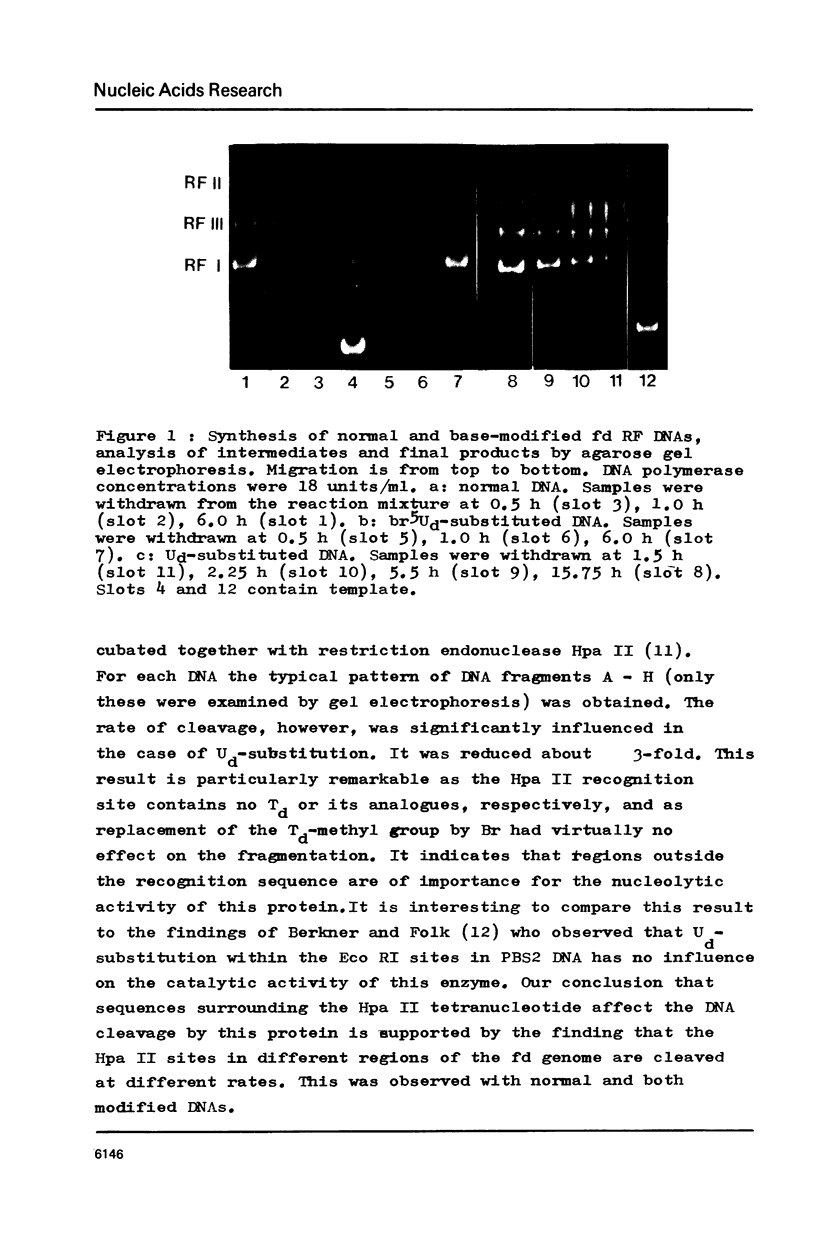
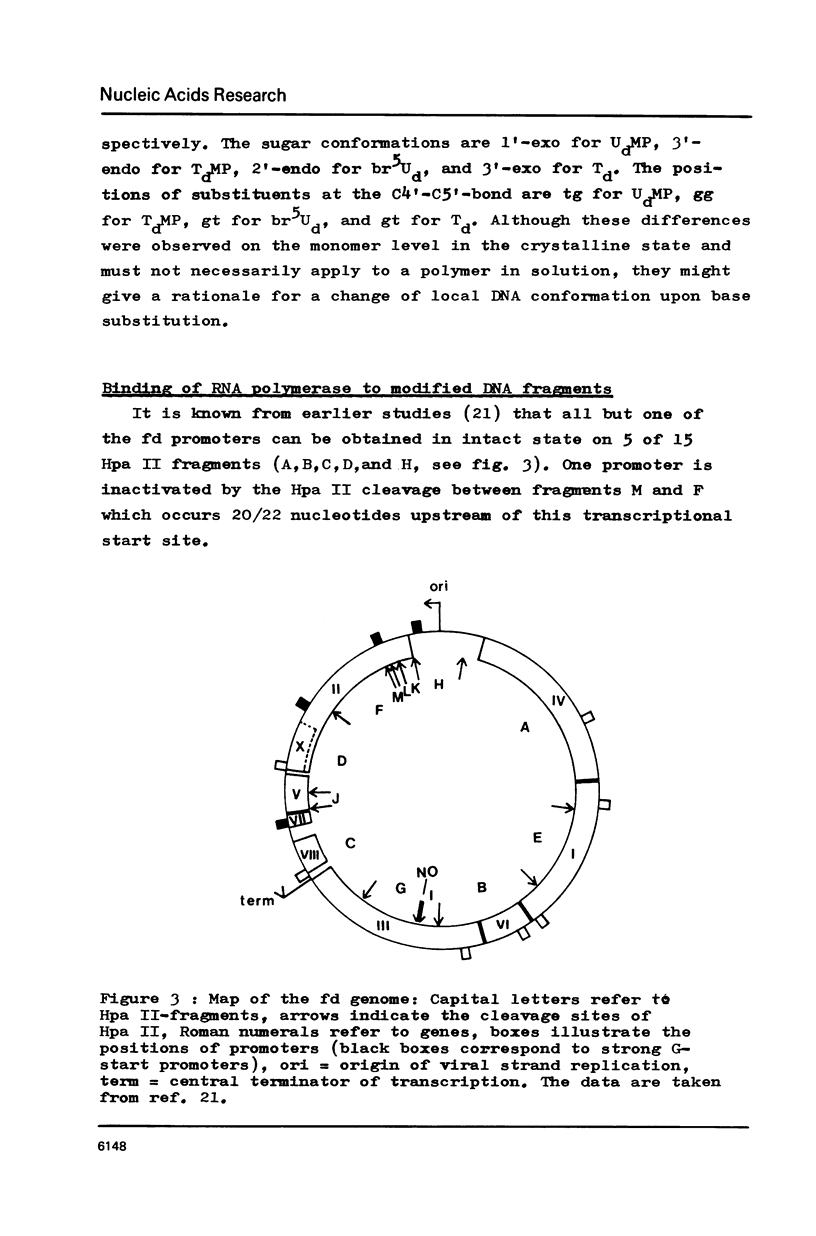
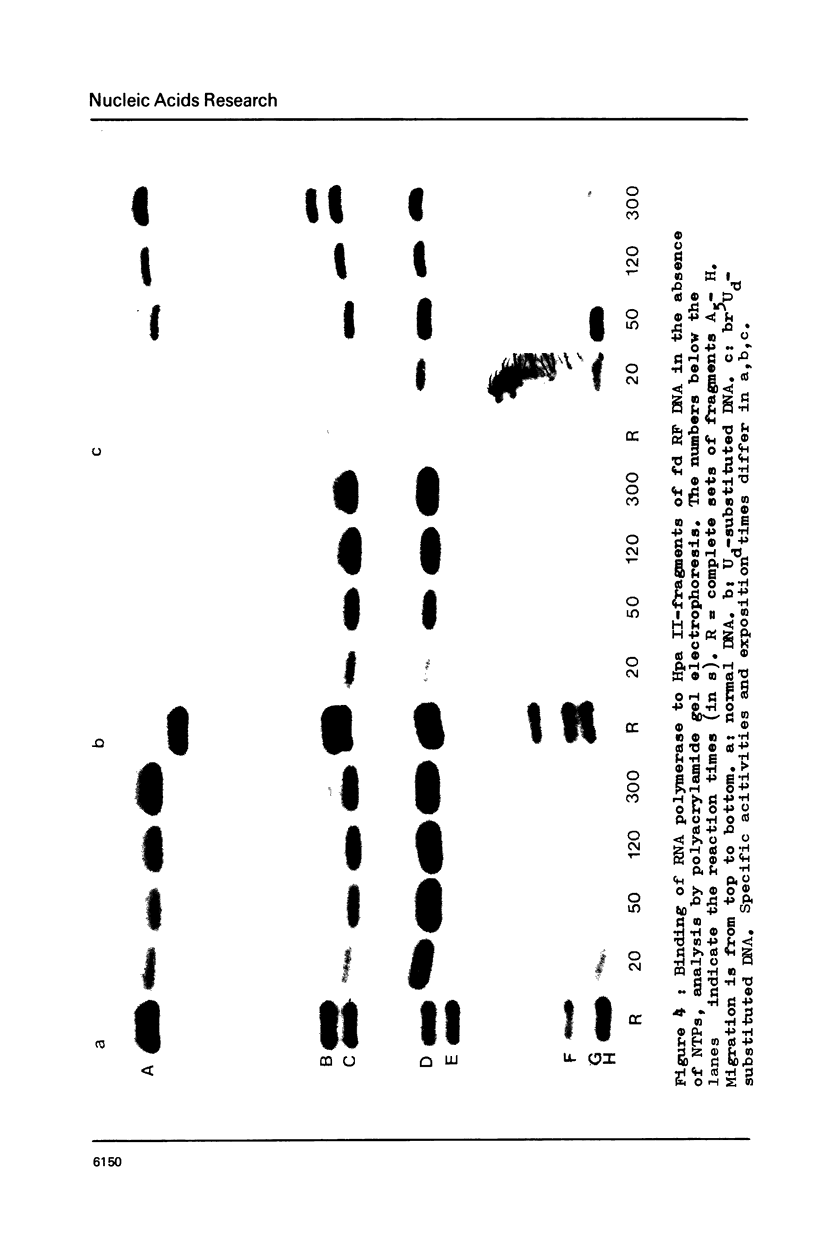
Abstract
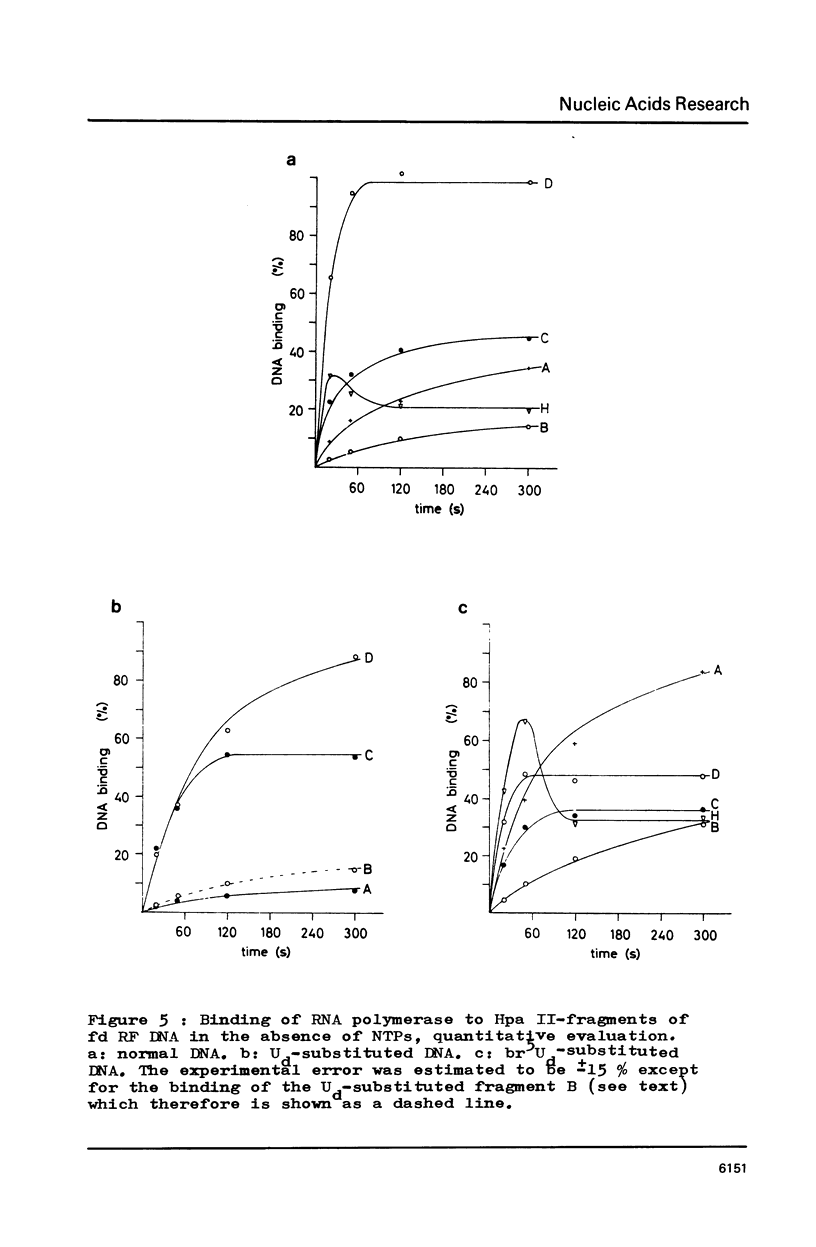
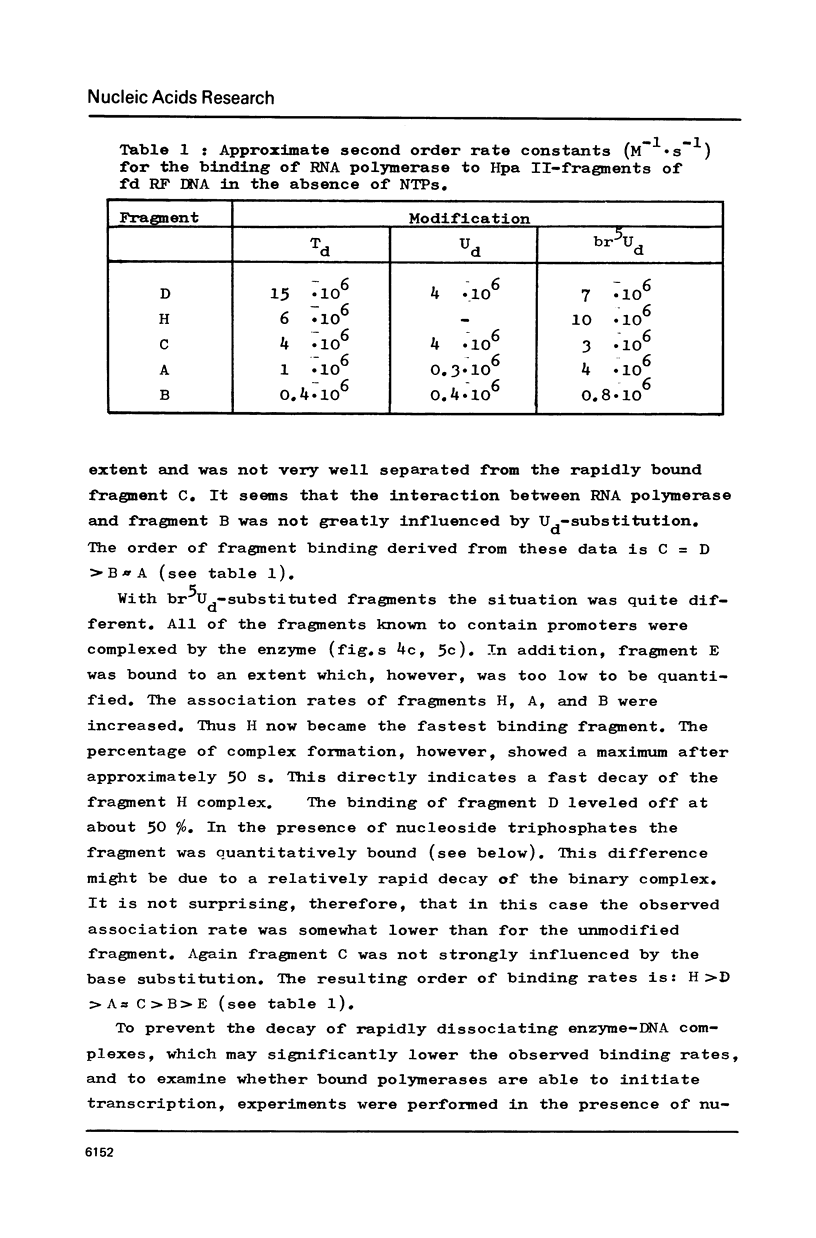
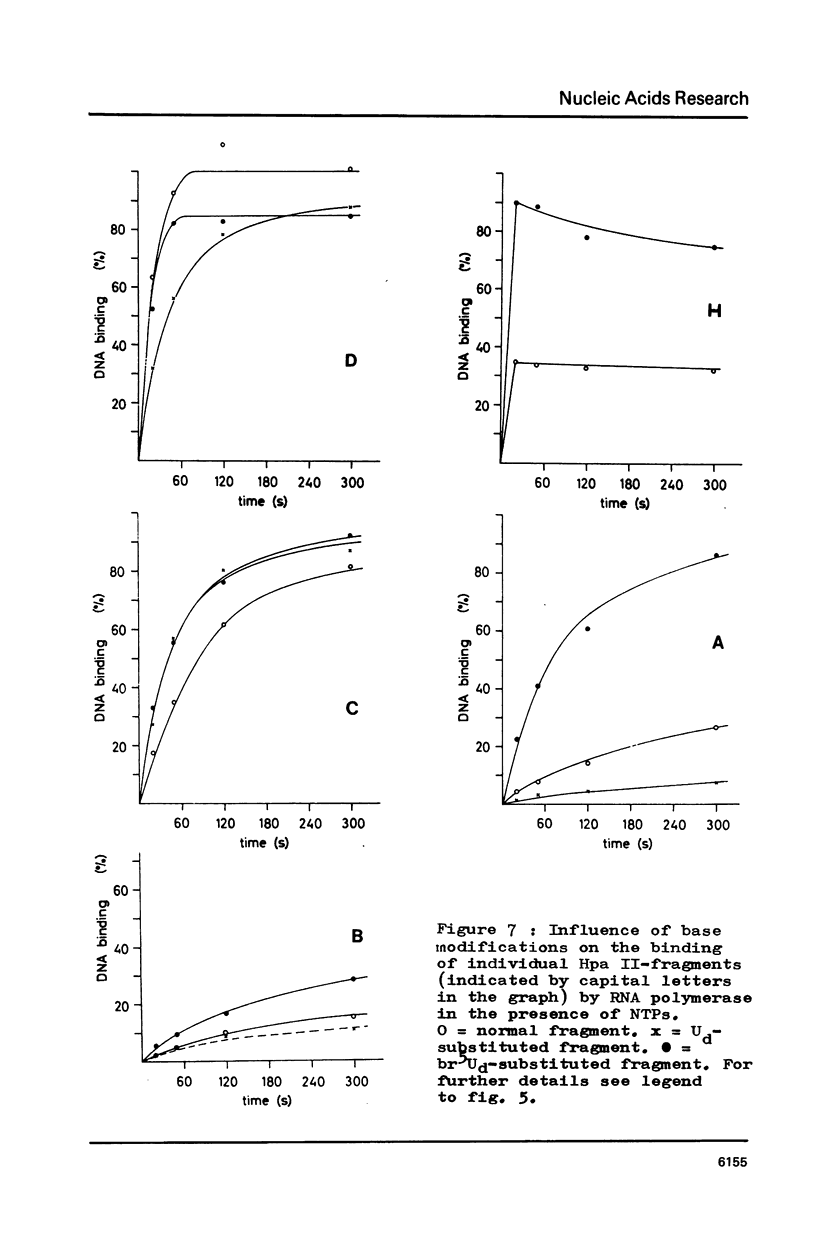
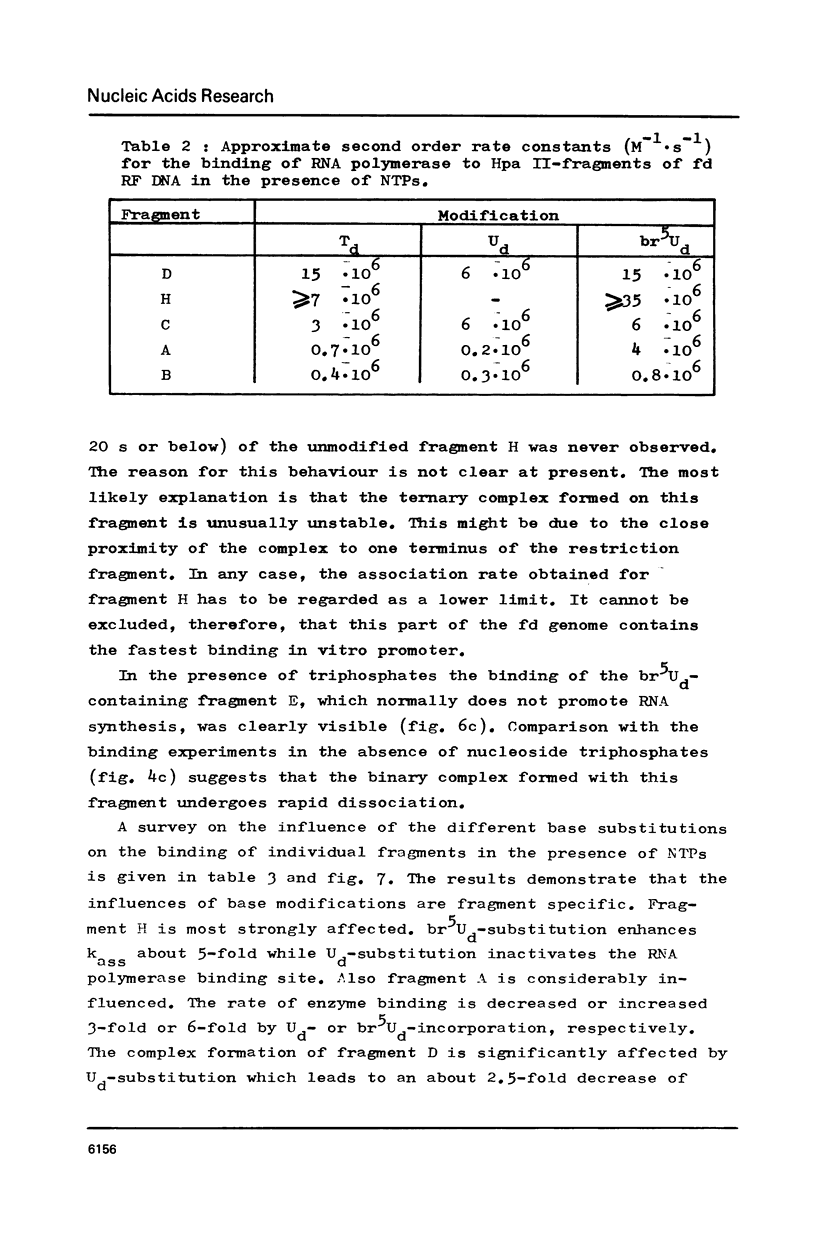
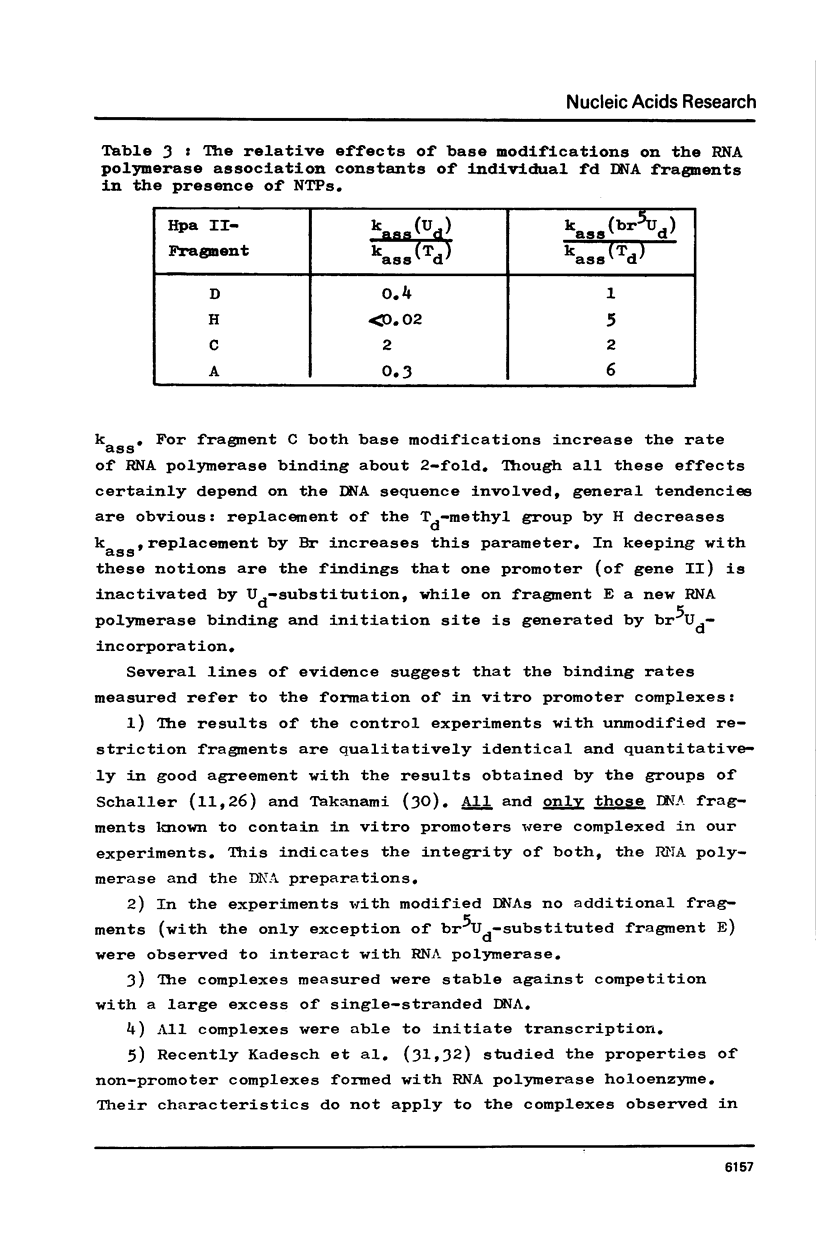
RF I DNA of phage fd containing 5-bromo-deoxyuridine (br5Ud) or deoxyuridine (Ud) instead of deoxythymidine (Td) inthe codogenic strand was synthesized in vitro. The modified genomes could be cleaved by restriction endonuclease Hpa II. Although the recognition site of Hpa II is CCGG, the cleavage rate was significantly reduced with Ud-containing DNA. Both base substitutions altered the mobilities of several DNA fragments under the conditions of polyacrylamide gel electrophoresis. The fragments containing binding sites for RNA polymerase were assayed for the rates of stable complex formation. The substitution of Td for both, Ud and br5Ud, strongly influenced this parameter. Thus the methyl group of Td has to be regarded as one of the sites in DNA which determine the rate of stable RNA polymerase binding and thereby possibly mediate promoter activity in vitro (24,25,26). In most cases the rate of complex formation was decreased by Ud, but increased by br5Ud.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beck E., Sommer R., Auerswald E. A., Kurz C., Zink B., Osterburg G., Schaller H., Sugimoto K., Sugisaki H., Okamoto T. Nucleotide sequence of bacteriophage fd DNA. Nucleic Acids Res. 1978 Dec;5(12):4495–4503. doi: 10.1093/nar/5.12.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkner K. L., Folk W. R. EcoRI cleavage and methylation of DNAs containing modified pyrimidines in the recogintion sequence. J Biol Chem. 1977 May 25;252(10):3185–3193. [PubMed] [Google Scholar]
- Bick M. D., Devine E. A. Interaction of chromosomal proteins with BrdU substituted DNA as determined by chromatin-DNA competition. Nucleic Acids Res. 1977 Nov;4(11):3687–3700. doi: 10.1093/nar/4.11.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutlag D., Atkinson M. R., Setlow P., Kornberg A. An active fragment of DNA polymerase produced by proteolytic cleavage. Biochem Biophys Res Commun. 1969 Dec 4;37(6):982–989. doi: 10.1016/0006-291x(69)90228-9. [DOI] [PubMed] [Google Scholar]
- CHAMBERLIN M., BALDWIN R. L., BERG P. AN ENZYMICALLY SYNTHESIZED RNA OF ALTERNATING BASE SEQUENCE: PHYSICAL AND CHEMICAL CHARACTERIZATION. J Mol Biol. 1963 Oct;7:334–349. doi: 10.1016/s0022-2836(63)80028-5. [DOI] [PubMed] [Google Scholar]
- Cullen B. R., Bick M. D. Thermal denaturation of DNA from bromodeoxyuridine substituted cells. Nucleic Acids Res. 1976 Jan;3(1):49–62. doi: 10.1093/nar/3.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dausse J. P., Sentenac A., Fromageot P. Interaction of RNA polymerase from Escherichia coli with DNA. Effect of temperature and ionic strength on selection of T7 DNA early promoters. Eur J Biochem. 1976 Jun 1;65(2):387–393. doi: 10.1111/j.1432-1033.1976.tb10352.x. [DOI] [PubMed] [Google Scholar]
- FOX J. J., SHUGAR D. Spectrophotometric studies of nucleic acid derivatives and related compounds as a function of pH. II. Natural and synthetic pyrimidine nucleosides. Biochim Biophys Acta. 1952 Oct;9(4):369–384. doi: 10.1016/0006-3002(52)90181-9. [DOI] [PubMed] [Google Scholar]
- Goeddel D. V., Yansura D. G., Caruthers M. H. Studies on gene control regions. VI. The 5- methyl of thymine, a lac repressor recognition site. Nucleic Acids Res. 1977 Sep;4(9):3039–3054. doi: 10.1093/nar/4.9.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J. S., Bell G. I., Martinson H. C., Rutter W. J. Selective interaction of 5'-bromodeoxyuridine substituted DNA with different chromosomal proteins. Biochemistry. 1976 Nov 2;15(22):4778–4785. doi: 10.1021/bi00667a005. [DOI] [PubMed] [Google Scholar]
- Gray C. P., Sommer R., Polke C., Beck E., Schaller H. Structure of the orgin of DNA replication of bacteriophage fd. Proc Natl Acad Sci U S A. 1978 Jan;75(1):50–53. doi: 10.1073/pnas.75.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyden B., Nüsslein C., Schaller H. Single RNA polymerase binding site isolated. Nat New Biol. 1972 Nov 1;240(96):9–12. doi: 10.1038/newbio240009a0. [DOI] [PubMed] [Google Scholar]
- Hofer B., Köster H. Elimination of promoter function by base modification of DNA. FEBS Lett. 1979 Jun 1;102(1):87–90. doi: 10.1016/0014-5793(79)80934-5. [DOI] [PubMed] [Google Scholar]
- INMAN R. B., BALDWIN R. L. Formation of hybrid molecules from two alternating DNA copolymers. J Mol Biol. 1962 Aug;5:185–200. doi: 10.1016/s0022-2836(62)80083-7. [DOI] [PubMed] [Google Scholar]
- INMAN R. B., BALDWIN R. L. Helix-random coil transitions in synthetic DNAs of alternating sequence. J Mol Biol. 1962 Aug;5:172–184. doi: 10.1016/s0022-2836(62)80082-5. [DOI] [PubMed] [Google Scholar]
- Jansz H. S., Pouwels P. H., Schiphorst J. Preparation of double-stranded DNA (replicative form) of bacteriophage phi-X174: a simplified method. Biochim Biophys Acta. 1966 Sep;123(3):626–627. doi: 10.1016/0005-2787(66)90233-4. [DOI] [PubMed] [Google Scholar]
- Johnsrud L. Contacts between Escherichia coli RNA polymerase and a lac operon promoter. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5314–5318. doi: 10.1073/pnas.75.11.5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones O. W., Berg P. Studies on the binding of RNA polymerase to polynucleotides. J Mol Biol. 1966 Dec 28;22(2):199–209. doi: 10.1016/0022-2836(66)90126-4. [DOI] [PubMed] [Google Scholar]
- Kadesch T. R., Williams R. C., Chamberlin M. J. Electron microscopic studies of the binding of Escherichia coli RNA polymerase to DNA. I. Characterization of the non-specific interactions of holoenzyme with a restriction fragment of bacteriophage T7 DNA. J Mol Biol. 1980 Jan 5;136(1):65–78. doi: 10.1016/0022-2836(80)90366-6. [DOI] [PubMed] [Google Scholar]
- Kadesch T. R., Williams R. C., Chamberlin M. J. Electron microscopic studies of the binding of Escherichia coli RNA polymerase to DNA. II. Formation of multiple promoter-like complexes at non-promoter sites. J Mol Biol. 1980 Jan 5;136(1):79–93. doi: 10.1016/0022-2836(80)90367-8. [DOI] [PubMed] [Google Scholar]
- Kallos J., Fasy T. M., Hollander V. P., Bick M. D. Estrogen receptor can distinguish among various halodeoxyuridine-substituted DNAs. FEBS Lett. 1979 Feb 15;98(2):347–350. doi: 10.1016/0014-5793(79)80214-8. [DOI] [PubMed] [Google Scholar]
- Klenow H., Henningsen I. Selective elimination of the exonuclease activity of the deoxyribonucleic acid polymerase from Escherichia coli B by limited proteolysis. Proc Natl Acad Sci U S A. 1970 Jan;65(1):168–175. doi: 10.1073/pnas.65.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A., Jack A., Viswamitra M. A., Kennard O., Shakked Z., Steitz T. A. A hypothesis on a specific sequence-dependent conformation of DNA and its relation to the binding of the lac-repressor protein. J Mol Biol. 1979 Jul 15;131(4):669–680. doi: 10.1016/0022-2836(79)90196-7. [DOI] [PubMed] [Google Scholar]
- Lin S. Y., Riggs A. D. Lac operator analogues: bromodeoxyuridine substitution in the lac operator affects the rate of dissociation of the lac repressor. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2574–2576. doi: 10.1073/pnas.69.9.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Lin D., Riggs A. D. Histones bind more tightly to bromodeoxyuridine-substituted DNA than to normal DNA. Nucleic Acids Res. 1976 Sep;3(9):2183–2191. doi: 10.1093/nar/3.9.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T., Sugimoto K., Sugisaki H., Takanami M. Studies on bacteriophage fd DNA. II. Localization of RNA initiation sites on the cleavage map of the fd genome. J Mol Biol. 1975 Jun 15;95(1):33–44. doi: 10.1016/0022-2836(75)90333-2. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Canuel L. L., Pohl F. M. "Alternating B-DNA" conformation for the oligo(dG-dC) duplex in high-salt solution. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2508–2511. doi: 10.1073/pnas.76.6.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J. P. Enzymic synthesis of RNA from T7 DNA. J Mol Biol. 1966 Oct 28;21(1):115–127. doi: 10.1016/0022-2836(66)90083-0. [DOI] [PubMed] [Google Scholar]
- Seeburg P. H., Nüsslein C., Schaller H. Interaction of RNA polymerase with promoters from bacteriophage fd. Eur J Biochem. 1977 Mar 15;74(1):107–113. doi: 10.1111/j.1432-1033.1977.tb11372.x. [DOI] [PubMed] [Google Scholar]
- Seeburg P. H., Schaller H. Mapping and characterization of promoters in bacteriophages fd, f1 and m13. J Mol Biol. 1975 Feb 25;92(2):261–277. doi: 10.1016/0022-2836(75)90226-0. [DOI] [PubMed] [Google Scholar]
- Siebenlist U. RNA polymerase unwinds an 11-base pair segment of a phage T7 promoter. Nature. 1979 Jun 14;279(5714):651–652. doi: 10.1038/279651a0. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Simpson R. B. Contacts between Escherichia coli RNA polymerase and thymines in the lac UV5 promoter. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3233–3237. doi: 10.1073/pnas.76.7.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura M., Okamoto T., Takanami M. Starting nucleotide sequences of RNA synthesized on the replicative form DNA of coliphage fd. J Mol Biol. 1969 Jul 28;43(2):299–315. doi: 10.1016/0022-2836(69)90269-1. [DOI] [PubMed] [Google Scholar]
- Sági J., Brahms S., Brahms J., Otvös L. Effect of 5-alkyl substitution of uracil on the thermal stability of poly [d(A-r5U)] copolymers. Nucleic Acids Res. 1979 Jun 25;6(8):2839–2848. doi: 10.1093/nar/6.8.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAKAHASHI I., MARMUR J. Replacement of thymidylic acid by deoxyuridylic acid in the deoxyribonucleic acid of a transducing phage for Bacillus subtilis. Nature. 1963 Feb 23;197:794–795. doi: 10.1038/197794a0. [DOI] [PubMed] [Google Scholar]
- Tabak H. F., Flavell R. A. A method for the recovery of DNA from agarose gels. Nucleic Acids Res. 1978 Jul;5(7):2321–2332. doi: 10.1093/nar/5.7.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswamitra M. A., Seshadri T. P., Post M. L. An uncommon nucleotide conformation shown by molecular structure of deoxyuridine-5'-phosphate and nucleic acid stereochemistry. Nature. 1975 Dec 11;258(5535):542–544. doi: 10.1038/258542a0. [DOI] [PubMed] [Google Scholar]
- WEIL R., VINOGRAD J. THE CYCLIC HELIX AND CYCLIC COIL FORMS OF POLYOMA VIRAL DNA. Proc Natl Acad Sci U S A. 1963 Oct;50:730–738. doi: 10.1073/pnas.50.4.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wovcha M. G., Warner H. R. Synthesis and nucleolytic degradation of uracil-containing deoxyribonucleic acid by Escherichia coli deoxyribonucleic acid polymerase. I. J Biol Chem. 1973 Mar 10;248(5):1746–1750. [PubMed] [Google Scholar]
- von Gabain A., Bujard H. Interaction of E. coli RNA polymerase with promotors of coliphage T5: the rates of complex formation and decay and their correlation with in vitro and in vivo transcriptional activity. Mol Gen Genet. 1977 Dec 9;157(3):301–311. doi: 10.1007/BF00268667. [DOI] [PubMed] [Google Scholar]