Abstract
Accumulating evidence has implicated the deregluation of miRNAs in tumorigenesis. Previous studies have reported that microRNA-195 (miR-195) is markedly down-regulated in human glioblastoma cells, compared with normal brain tissue, but the biological role of miR-195 in glioblastoma development is currently unknown. In this study, we define a tumor-suppressor role for miR-195 in human glioblastoma cells. Over-expression of miR-195 in glioblastoma cell lines robustly arrested cell cycle progression and significantly repressed cellular invasion. We identified E2F3 and CCND3 as functional downstream targets of miR-195 in glioblastoma cells. Through knockdown studies, we demonstrated that E2F3 was the dominant effector of miR-195-mediated cell cycle arrest and that CCND3 was a key mediator of miR-195–induced inhibition of glioblastoma cell invasion. Furthermore, we showed that p27Kip1 was an important regulator downstream of CCND3 and that the accumulation of p27Kip1 in the cytoplasm might be responsible for the miR-195–mediated cell invasion inhibition in glioblastoma cells. This work provides evidence for the initial mechanism by which miR-195 negatively regulates both the proliferation and invasion of glioblastoma cells, suggesting that the down-regulation of miR-195 might contribute to the malignant transformation of glioblastoma cells and could be a molecular signature associated with glioblastoma progression.
Keywords: glioblastoma, invasion, microRNA-195, proliferation, tumor-suppressor
Glioblastoma (World Health Organization grade IV) is one of the most common and aggressive primary brain tumors and is among the most lethal types of human cancer.1 It is characterized by rapid infiltrative growth and cellular heterogeneity, which contribute to its formidable resistance to combination therapy.2–4 Moreover, because of our poor understanding of this cancer's pathogenesis, therapeutic strategies for glioblastoma are limited.5–7 Studying the molecular implications of genetic alterations underlying the malignant phenotype of glioblastoma should aid in the development of new diagnostic, prognostic, and therapeutic approaches.
MicroRNAs (miRNAs) have emerged as key posttranscriptional regulators of gene expression in the past decade, and they have been shown to be involved in a number of diverse physiological and pathological processes.8,9 miRNAs are particularly notable because of their important roles in cancer pathogenesis.10–12 Virtually all examined tumor types are characterized by globally abnormal miRNA expression patterns, and the evidence that miRNAs frequently function as oncogenes or tumor suppressors in various cancers has generated great interest worldwide.
The miR-15/16 family encompasses a group of miRNAs with the same seed sequence (including miR-15, miR-16, miR-103, miR-107, miR-195, miR-424, miR-497, miR-503, and miR-646).13 There are 2 miR-15/16 clusters (miR-15a/16-1 cluster and miR-15b/16-2 cluster) in mammals, and the miR-15a/16-1 cluster was the first identified example of frequently deleted or down-regulated miRNAs in human cancer (chronic lymphocytic leukemia [CLL]).14 A subsequent report showed that miR-15 and miR-16 could induce apoptosis by directly targeting Bcl-2, an antiapoptotic oncogene that is commonly overexpressed in CLL.15 miR-15 and miR-16 are therefore considered to be representative examples of tumor-suppressor miRNAs. Of all the miR-15/16 family members, miR-195 was identified as the most significantly down-regulated miRNA in glioblastoma cell lines, compared with normal brain tissues.16,17 Because of its high degree of sequence homology with miR-15/16, miR-195 is expected to regulate a group of targets similar to those of miR-15/16,18 thus making it a likely candidate tumor suppressor in glioblastoma cells. However, whether deregulation of miR-195 contributes to glioblastoma pathobiology is currently undetermined.
In this study, we attempted to define the role of miR-195 in human glioblastoma cells. First, we over-expressed miR-195 in glioblastoma cells and found that miR-195 strongly arrested cell cycle progression but did not induce apoptosis. Next, we used luciferase reporter assays and Western blotting to identify targets of miR-195. Then, we performed knockdown of the identified targets by synthesized small interfering RNAs (siRNAs) and compared the effect of siRNA knockdown with that of miR-195 over-expression to confirm whether these genes were functional miR-195 targets in glioblastoma cells. Of note, we found that E2F3 and CCND3 were 2 in vivo targets through which miR-195 regulated cellular proliferation and invasion, respectively. We then analyzed the possible mechanisms by which CCND3 promoted cell invasion. We found that CCND3 influenced glioblastoma invasion by negatively regulating the expression of p27Kip1 in the cytoplasm. These data demonstrate that miR-195 may function as an important tumor suppressor in glioblastoma cells by modulating the signaling pathways controlling both cellular proliferation and invasion, which implies that the down-regulation of miR-195 may be a significant event associated with glioblastoma progression.
Materials and Methods
Cell Culture and Transfection
Glioblastoma cell lines (U87MG, LN-308, LN-229, A172, T98G, U251, LN-428, U373, and U138) and the 293T cell line were maintained as adherent cultures in Dulbecco's Modified Eagle's Medium (DMEM; GIBCO) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin and were incubated at 37°C in a humidified chamber supplemented with 5% CO2. Cells transfection with RNA mimics and plasmids were performed using Lipofectamine 2000 (Invitrogen). All RNA transfections were performed at a final concentration of 50 nM, unless otherwise indicated.
Total RNA, RNA Oligoribonucleotides, and Antibodies
Human brain total RNA was purchased from Ambion (AM7962). miRNA mimics of miR-195, negative control RNA mimics (denoted as NC), and siRNA duplexes for E2F3, CCND1, and CCND3 (Supplementary Table) were obtained from GenePharma. The following primary antibodies were used for Western blotting: E2F3 (05-551) and Histone H1 (05-457) were obtained from Millipore; CCND1 (2926), CCND3 (2936), p27Kip1 (2552), and β-actin (4970) were obtained from Cell Signaling Technology; p27Kip1 (sc-528) was obtained from Santa Cruz Biotechnology; and STMN1 antibody (00138) was obtained from Sigma-Aldrich.
Construction of miR-195 Expression Plasmids and Luciferase Reporter Plasmids
To construct a plasmid expressing miR-195, we amplified a 160-bp DNA fragment containing a miR-195 precursor from human genomic DNA (293T) and cloned the amplified fragment into a modified pcDNA6.2-GW/EmGFP vector.19 TargetScan 4.1 (http://www.targetscan.org), a miRNA target prediction program, was used to search for putative miR-195 targets. The 3′UTR regions of the predicted miR-195 targets were cloned into the psiCHECK-2 vector (Promega) downstream of the Renilla luciferase gene (Xho I/Not I sites).19 Mutant plasmids corresponding to wild-type 3′ UTR regions were also constructed (all primers and sequences are listed in Supplementary Table).
Northern Blot
Total RNA (30 μg) extracted from glioblastoma cells was electrophoresed on a 12% polyacrylamide-urea gel and transferred to a Hybond-N+ membrane (Amersham Biosciences). Membranes were hybridized with miR-195 oligonucleotide probes labeled with γ-32P-ATP at the 5′-ends. U6 snRNA was used as a loading control (all probes are listed in Supplementary Table).
Luciferase Assays
293T cells were seeded into 48-well plates (6.0 × 104 per well). After 24 h, the cells were cotransfected with the reporter vectors and the miRNA-expressing plasmid (pcDNA6.2-miR-195 or pcDNA6.2-miR-neg) at a ratio of 0.1 (µg): 0.1 (µg). Luciferase activity was measured 48 h posttransfection using the Dual-Luciferase Reporter Assay System, according to the manufacturer's instructions (Promega). For each sample, Renilla luciferase activity was normalized to firefly luciferase expression.
Western Blotting
Cells were transfected with 100 nM NC, miR-195 mimics, or siRNA duplexes in 24-well plates. Cell samples and nuclear/cytoplasmic extracts (prepared according to the manufacturer's instructions [Thermo, 78833]) were collected 48 h later and analyzed using Western blotting. β-actin and histone H1 were used as loading controls. Equivalent total protein extracts were separated by SDS-polyacrylamide gel electrophoresis and transferred onto PVDF transfer membranes (Amersham Biosciences). The membranes were incubated with a primary antibody and an HRP-conjugated secondary antibody and were then exposed to X-ray films.
Flow Cytometry Assays
Cells were transfected with mimics or siRNAs. Nocodazole (100 ng/mL; Sigma-Aldrich) was added 24 h posttransfection, and cells were incubated for an additional 20 h. Floating and adherent cells were harvested, combined, washed twice in phosphate buffered saline, lysed, and stained with NP40/PI solution (0.3% NP40, 10 μg/mL propidium iodide, 0.03 mg/mL DNase-free RNase A, 10 mM NaCl, 3 mM trisodium citrate) at 37°C for 30 min. The analysis was performed using a FACSCalibur flow cytometer (Becton Dickinson) with Cell Quest Pro software. Cell cycle modeling was performed using Modfit software, version 3.0 (Verity Software House).
Invasion Assays
Transwell 24-well chambers with 8-μm pore size (Costar) were used for the invasion assay. The well was precoated with basement membrane extract (Trevigen) diluted to 1 μg/μL in DMEM without FBS. U87MG, LN-308, and U251 cells were transfected with NC mimics or miR-195 mimics or siRNAs. To assess the invasive ability of the cells, 5 × 104 transfected cells were harvested and resuspended in 0.2 mL DMEM without FBS. Cells were then loaded into the upper compartment of the chambers, and 0.6 mL DMEM containing 10% FBS was placed in the lower compartment. The transwell chambers were incubated for 20 h at 37°C in a 5% CO2 incubator. The upper surface of the membrane was wiped with a cotton tip to remove noninvasive cells, and the invasive cells attached to the membrane's lower surface were stained for 20 min with crystal violet. All assays were performed in triplicate.
Statistical Analysis
Data are presented as the mean ± standard deviation from at least 3 separate experiments. Unless otherwise noted, the differences between groups were analyzed using a Student's t test when only 2 groups were compared or by 1-way analysis of variance when more than 2 groups were compared. All tests were 2-sided. Differences were considered to be statistically significant at P< .05.
Results
Over-Expression of miR-195 in Glioblastoma Cells Potently Arrests Cell Cycle Progression at the G1/S Transition
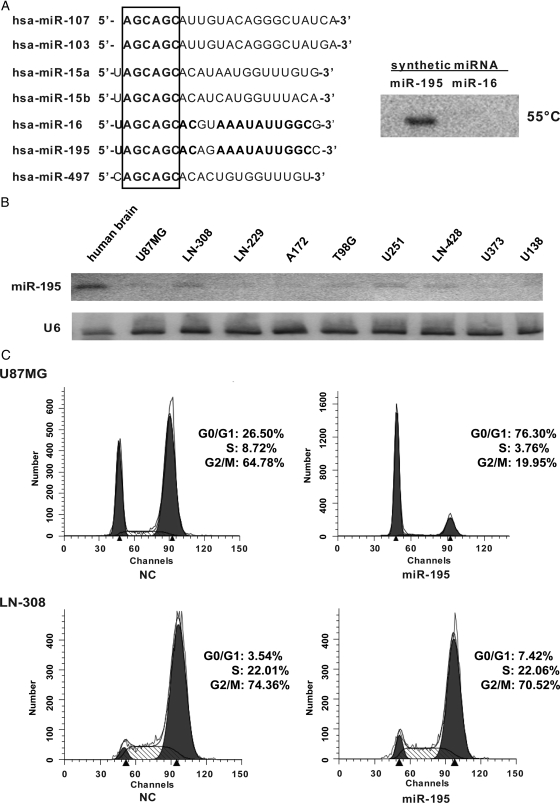
To explore the biological role of miR-195 in glioma cells, we analyzed miR-195 expression in 9 cell lines derived from human glioblastoma (U87MG, LN-308, LN-229, A172, T98G, U251, LN-428, U373, and U138) and a normal human brain RNA sample. To exclude nonspecific cross-reactions with other miR-15/16 family members, we used synthetic RNA mimics of miR-195 and miR-16 (which differs from miR-195 at only 3 bases, as shown in Fig. 1A) to test the specificity of the miR-195 probe and the effectiveness of the Northern blot experiment. As shown in Fig. 1A, Northern blot analysis showed that our assay was specific for miR-195 expression. The results showed that miR-195 was markedly down-regulated in all glioblastoma cell lines, compared with the normal human brain (Fig. 1B). Because miR-195 expression was lowest in U87MG cells and highest in LN-308 cells, these 2 cell lines were selected for further study.
Fig. 1.
Northern blot detected miR-195 expression and miR-195 triggered G1 arrest in glioblastoma cell lines. A, The difference between miR-195 and other miR-16 family members was shown, and the expression signature was detected at 55°C to demonstrate the specificity of the Northern blot. B, Relative expression of endogenous miR-195 in human brain tissues and several glioblastoma cells (U87MG, LN-308, LN-229, A172, T98G, U251, LN-428, U373, and U138), as shown by Northern blot. The miR-195 level was normalized by the intensity of U6 in each sample. C, miR-195 arrested U87MG cells and LN-308 cells at the G1/S transition. Cells were treated with nocodazole 24 h posttransfection, and cell cycle distributions were detected 20 h later.
Because miR-195 is down-regulated in glioblastoma cells, compared with normal brain tissues, we performed miR-195 over-expression studies in U87MG and LN-308 cells. Because many members of the miR-15/16/195 family are involved in apoptosis and/or cell cycle control, we first investigated whether transfection of miR-195 mimics into glioblastoma cells could affect cell cycle progression or induce cell apoptosis. Flow cytometry was used for cell cycle and apoptosis analyses. Nocodazole was added 24 h posttransfection to synchronize the cell cycle. As shown in Fig. 1C, compared with the negative control (NC) groups transfected with an unrelated mimic, introduction of miR-195 mimics into the 2 glioblastoma cell lines significantly arrested the cell cycle at the G1/S transition, as indicated by a marked accumulation of cells in the G0/G1 peak. No significant change in the number of apoptotic cells was observed, as evaluated by counting cells in the sub-G1 peak. These results indicate that miR-195 may be primarily involved in regulating glioblastoma cell cycle progression, rather than apoptosis.
CCND3 and E2F3 Were Identified as Functional Targets of miR-195 in Glioblastoma Cells
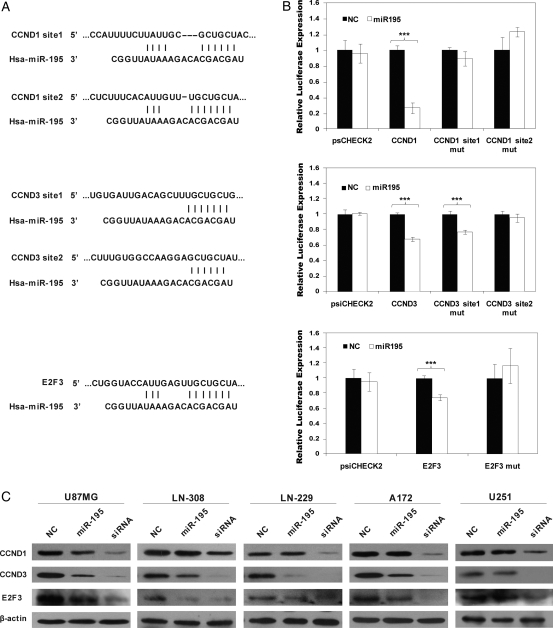
Growth factors and mitogenic signals stimulate the expression of D-cyclins and E2F activity to drive cell cycle progression from the G0/G1 to S phase.20 Over the past few years, studies have detected over-expression of cyclin D1 (CCND1), cyclin D3 (CCND3), and E2F3 in gliomas.21,22 These same genes are predicted to be candidate miR-195 targets by TargetScan4.2 (Fig. 2A). We therefore performed luciferase reporter assays to determine whether CCND1, CCND3, and E2F3 are direct targets of miR-195 in glioblastoma cells. As shown in Fig. 2B, miR-195 significantly suppressed the expression of the Renilla luciferase reporter carrying the wild-type putative target sites of CCND1 (∼70% reduction), CCND3 (∼30%), and E2F3 (∼20%), compared with controls (P < .05), suggesting that all 3 genes contain effective miR-195 binding sites in their 3′UTRs. Moreover, these effects were abolished when we introduced mutations into the seed sequences complementary to the 5′ end of miR-195 (Fig. 2B).
Fig. 2.
CCND3 and E2F3 are direct primary functional targets of miR-195. CCND1, CCND3, and E2F3 possess binding sites for miR-195 in their 3′UTRs (A), and luciferase assays indicated that miR-195 directly down-regulated the expression of CCND1, CCND3, and E2F3 (B). Relative luciferase values were normalized to cotransfections with the pcDNA6.2-control and the psiCHECK-2-control. Data represent the mean ± standard deviation from 2 separate determinations performed in triplicate. *** P < .001, using a 2-tailed t test. C, Western blotting shows the endogenous expression levels of CCND1, CCND3, and E2F3 after transfection with miR-195 or their cognate siRNA in glioblastoma cells. β-actin served as an internal control.
We then performed Western blotting to determine whether transfection of miR-195 into glioblastoma cells reduced the endogenous protein expression of these genes. Five glioblastoma cell lines were used. Specific siRNAs for each gene were used as positive controls. As shown in Fig. 2C, CCND3 and E2F3 were significantly down-regulated by their cognate siRNAs and miR-195 in 4 glioblastoma cell lines (U87MG, LN-308, A172, and LN-229, but not U251), whereas CCND1 protein levels were only slightly reduced by miR-195 in U87MG cells. In the other 4 cell lines, miR-195 transfection had no significant effect on CCND1 protein expression. These data suggest that CCND3 and E2F3 may be functional targets of miR-195 in glioblastoma cells.
E2F3 is the Dominant Target of miR-195 in Regulating Glioblastoma Cell Cycle Progression
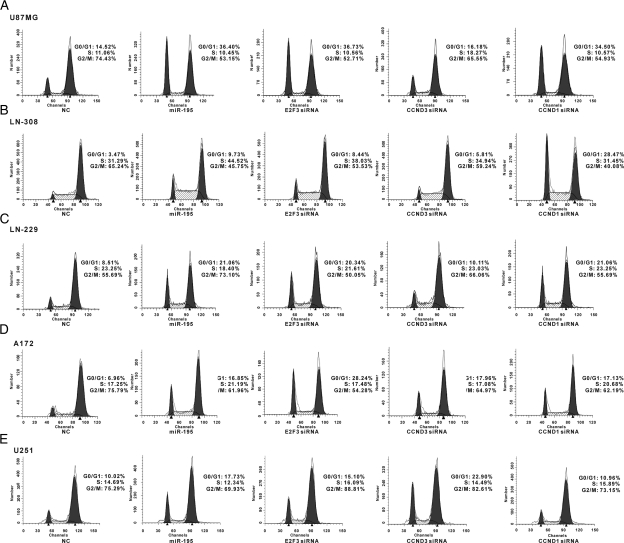
To confirm whether CCND3, E2F3, and CCND1 were responsible for the miR-195–mediated cell cycle arrest effect, we used siRNAs to knockdown endogenous CCND3, E2F3, and CCND1 expression and determined whether the resulting effect on cell cycle progression was similar to that of miR-195 restoration. siRNAs were transfected into the 5 glioblastoma cell lines, and cell cycle assays were performed as described above. As shown in Fig. 3, knockdown of E2F3 arrested glioblastoma cells at the G1/S transition in a similar manner to miR-195 expression, but knockdown of CCND3 did not cause significant cell cycle arrest on the majority of glioblastoma cell lines, except for A172 cells and U251 cells (Fig. 3D and E). Of note, although CCND1 may not be a functional target of miR-195 in most glioblastoma cells, CCND1 knockdown significantly blocked cells at the G1/S transition. These data indicate that E2F3 is likely to be the dominant target of miR-195 in regulating glioblastoma cell cycle progression (except for U251 cells), whereas CCND3 may be involved in regulating different aspects of glioblastoma cell behavior.
Fig. 3.
E2F3 is involved in miR-195-mediated G1/S transition arrest. Cell cycle distributions are detected in U87MG (A), LN-308 (B), LN-229 (C), A172 (D), and U251 (E) glioblastoma cells. Over-expression of miR-195 and knockdown of CCND1, CCND3, and E2F3 indicated that knockdown of E2F3 promotes the accumulation of cells at the G0/G1 stage and arrests cells at the G1/S transition in a similar manner to miR-195; however, knockdown of CCND3 did not inhibit cell cycle progression, except for A172 and U251 cells.
miR-195 Likely Regulates Glioblastoma Cell Invasion by Modulating the CCND3/p27Kip1 Pathway
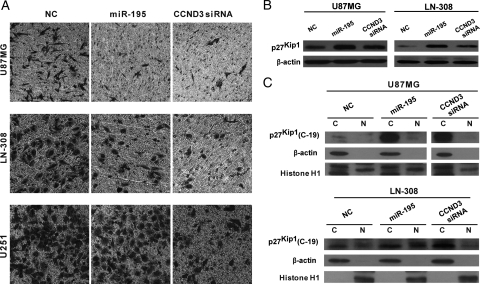
In addition to their role in cell cycle control, D-cyclins are also involved in tumor cell migration.20,23,24 However, their role in regulating cell migration or invasion in glioma cells is currently unknown. To address this question, we investigated whether silencing CCND3 impacts glioblastoma cell mobility. The U87MG, LN-308, and U251 cell lines were used in these experiments. As shown in Fig. 4A, knockdown of CCND3 significantly inhibited the invasive ability of glioblastoma cells through extracellular matrix substrates. Moreover, over-expression of miR-195 in glioblastoma cells similarly inhibited their invasion, except for U251 cells. This result suggests that CCND3 is primarily responsible for modulating glioblastoma cell invasion and that miR-195 may repress glioma cell invasion by targeting CCND3. Silencing of CCND1 also substantially reduced the migration of U87MG cells, in which miR-195 only slightly down-regulated CCND1 protein expression (Supplementary Fig. S1A). This result suggests that CCND3 might regulate cell invasion in a similar way to CCND1.
Fig. 4.
miR-195 impairs cell invasion through extracellular matrix substrates and promotes the expression of p27Kip1 in U87MG, LN-308, and U251 cells. A, Over-expression of miR-195 and knockdown of CCND3 inhibit cell invasion through ECM in U87MG cells and LN-308 cells, except for U251 cells. Forty-eighthours posttransduction, cells were collected and seeded in 8-μm pore size transwell chambers. After 20 h, the cells on the bottom side of each chamber were fixed and stained. Rescue of miR-195 expression and knockdown of CCND3 up-regulated both total expression of p27Kip1 (B) and cytoplasmic expression of p27Kip1 (with the COOH-terminal) in U87MG and LN-308 cells (C).
Previous studies have shown that CCND1 promotes cellular migration by inducing the expression of p27Kip1;23 therefore, we analyzed protein expression of p27Kip1 in U87MG and LN-308 cells transfected with miR-195 or si-CCND3. Of surprise, the expression of p27Kip1 was significantly elevated by miR-195 over-expression and by knockdown of CCND3 in U87MG and LN-308 cells, compared with the NC group (Fig. 4B). In addition, silencing of CCND1 also increased the expression of p27Kip1 in U87 cells (Supplementary Fig. S1B). This result indicates that p27Kip1 is repressed by CCND1 and CCND3 in U87MG cells, suggesting that CCND1 and CCND3 promote cellular invasion in glioblastoma cells through a mechanism distinct from that in mammary epithelial cells.23
p27Kip1 has been shown to either inhibit or promote cell migration in a cell type-dependent manner.23,25,26 In U87MG and other glioblastoma cells, p27Kip1 has been shown to inhibit cell invasion.26 In addition, the cytoplasmic abundance of p27Kip1 has been inversely correlated with glioblastoma cell motility. To assess whether down-regulation of CCND3 could increase the cytoplasmic protein levels of p27Kip1 in glioblastoma cells, we analyzed the abundance of p27Kip1 in the cytoplasm and nucleus by differential extraction of cytoplasmic and nuclear proteins. As expected, the cytoplasmic expression of p27Kip1 was substantially elevated by knockdown of CCND3 in U87MG and LN-308 cells (Fig. 4C). In addition, knockdown of CCND1 also increased the expression of p27Kip1 in U87MG cells (Supplementary Fig. S1C). Of note, knockdown of CCND3 increased p27Kip1 expression only in the cytoplasm, but CCND1 knockdown resulted in a significant up-regulation of both nuclear and cytoplasmic p27Kip1 (Fig. 4C and Supplementary Fig. S1C). Of interest, over-expression of miR-195 considerably increased the expression of p27Kip1 in the cytoplasm, but only slightly up-regulated p27Kip1 nuclear expression (Fig. 4C). This outcome may be attributable to the fact that over-expression of miR-195 significantly reduces the expression of CCND3 but only slightly down-regulates CCND1 in U87MG cells. Therefore, these data suggest that miR-195 inhibits glioblastoma cell invasion primarily by targeting CCND3 to up-regulate the cytoplasmic expression of p27Kip1.
Discussion
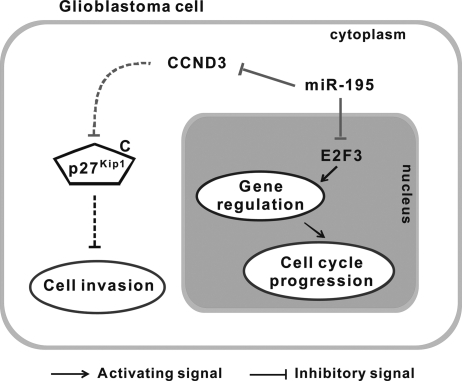
This study demonstrates that miR-195 plays a tumor-suppressor role in glioblastoma cells by inhibiting both cell proliferation and invasion, likely by directly targeting E2F3 and CCND3, respectively. These data support a model (illustrated in Fig. 5) in which miR-195 is down-regulated during glioblastoma progression, directly resulting in the deregulation of 2 miR-195 targets: E2F3 and CCND3. High levels of E2F3 protein activate the transcription of genes promoting cell cycle progression, which leads to abnormal cell proliferation. In addition, constitutive expression of CCND3 may reduce the expression of p27Kip1 in the cytoplasm and affect downstream signaling pathways that normally repress cell invasion, resulting in increased glioma cell invasion.
Fig. 5.
miR-195 regulates cell cycle progression and cell invasion in glioblastoma cells. In glioblastoma cells, miR-195 is down-regulated. E2F3 and CCND3 protein levels inversely correlate with miR-195 expression in glioblastoma cell lines. Moreover, E2F3 and CCND3 are direct targets of miR-195. The down-regulation of E2F3 can suppress the transcription of cell cycle-related genes and can cause accumulation of cells in G1 phase and inhibition of cell cycle progression. However, the miR-195–mediated CCND3 suppression can up-regulate the expression of cytoplasmic p27Kip1 and affect the expression of other proteins controlling cell migration, resulting in repression of glioblastoma cell invasion.
The miR-15/16 family is involved in regulating a wide variety of biological and pathological processes, including cell division, apoptosis, metabolism, the stress response, and angiogenesis.12 Aberrant expression of miR-15/16 family members has been widely implicated in cancer; however, whether they are involved in cellular migration or invasion is unclear. In this work, we provide conclusive evidence that miR-195, one member of the miR-15/16 family, not only arrests the cell cycle progression but also strongly inhibits glioblastoma cell invasion. These results suggest that miR-15/16 family members play more complicated roles in tumorigenesis than currently appreciated.
While our study was under way, other groups have reported that miR-195 arrests cell cycle progression in a lung cancer cell line by targeting CCND1 and CDK6 or by repressing CCND1, CDK6, and E2F3 in human hepatocellular carcinoma cell lines.27,28 In our experiments, we showed that miR-195 did not suppress CCND1 expression in 4 of 5 glioblastoma cell lines tested (A172, LN-229, U251, and LN-308, but not U87MG). We analyzed the 3′UTR of CCND1 mRNA from the 4 glioblastoma cell lines by reverse-transcription polymerase chain reaction and sequencing but did not find any mutations or deletions in the putative miR-195 binding site (data not shown). We therefore surmised that the inability of miR-195 to suppress CCND1 expression may be mediated by an unclear mechanism in which unknown factors may interfere with the interaction between miR-195 and CCND1 mRNA. For example, previous studies have reported that some RNA-binding proteins (RBPs) can bind to the 3′UTR of mRNAs to protect them from miRNA-mediated repression.29,30 We analyzed the high-throughput CLIP-seq data from other researchers using the starBase computational platform31 (developed by our laboratory, available on http://starbase.sysu.edu.cn/deepView.php) to investigate whether any RBP could bind to the miR-195 binding sites in the 3′UTR of CCND1, CCND3, and E2F3. Of interest, we find that IGF2BP1 or IGF2BP2 could bind to the 3′UTR of CCND1 and CCND3, and the binding regions happen to overlap with the binding sites for miR-195 (as shown in the Supplementary Fig. S2). Therefore, these RBPs may be over-expressed in the corresponding glioblastoma cells and protect the targets from miR-195–mediated repression.
In this work, we provide the first evidence that CCND3 is an important regulator of glioblastoma invasion in addition to its role as a cell cycle regulator. Further supporting this role in invasion, other studies have shown that CCND3 is mainly expressed in high-grade malignant gliomas and associated with transformation to a malignant phenotype.32 Moreover, we reveal a novel mechanism of CCND3- and CCND1-mediated facilitation of glioblastoma cell invasion through their inhibition of p27Kip1 cytoplasmic expression. Previous studies support our model by showing that p27Kip1 inhibits glioblastoma cell invasion.26 This is distinct from previous studies in mammary epithelial cells and fibroblast cells, which have shown that CCND1 induces p27Kip1 expression to promote cell migration.23,25
Stathmin/oncoprotein 18, also known as STMN1, is a microtubule regulatory protein that was recently shown to play an important role in regulation of malignant glioma cell migration and invasion.33 Moreover, STMN1 may be an important downstream target of p27Kip1 in mediating cell migration and invasion.34,35 We found that over-expression of miR-195 can reduce STMN1 expression in U87MG and LN-308 cells (Supplementary Fig. S3A), implying that miR-195 might regulate the migration and invasion of glioblastoma cells through modulating the CCND3-p27-stathmin pathway (Supplementary Fig. S3B).
miR-15/16 family members usually display reduced expression in multiple forms of human cancer, and it is likely that they play a tumor-suppressive role.14,15,27,28,36,37 However, certain members of this family have been shown to be up-regulated in some types of cancers, thus suggesting they might function as oncomiRs.37 Of note, miR-195 has always shown down-regulated expression in all cancer cell types studied when compared with related normal tissues,37 which suggests that down-regulation of miR-195 may be a common event during tumor development. In support of this hypothesis, miR-195 has been shown to suppress cell proliferation and/or tumorigenicity of all tested cancers, including lung cancer,28 hepatocellular carcinoma,27 colorectal cancer,36 and here, in glioblastoma. A recent report showed that down-regulation of miR-195 in adrenocortical carcinomas is significantly associated with poor prognosis.38 Because miR-195 is down-regulated in glioblastoma cells and suppresses both cell proliferation and invasion, we speculate that down-regulation of miR-195 may be similarly associated with a poorer prognosis for patients with glioblastoma.
Supplementary Material
Funding
This work was supported by the National Natural Science Foundation of China (30870530, 81070589, and 30830066) and the National Basic Research Program (2011CB811300) from the Ministry of Science and Technology of China.
Supplementary Material
Acknowledgments
We thank Dr. Shi-Yuan Cheng (the University of Pittsburgh Cancer Institute) for providing the glioblastoma cell lines; Zhi Rao, Xiao-Hong Chen, and Yi-Ling Chen for technical assistance; and Dr. Meng-Feng Li (Sun Yat-Sen University) for helpful discussion. Q.-Q.Z. and H.X. contributed equally to this work.
Conflict of interest statement. None declared.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. doi:10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–2710. doi: 10.1101/gad.1596707. doi:10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 3.Lefranc F, Brotchi J, Kiss R. Possible future issues in the treatment of glioblastomas: special emphasis on cell migration and the resistance of migrating glioblastoma cells to apoptosis. J Clin Oncol. 2005;23(10):2411–2422. doi: 10.1200/JCO.2005.03.089. doi:10.1200/JCO.2005.03.089. [DOI] [PubMed] [Google Scholar]
- 4.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126. doi:10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 5.Holland EC. Gliomagenesis: genetic alterations and mouse models. Nat Rev Genet. 2001;2(2):120–129. doi: 10.1038/35052535. doi:10.1038/35052535. [DOI] [PubMed] [Google Scholar]
- 6.Louis DN. Molecular pathology of malignant gliomas. Annu Rev Pathol. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. doi:10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- 7.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170(5):1445–1453. doi: 10.2353/ajpath.2007.070011. doi:10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. doi:10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 9.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. doi:10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 10.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. doi:10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 11.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. doi:10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 12.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. doi:10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths-Jones S, Saini HK, van Dongen S, et al. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. doi:10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. doi:10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Applied Biosystems. MicroRNA expression signature in human glioblastoma multiforme brain tumor. 2005. Available from: http://www.appliedbiosystems.com .
- 17.Gaur A, Jewell DA, Liang Y, et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67(6):2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. doi:10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 18.Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–1801. doi: 10.1056/NEJMoa050995. doi:10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 19.Xu H, He JH, Xiao ZD, et al. Liver-Enriched Transcription Factors Regulate MicroRNA-122 That Targets CUTL1 During Liver Development. Hepatology. 2010;52(4):1431–1442. doi: 10.1002/hep.23818. doi:10.1002/hep.23818. [DOI] [PubMed] [Google Scholar]
- 20.Radulovich N, Pham NA, Strumpf D, et al. Differential roles of cyclin D1 and D3 in pancreatic ductal adenocarcinoma. Mol Cancer. 2010;9:24. doi: 10.1186/1476-4598-9-24. doi:10.1186/1476-4598-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buschges R, Weber RG, Actor B, et al. Amplification and expression of cyclin D genes (CCND1, CCND2 and CCND3) in human malignant gliomas. Brain Pathology. 1999;9(3):435–442. doi: 10.1111/j.1750-3639.1999.tb00532.x. doi:10.1111/j.1750-3639.1999.tb00532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui JG, Zhao Y, Sethi P, et al. Micro-RNA-128 (miRNA-128) down-regulation in glioblastoma targets ARP5 (ANGPTL6), Bmi-1 and E2F-3a, key regulators of brain cell proliferation. J Neurooncol. 2010;98(3):297–304. doi: 10.1007/s11060-009-0077-0. doi:10.1007/s11060-009-0077-0. [DOI] [PubMed] [Google Scholar]
- 23.Li Z, Jiao X, Wang C, et al. Cyclin D1 induction of cellular migration requires p27(KIP1) Cancer Res. 2006;66(20):9986–9994. doi: 10.1158/0008-5472.CAN-06-1596. doi:10.1158/0008-5472.CAN-06-1596. [DOI] [PubMed] [Google Scholar]
- 24.Li Z, Wang C, Prendergast GC, et al. Cyclin D1 functions in cell migration. Cell Cycle. 2006;5(21):2440–2442. doi: 10.4161/cc.5.21.3428. doi:10.4161/cc.5.21.3428. [DOI] [PubMed] [Google Scholar]
- 25.Besson A, Gurian-West M, Schmidt A, et al. p27Kip1 modulates cell migration through the regulation of RhoA activation. Genes Dev. 2004;18(8):862–876. doi: 10.1101/gad.1185504. doi:10.1101/gad.1185504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schiappacassi M, Lovat F, Canzonieri V, et al. p27Kip1 expression inhibits glioblastoma growth, invasion, and tumor-induced neoangiogenesis. Mol Cancer Ther. 2008;7(5):1164–1175. doi: 10.1158/1535-7163.MCT-07-2154. doi:10.1158/1535-7163.MCT-07-2154. [DOI] [PubMed] [Google Scholar]
- 27.Xu T, Zhu Y, Xiong Y, et al. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology. 2009;50(1):113–121. doi: 10.1002/hep.22919. doi:10.1002/hep.22919. [DOI] [PubMed] [Google Scholar]
- 28.Liu Q, Fu H, Sun F, et al. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36(16):5391–5404. doi: 10.1093/nar/gkn522. doi:10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kedde M, Strasser MJ, Boldajipour B, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131(7):1273–1286. doi: 10.1016/j.cell.2007.11.034. doi:10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 30.Bhattacharyya SN, Habermacher R, Martine U, et al. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125(6):1111–1124. doi: 10.1016/j.cell.2006.04.031. doi:10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 31.Yang JH, Li JH, Shao P, et al. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39(Database issue):D202–D209. doi: 10.1093/nar/gkq1056. doi:10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Zhao M, Huang AY, et al. The effect of cyclin D expression on cell proliferation in human gliomas. J Clin Neurosci. 2005;12(2):166–168. doi: 10.1016/j.jocn.2004.03.036. doi:10.1016/j.jocn.2004.03.036. [DOI] [PubMed] [Google Scholar]
- 33.Liang XJ, Choi Y, Sackett DL, et al. Nitrosoureas inhibit the stathmin-mediated migration and invasion of malignant glioma cells. Cancer Res. 2008;68(13):5267–5272. doi: 10.1158/0008-5472.CAN-07-6482. doi:10.1158/0008-5472.CAN-07-6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iancu-Rubin C, Atweh GF. p27(Kip1) and stathmin share the stage for the first time. Trends Cell Biol. 2005;15(7):346–348. doi: 10.1016/j.tcb.2005.05.008. doi:10.1016/j.tcb.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 35.Baldassarre G, Belletti B, Nicoloso MS, et al. p27(Kip1)-stathmin interaction influences sarcoma cell migration and invasion. Cancer Cell. 2005;7(1):51–63. doi: 10.1016/j.ccr.2004.11.025. doi:10.1016/j.ccr.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 36.Liu L, Chen L, Xu Y, et al. microRNA-195 promotes apoptosis and suppresses tumorigenicity of human colorectal cancer cells. Biochem Biophys Res Commun. 2010;400(2):236–240. doi: 10.1016/j.bbrc.2010.08.046. doi:10.1016/j.bbrc.2010.08.046. [DOI] [PubMed] [Google Scholar]
- 37.Finnerty JR, Wang WX, Hebert SS, et al. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402(3):491–509. doi: 10.1016/j.jmb.2010.07.051. doi:10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soon PS, Tacon LJ, Gill AJ, et al. miR-195 and miR-483-5p Identified as Predictors of Poor Prognosis in Adrenocortical Cancer. Clin Cancer Res. 2009;15(24):7684–7692. doi: 10.1158/1078-0432.CCR-09-1587. doi:10.1158/1078-0432.CCR-09-1587. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.