspase 4 activation [51]. BOR has also been shown to trigger ERSR mediated apoptosis in glioma cells [51, 52] (Fig. 2). Results from in vitro studies have shown cytostatic [51] and significant tumoricidal effects in BOR treated glioma cells [110].
HIV-PIs, developed primarily to inhibit the HIV proteases, also have the ability to inhibit the proteasome. Nelfinavir (NFZ) and atazanavir (ATZ) have recently been investigated for their role in ERSR mediated apoptosis in glioma cells. It has been reported that NFZ and ATZ induced ERSR via proteasome inhibition [52] (Fig. 2). In this condition, ERSR classic markers, such as GRP78 and CHOP, were upregulated and caspase 4 was activated, clearly indicating ERSR induction. Downregulation of caspase 4 by siRNA decreased the HIV-PIs’ cytotoxic effect in glioma cells [52]. Furthermore, U87MG glioma tumor skin xenograft models exhibited reduced tumor growth following NFZ treatment [52].
7. ROLE OF APOPTOSIS IN COMBINATORIAL THERAPY TARGETING ERSR
Cancers cells are widely considered to live in a state of low and chronic ERSR, which is thought to promote tumor cell survival [66]. Such an adaptation, triggered by the harsh environment, can also play a role in chemoresistance [69]. BOR, a proteasome inhibitor, in combination with CELE or DMC, SERCA inhibitors, potently triggered ERSR as indicated by increased expression of GRP78, CHOP, JNK, and also strongly activated caspases 4 and 7, thus causing an enhancement of apoptotic glioblastoma cell death. Likewise, BOR in combination with TMZ (DNA methylating agent [111] and CHOP inducer [53]) and radiotherapy increased patient survival to a median survival of 17.4 months in a Phase I clinical trial [112].
8. CONCLUSION AND FUTURE DIRECTIONS
This review summarizes the current evidences indicating that ERSR induction can be utilized to activate apoptotic cell death preferentially in cancer cells. Moreover, ERSR mediated apoptosis can be a selective glioma target to develop novel chemotherapeutic agents. ERSR inducing agents have the potential to become powerful anticancer agents for gliomas and other cancer cells. Numerous reports suggest that ERSR mediated apoptosis is able to cause selective glioma cell death. A recent Phase I clinical trial result indicates that combination of ERSR inducing compounds, BOR and TMZ, reduce tumor growth in patients diagnosed with malignant glioma [112]. These data indicate that ERSR induction in glioma cells could be a valuable option to develop novel and effective anti-neoplastic agents; however more studies are necessary to completely unravel the potential of such an approach.
References
- 1.Hegi ME, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 2.Furnari FB, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 3.Schroder M, Kaufman RJ. ER stress and the unfolded protein response. Mutat Res. 2005;569(1–2):29–63. doi: 10.1016/j.mrfmmm.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 4.Rao RV, Ellerby HM, Bredesen DE. Coupling endoplasmic reticulum stress to the cell death program. Cell Death Differ. 2004;11(4):372–80. doi: 10.1038/sj.cdd.4401378. [DOI] [PubMed] [Google Scholar]
- 5.Chevalier M, et al. Interaction of murine BiP/GRP78 with the DnaJ homologue MTJ1. J Biol Chem. 2000;275(26):19620–7. doi: 10.1074/jbc.M001333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Lee AS. Stress induction of GRP78/BiP and its role in cancer. Curr Mol Med. 2006;6(1):45–54. doi: 10.2174/156652406775574523. [DOI] [PubMed] [Google Scholar]
- 7.Freiden PJ, Gaut JR, Hendershot LM. Interconversion of three differentially modified and assembled forms of BiP. Embo J. 1992;11(1):63–70. doi: 10.1002/j.1460-2075.1992.tb05028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendershot LM, Ting J, Lee AS. Identity of the immunoglobulin heavy-chain-binding protein with the 78,000-dalton glucose-regulated protein and the role of posttranslational modifications in its binding function. Mol Cell Biol. 1988;8(10):4250–6. doi: 10.1128/mcb.8.10.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laitusis AL, Brostrom MA, Brostrom CO. The dynamic role of GRP78/BiP in the coordination of mRNA translation with protein processing. J Biol Chem. 1999;274(1):486–93. doi: 10.1074/jbc.274.1.486. [DOI] [PubMed] [Google Scholar]
- 10.Shen J, et al. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3(1):99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 11.Ye J, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6(6):1355–64. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 12.Haze K, et al. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10(11):3787–99. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshida H, et al. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem. 1998;273(50):33741–9. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 14.Okada T, et al. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem J. 2002;366(Pt 2):585–94. doi: 10.1042/BJ20020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23(21):7448–59. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu J, et al. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13(3):351–64. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7(12):1013–30. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 18.Tirasophon W, et al. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 2000;14(21):2725–36. doi: 10.1101/gad.839400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida H, et al. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–91. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 20.Lee K, et al. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16(4):452–66. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sood R, et al. Pancreatic eukaryotic initiation factor-2alpha kinase (PEK) homologues in humans, Drosophila melanogaster and Caenorhabditis elegans that mediate translational control in response to endoplasmic reticulum stress. Biochem J. 2000;346(Pt 2):281–93. [PMC free article] [PubMed] [Google Scholar]
- 22.Bertolotti A, et al. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2(6):326–32. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 23.Prostko CR, et al. Phosphorylation of eukaryotic initiation factor (eIF) 2 alpha and inhibition of eIF-2B in GH3 pituitary cells by perturbants of early protein processing that induce GRP78. J Biol Chem. 1992;267(24):16751–4. [PubMed] [Google Scholar]
- 24.Harding HP, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6(5):1099–108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 25.Luo S, et al. Induction of Grp78/BiP by translational block: activation of the Grp78 promoter by ATF4 through and upstream ATF/CRE site independent of the endoplasmic reticulum stress elements. J Biol Chem. 2003;278(39):37375–85. doi: 10.1074/jbc.M303619200. [DOI] [PubMed] [Google Scholar]
- 26.Harding HP, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11(3):619–33. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 27.Jiang HY, et al. Phosphorylation of the alpha subunit of eukaryotic initiation factor 2 is required for activation of NF-kappaB in response to diverse cellular stresses. Mol Cell Biol. 2003;23(16):5651–63. doi: 10.1128/MCB.23.16.5651-5663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang CY, et al. NF-kappaB induces expression of the Bcl-2 homologue A1/Bfl-1 to preferentially suppress chemotherapy-induced apoptosis. Mol Cell Biol. 1999;19(9):5923–9. doi: 10.1128/mcb.19.9.5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Healy SJ, et al. Targeting the endoplasmic reticulum-stress response as an anticancer strategy. Eur J Pharmacol. 2009;625(1–3):234–46. doi: 10.1016/j.ejphar.2009.06.064. [DOI] [PubMed] [Google Scholar]
- 30.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–89. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 31.Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11(4):381–9. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- 32.Ma Y, et al. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J Mol Biol. 2002;318(5):1351–65. doi: 10.1016/s0022-2836(02)00234-6. [DOI] [PubMed] [Google Scholar]
- 33.Wang XZ, et al. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. Embo J. 1998;17(19):5708–17. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida H, et al. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20(18):6755–67. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oyadomari S, et al. Nitric oxide-induced apoptosis in pancreatic beta cells is mediated by the endoplasmic reticulum stress pathway. Proc Natl Acad Sci U S A. 2001;98(19):10845–50. doi: 10.1073/pnas.191207498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCullough KD, et al. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol. 2001;21(4):1249–59. doi: 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma Y, Hendershot LM. The role of the unfolded protein response in tumour development: friend or foe? Nat Rev Cancer. 2004;4(12):966–77. doi: 10.1038/nrc1505. [DOI] [PubMed] [Google Scholar]
- 38.Puthalakath H, et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007;129(7):1337–49. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 39.Yamaguchi H, Wang HG. CHOP is involved in endoplasmic reticulum stress-induced apoptosis by enhancing DR5 expression in human carcinoma cells. J Biol Chem. 2004;279(44):45495–502. doi: 10.1074/jbc.M406933200. [DOI] [PubMed] [Google Scholar]
- 40.Thastrup O, et al. Thapsigargin, a tumor promoter, discharges intracellular Ca2+ stores by specific inhibition of the endoplasmic reticulum Ca2(+)-ATPase. Proc Natl Acad Sci U S A. 1990;87(7):2466–70. doi: 10.1073/pnas.87.7.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang CC, et al. Inhibition of MEK sensitizes human melanoma cells to endoplasmic reticulum stress-induced apoptosis. Cancer Res. 2007;67(20):9750–61. doi: 10.1158/0008-5472.CAN-07-2047. [DOI] [PubMed] [Google Scholar]
- 42.Rao RV, et al. Coupling endoplasmic reticulum stress to the cell death program: role of the ER chaperone GRP78. FEBS Lett. 2002;514(2–3):122–8. doi: 10.1016/s0014-5793(02)02289-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakagawa T, Yuan J. Cross-talk between two cysteine protease families. Activation of caspase-12 by calpain in apoptosis. J Cell Biol. 2000;150(4):887–94. doi: 10.1083/jcb.150.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Imaizumi K, Katayama T, Tohyama M. Presenilin and the UPR. Nat Cell Biol. 2001;3(5):E104. doi: 10.1038/35074613. [DOI] [PubMed] [Google Scholar]
- 45.Rao RV, et al. Coupling endoplasmic reticulum stress to the cell death program. Mechanism of caspase activation. J Biol Chem. 2001;276(36):33869–74. doi: 10.1074/jbc.M102225200. [DOI] [PubMed] [Google Scholar]
- 46.Nakagawa T, et al. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature. 2000;403(6765):98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- 47.Rao RV, et al. Coupling endoplasmic reticulum stress to the cell death program. An Apaf-1-independent intrinsic pathway. J Biol Chem. 2002;277( 24):21836–42. doi: 10.1074/jbc.M202726200. [DOI] [PubMed] [Google Scholar]
- 48.Fischer H, et al. Human caspase 12 has acquired deleterious mutations. Biochem Biophys Res Commun. 2002;293(2):722–6. doi: 10.1016/S0006-291X(02)00289-9. [DOI] [PubMed] [Google Scholar]
- 49.Hitomi J, et al. Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Abeta-induced cell death. J Cell Biol. 2004;165(3):347–56. doi: 10.1083/jcb.200310015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsuzaki S, et al. Caspase-4 is partially cleaved by calpain via the impairment of Ca2+ homeostasis under the ER stress. Neurochem Int. 2010;56(2):352–6. doi: 10.1016/j.neuint.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 51.Kardosh A, et al. Aggravated endoplasmic reticulum stress as a basis for enhanced glioblastoma cell killing by bortezomib in combination with celecoxib or its non-coxib analogue, 2,5-dimethyl-celecoxib. Cancer Res. 2008;68(3):843–51. doi: 10.1158/0008-5472.CAN-07-5555. [DOI] [PubMed] [Google Scholar]
- 52.Pyrko P, et al. HIV-1 protease inhibitors nelfinavir and atazanavir induce malignant glioma death by triggering endoplasmic reticulum stress. Cancer Res. 2007;67(22):10920–8. doi: 10.1158/0008-5472.CAN-07-0796. [DOI] [PubMed] [Google Scholar]
- 53.Pyrko P, et al. The unfolded protein response regulator GRP78/BiP as a novel target for increasing chemosensitivity in malignant gliomas. Cancer Res. 2007;67(20):9809–16. doi: 10.1158/0008-5472.CAN-07-0625. [DOI] [PubMed] [Google Scholar]
- 54.Tan Y, et al. Ubiquitous calpains promote caspase-12 and JNK activation during endoplasmic reticulum stress-induced apoptosis. J Biol Chem. 2006;281(23):16016–24. doi: 10.1074/jbc.M601299200. [DOI] [PubMed] [Google Scholar]
- 55.Nishitoh H, et al. ASK1 is essential for JNK/SAPK activation by TRAF2. Mol Cell. 1998;2(3):389–95. doi: 10.1016/s1097-2765(00)80283-x. [DOI] [PubMed] [Google Scholar]
- 56.Urano F, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287(5453):664–6. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 57.Hetz C, et al. The proapoptotic BCL-2 family member BIM mediates motoneuron loss in a model of amyotrophic lateral sclerosis. Cell Death Differ. 2007;14(7):1386–9. doi: 10.1038/sj.cdd.4402166. [DOI] [PubMed] [Google Scholar]
- 58.Lei K, Davis RJ. JNK phosphorylation of Bim-related members of the Bcl2 family induces Bax-dependent apoptosis. Proc Natl Acad Sci U S A. 2003;100(5):2432–7. doi: 10.1073/pnas.0438011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamamoto K, Ichijo H, Korsmeyer SJ. BCL-2 is phosphorylated and inactivated by an ASK1/Jun N-terminal protein kinase pathway normally activated at G(2)/M. Mol Cell Biol. 1999;19(12):8469–78. doi: 10.1128/mcb.19.12.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nishitoh H, et al. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 2002;16(11):1345–55. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim I, et al. Chemical biology investigation of cell death pathways activated by endoplasmic reticulum stress reveals cytoprotective modulators of ASK1. J Biol Chem. 2009;284(3):1593–603. doi: 10.1074/jbc.M807308200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Auf G, et al. Inositol-requiring enzyme 1alpha is a key regulator of angiogenesis and invasion in malignant glioma. Proc Natl Acad Sci U S A. 2010;107(35):15553–8. doi: 10.1073/pnas.0914072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 64.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136(5):823–37. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsiao JR, et al. Endoplasmic reticulum stress triggers XBP-1-mediated up-regulation of an EBV oncoprotein in nasopharyngeal carcinoma. Cancer Res. 2009;69(10):4461–7. doi: 10.1158/0008-5472.CAN-09-0277. [DOI] [PubMed] [Google Scholar]
- 66.Schonthal AH. Endoplasmic reticulum stress and autophagy as targets for cancer therapy. Cancer Lett. 2009;275(2):163–9. doi: 10.1016/j.canlet.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 67.Dong D, et al. Critical role of the stress chaperone GRP78/BiP in tumor proliferation, survival, and tumor angiogenesis in transgene-induced mammary tumor development. Cancer Res. 2008;68(2):498–505. doi: 10.1158/0008-5472.CAN-07-2950. [DOI] [PubMed] [Google Scholar]
- 68.Wang XZ, et al. Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein (CHOP/GADD153) Mol Cell Biol. 1996;16(8):4273–80. doi: 10.1128/mcb.16.8.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee AS. GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res. 2007;67(8):3496–9. doi: 10.1158/0008-5472.CAN-07-0325. [DOI] [PubMed] [Google Scholar]
- 70.Doroudgar S, et al. Ischemia activates the ATF6 branch of the endoplasmic reticulum stress response. J Biol Chem. 2009;284(43):29735–45. doi: 10.1074/jbc.M109.018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Romero-Ramirez L, et al. XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Res. 2004;64(17):5943–7. doi: 10.1158/0008-5472.CAN-04-1606. [DOI] [PubMed] [Google Scholar]
- 72.Romero-Ramirez L, et al. X box-binding protein 1 regulates angiogenesis in human pancreatic adenocarcinomas. Transl Oncol. 2009;2(1):31–8. doi: 10.1593/tlo.08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Drogat B, et al. IRE1 signaling is essential for ischemia-induced vascular endothelial growth factor-A expression and contributes to angiogenesis and tumor growth in vivo. Cancer Res. 2007;67(14):6700–7. doi: 10.1158/0008-5472.CAN-06-3235. [DOI] [PubMed] [Google Scholar]
- 74.Fels DR, Koumenis C. The PERK/eIF2alpha/ATF4 module of the UPR in hypoxia resistance and tumor growth. Cancer Biol Ther. 2006;5(7):723–8. doi: 10.4161/cbt.5.7.2967. [DOI] [PubMed] [Google Scholar]
- 75.Blais JD, et al. Perk-dependent translational regulation promotes tumor cell adaptation and angiogenesis in response to hypoxic stress. Mol Cell Biol. 2006;26(24):9517–32. doi: 10.1128/MCB.01145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koomagi R, Mattern J, Volm M. Glucose-related protein (GRP78) and its relationship to the drug-resistance proteins P170, GST-pi, LRP56 and angiogenesis in non-small cell lung carcinomas. Anticancer Res. 1999;19(5B):4333–6. [PubMed] [Google Scholar]
- 77.Dong D, et al. Vascular targeting and antiangiogenesis agents induce drug resistance effector GRP78 within the tumor microenvironment. Cancer Res. 2005;65(13):5785–91. doi: 10.1158/0008-5472.CAN-05-0754. [DOI] [PubMed] [Google Scholar]
- 78.Bilmen JG, et al. Inhibition of the SERCA Ca2+ pumps by curcumin. Curcumin putatively stabilizes the interaction between the nucleotide-binding and phosphorylation domains in the absence of ATP. Eur J Biochem. 2001;268(23):6318–27. doi: 10.1046/j.0014-2956.2001.02589.x. [DOI] [PubMed] [Google Scholar]
- 79.Das A, Banik NL, Ray SK. Flavonoids activated caspases for apoptosis in human glioblastoma T98G and U87MG cells but not in human normal astrocytes. Cancer. 2010;116(1):164–76. doi: 10.1002/cncr.24699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ermakova SP, et al. (−)-Epigallocatechin gallate overcomes resistance to etoposide-induced cell death by targeting the molecular chaperone glucose-regulated protein 78. Cancer Res. 2006;66(18):9260–9. doi: 10.1158/0008-5472.CAN-06-1586. [DOI] [PubMed] [Google Scholar]
- 81.Johnson AJ, et al. The cyclo-oxygenase-2 inhibitor celecoxib perturbs intracellular calcium by inhibiting endoplasmic reticulum Ca2+-ATPases: a plausible link with its anti-tumour effect and cardiovascular risks. Biochem J. 2002;366(Pt 3):831–7. doi: 10.1042/BJ20020279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pyrko P, et al. Calcium-activated endoplasmic reticulum stress as a major component of tumor cell death induced by 2,5-dimethyl-celecoxib, a non-coxib analogue of celecoxib. Mol Cancer Ther. 2007;6(4):1262–75. doi: 10.1158/1535-7163.MCT-06-0629. [DOI] [PubMed] [Google Scholar]
- 83.Berridge MJ. The endoplasmic reticulum: a multifunctional signaling organelle. Cell Calcium. 2002;32(5–6):235–49. doi: 10.1016/s0143416002001823. [DOI] [PubMed] [Google Scholar]
- 84.Flourakis M, et al. Passive calcium leak via translocon is a first step for iPLA2-pathway regulated store operated channels activation. Faseb J. 2006;20(8):1215–7. doi: 10.1096/fj.05-5254fje. [DOI] [PubMed] [Google Scholar]
- 85.Hajnoczky G, Davies E, Madesh M. Calcium signaling and apoptosis. Biochem Biophys Res Commun. 2003;304(3):445–54. doi: 10.1016/s0006-291x(03)00616-8. [DOI] [PubMed] [Google Scholar]
- 86.Van Coppenolle F, et al. Ribosome-translocon complex mediates calcium leakage from endoplasmic reticulum stores. J Cell Sci. 2004;117(Pt 18):4135–42. doi: 10.1242/jcs.01274. [DOI] [PubMed] [Google Scholar]
- 87.Michalak M, Robert Parker JM, Opas M. Ca2+ signaling and calcium binding chaperones of the endoplasmic reticulum. Cell Calcium. 2002;32(5–6):269–78. doi: 10.1016/s0143416002001884. [DOI] [PubMed] [Google Scholar]
- 88.Thastrup O, et al. Thapsigargin, a novel molecular probe for studying intracellular calcium release and storage. Agents Actions. 1989;27(1–2):17–23. doi: 10.1007/BF02222186. [DOI] [PubMed] [Google Scholar]
- 89.Hakii H, et al. Thapsigargin, a histamine secretagogue, is a non-12-O-tetradecanoylphorbol-13-acetate (TPA) type tumor promoter in two-stage mouse skin carcinogenesis. J Cancer Res Clin Oncol. 1986;111(3):177–81. doi: 10.1007/BF00389230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Denmeade SR, Isaacs JT. The SERCA pump as a therapeutic target: making a “smart bomb” for prostate cancer. Cancer Biol Ther. 2005;4(1):14–22. doi: 10.4161/cbt.4.1.1505. [DOI] [PubMed] [Google Scholar]
- 91.Denmeade SR, et al. Prostate-specific antigen-activated thapsigargin prodrug as targeted therapy for prostate cancer. J Natl Cancer Inst. 2003;95(13):990–1000. doi: 10.1093/jnci/95.13.990. [DOI] [PubMed] [Google Scholar]
- 92.Furuya Y, et al. The role of calcium, pH, and cell proliferation in the programmed (apoptotic) death of androgen-independent prostatic cancer cells induced by thapsigargin. Cancer Res. 1994;54(23):6167–75. [PubMed] [Google Scholar]
- 93.Wu SH, et al. Curcumin induces apoptosis in human non-small cell lung cancer NCI-H460 cells through ER stress and caspase cascade- and mitochondria-dependent pathways. Anticancer Res. 2010;30(6):2125–33. [PubMed] [Google Scholar]
- 94.Karmakar S, Banik NL, Ray SK. Curcumin suppressed anti-apoptotic signals and activated cysteine proteases for apoptosis in human malignant glioblastoma U87MG cells. Neurochem Res. 2007;32(12):2103–13. doi: 10.1007/s11064-007-9376-z. [DOI] [PubMed] [Google Scholar]
- 95.Purkayastha S, et al. Curcumin Blocks Brain Tumor Formation. Brain Res. 2009 doi: 10.1016/j.brainres.2009.01.066. [DOI] [PubMed] [Google Scholar]
- 96.Contessa JN, et al. Inhibition of N-linked glycosylation disrupts receptor tyrosine kinase signaling in tumor cells. Cancer Res. 2008;68(10):3803–9. doi: 10.1158/0008-5472.CAN-07-6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Contessa JN, et al. Molecular imaging of N-linked glycosylation suggests glycan biosynthesis is a novel target for cancer therapy. Clin Cancer Res. 2010;16(12):3205–14. doi: 10.1158/1078-0432.CCR-09-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wong AJ, et al. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc Natl Acad Sci U S A. 1987;84(19):6899–903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lippincott-Schwartz J, et al. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56(5):801–13. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Klausner RD, Donaldson JG, Lippincott-Schwartz J. Brefeldin A: insights into the control of membrane traffic and organelle structure. J Cell Biol. 1992;116(5):1071–80. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lippincott-Schwartz J, et al. Brefeldin A’s effects on endosomes, lysosomes, and the TGN suggest a general mechanism for regulating organelle structure and membrane traffic. Cell. 1991;67(3):601–16. doi: 10.1016/0092-8674(91)90534-6. [DOI] [PubMed] [Google Scholar]
- 102.Pommepuy I, et al. Brefeldin A induces apoptosis and cell cycle blockade in glioblastoma cell lines. Oncology. 2003;64(4):459–67. doi: 10.1159/000070307. [DOI] [PubMed] [Google Scholar]
- 103.Nakada M, et al. Roles of membrane type 1 matrix metalloproteinase and tissue inhibitor of metalloproteinases 2 in invasion and dissemination of human malignant glioma. J Neurosurg. 2001;94(3):464–73. doi: 10.3171/jns.2001.94.3.0464. [DOI] [PubMed] [Google Scholar]
- 104.Proulx-Bonneau S, Pratt J, Annabi B. A role for MT1-MMP as a cell death sensor/effector through the regulation of endoplasmic reticulum stress in U87 glioblastoma cells. J Neurooncol. 2010 doi: 10.1007/s11060-010-0468-2. [DOI] [PubMed] [Google Scholar]
- 105.Hosokawa N, et al. A novel ER alpha-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2001;2(5):415–22. doi: 10.1093/embo-reports/kve084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Molinari M, et al. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 2003;299(5611):1397–400. doi: 10.1126/science.1079474. [DOI] [PubMed] [Google Scholar]
- 107.Oda Y, et al. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science. 2003;299(5611):1394–7. doi: 10.1126/science.1079181. [DOI] [PubMed] [Google Scholar]
- 108.Yoshida H, et al. A time-dependent phase shift in the mammalian unfolded protein response. Dev Cell. 2003;4(2):265–71. doi: 10.1016/s1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 109.Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79(1):13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 110.Yin D, et al. Proteasome inhibitor PS-341 causes cell growth arrest and apoptosis in human glioblastoma multiforme (GBM) Oncogene. 2005;24(3):344–54. doi: 10.1038/sj.onc.1208225. [DOI] [PubMed] [Google Scholar]
- 111.Friedman HS, Kerby T, Calvert H. Temozolomide and treatment of malignant glioma. Clin Cancer Res. 2000;6(7):2585–97. [PubMed] [Google Scholar]
- 112.Kubicek GJ, et al. Phase I trial using proteasome inhibitor bortezomib and concurrent temozolomide and radiotherapy for central nervous system malignancies. Int J Radiat Oncol Biol Phys. 2009;74(2):433–9. doi: 10.1016/j.ijrobp.2008.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]