Abstract
The human cytomegalovirus (HCMV) and glioma symposium was convened on April 17, 2011 in Washington, DC, and was attended by oncologists and virologists involved in studying the relationship between HCMV and gliomas. The purpose of the meeting was to reach a consensus on the role of HCMV in the pathology of gliomas and to clarify directions for future research. First, the group summarized data that describe how HCMV biology overlaps with the key pathways of cancer. Then, on the basis of published data and ongoing research, a consensus was reached that there is sufficient evidence to conclude that HCMV sequences and viral gene expression exist in most, if not all, malignant gliomas, that HCMV could modulate the malignant phenotype in glioblastomas by interacting with key signaling pathways; and that HCMV could serve as a novel target for a variety of therapeutic strategies. In summary, existing evidence supports an oncomodulatory role for HCMV in malignant gliomas, but future studies need to focus on determining the role of HCMV as a glioma-initiating event.
Keywords: cancer, DNA virus, gliomas, herpesvirus, human cytomegalovirus
This consensus statement is the culmination of a series of discussions held at an open meeting among researchers studying the impact of human cytomegalovirus (HCMV) in gliomas. Sponsored by Accelerate Brain Cancer Cure and the National Brain Tumor Society, this meeting convened in Washington, DC, in April 2011, and provided the opportunity for oncologists and virologists to freely discuss their most current data addressing this topic. Here, we report the consensus position in 4 major areas:
(1) existence of HCMV in gliomas,
(2) role of HCMV in gliomas,
(3) HCMV as a therapeutic target, and
(4) key future investigative directions.
Existence of HCMV in Gliomas
Detection of HCMV Proteins, Genes, and Nucleotides
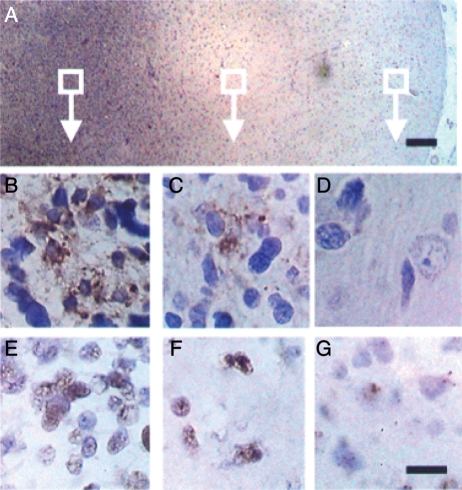
The expression of HCMV proteins and oligonucleotides in a high percentage of gliomas was first reported by Cobbs et al. in 2002.1 Since that time, controversy regarding the existence and role of HCMV in gliomas has been debated in the literature. An equal number of studies that specifically address the presence or absence of the virus in this disease have been published.1–8 Documenting the presence of HCMV in gliomas has been confounded by the lack of a uniform operational definition of positivity in tumor tissues and the use of different methodological approaches. The approaches used in these various studies were categorically addressed by Scheurer et al. in 2008. The authors described the necessity of optimizing sample preparation and detection techniques when extracting from paraffin-embedded tissues to adjust for low levels of infection. By doing so, they were able to detect the HCMV immediate early 1 (IE1) protein in 100% of glioblastomas and 82% of low-grade gliomas with use of immunohistochemistry.7 They further reported detection of HCMV-specific oligonucleotides in the same areas of IE1 expression in the tumor, as determined by in situ hybridization. Their conclusion, as initially described by Cobbs et al. in 2002, is that HCMV IE1 and virus-specific oligonucleotides could be readily detected by optimizing these techniques. Moreover, they validated another of the findings by Cobbs and colleagues, which was the lack of detection of virus expression (or virus-specific oligonucleotides) in areas of necrosis or outside the tumor margin. These findings (Fig. 1) are consistent among studies that have described the ability to detect HCMV proteins and oligonucleotides.1,3,7
Fig. 1.
Correlation of patterns of immunohistochemical localization of human cytomegalovirus (HCMV) immediate early 1 (IE1) protein with in situ hybridization for HCMV DNA in a glioblastoma (GBM) that invades normal brain. (A) Low-power view of anti-IE1 immunostain demonstrates GBM invading normal brain cortex (cortical surface at far right; bar, 200 µm). (B–D) Boxed areas in (A) at higher power demonstrate IE1 immunoreactivity moving from an area of frank tumor (B) to an area of invading tumor (C) to an area of normal brain (D). Detection of HCMV DNA by in situ hybridization using an HCMV total genome probe (in an adjacent section and similar regions of the same tumor in B–D) reveals a similar pattern, moving from malignant (E) to invasive (F) to normal (G) brain. Bar, 10 µm.
To date, immunohistochemistry, in situ hybridization, electron microscopy, polymerase chain reaction (PCR) coupled with DNA sequencing, enzyme-linked immunosorbent assay, and flow cytometry have been used to detect HCMV proteins and DNA in human glioblastoma tissue samples.1,3,5,7–11 Collectively, these studies have identified the presence of the HCMV proteins IE1, US28, pp65, gB, HCMV IL-10, and pp28 and the HCMV genes IE1 and gB. The most commonly studied protein has been IE1. Among studies with positive findings that used immunohistochemistry for the detection of IE1, one reported 16% of samples positive for this protein,8 and the remainder were in the range of 93%–100%.1,3,7,10 A control probe specific for detection of a herpes simplex virus oligonucleotide did not show positivity in glioblastoma samples.1 No HCMV proteins or nucleotides were detected in normal brain controls, areas of normal brain adjacent to tumor, or in one HCMV-negative glioblastoma.1,3,7,10
Sequencing the HCMV genome found in DNA isolated from human glioblastoma samples has proven to be challenging. Unpublished data presented by T. F. Kowalik demonstrated difficulty in sequencing complete genomic HCMV DNA from individual samples, possibly as a result of low copy numbers of viral DNA or of fragmented, discontinuous viral genomes. However, HCMV genomic DNA was detected in 94% of samples using a combined PCR-DNA sequencing methodology, a technique that revealed polymorphisms in certain regions of the glioblastoma-associated viral genomes, such as those encoding the tegument protein pp65. Because HCMV populations have been shown to be highly diverse in clinical specimens,12 a possible explanation is that the HCMV genomes associated with glioblastoma are tumor-specific and derive from the diverse viral populations that exist in individuals. Cumulatively, on the basis of these findings and the results of studies describing activity of HCMV proteins in gliomas and glioma cell lines,9–11,13,14 a consensus was reached that there is sufficient evidence to conclude that HCMV sequences and viral gene expression exist in most, if not all, malignant gliomas.
Lytic Versus Latent Disease
A second area of controversy is whether or not HCMV exists in a lytic or latent state in gliomas. Lytic HCMV infection is characterized by intranuclear expression of the IE genes IE1 and IE2. Their gene products, along with those of the delayed early (DE) genes, regulate transcription of viral and host genes, which, in turn, drive viral replication. This process is facilitated by the inhibition of apoptosis mediated by these gene products.15 When seen histologically, the presence of intranuclear viral protein inclusions, the classic “owl's eyes,” confirms this diagnosis. This finding has not been observed in glioma specimens or in glioma cancer stem cells (gCSCs) (A.B. Heimberger, unpublished data). To date, no investigator has demonstrated the production of infectious HCMV virions by gliomas.
In contrast, latency is characterized by the carriage of the viral genome in the absence of both lytic gene expression (which include the IE genes) and production of infectious virions.16 The mechanisms that govern latency have not yet been explicitly defined, and only a small number of HCMV latency-associated transcripts have been identified.16 Expression of HCMV latency-associated transcripts has not yet been measured in gliomas, but the ubiquitous expression of IE1 implies that HCMV does not reside in these tumors in a truly latent state.
The existence of HCMV in gliomas does not appear to fit classic definitions of lytic or latent disease. Typically, HCMV lytic infection results in productive replication, significant cellular damage, and frequently, cell lysis—referred to as the cytopathic effect. However, known tumor viruses are typically latent, and some replicate in the host cell using host cellular proteins without producing infectious virions.17 Given these facts, a model similar to that proposed for the role of HCMV in the development of cardiovascular disease could be considered for gliomas. Here, it was postulated that persistent infection of endothelial cells by HCMV plays a role in the development of hypertension.18 Persistent infection, as demonstrated by viral gene expression without cytopathic effect, led to production of inflammatory cytokines and renin, which resulted in the development of hypertension in an in vivo model. Applying this model to gliomas, persistent infection could result in the expression of HMCV genes, leading to production of cytokines that contribute to pathogenesis or of proteins known to disrupt cell-cycle regulation. Such candidate genes and products are outlined in more detail in later sections of this article. Furthermore, HCMV can encode ≥166 genes, not all of which have been extensively studied.15 In the context of tumor viruses, those expressed during latency could be the most significant, relative to gliomagenesis.
Epidemiology
The seroprevalence of HCMV in the general population is up to 80%,19 in contrast with the prevalence of glioblastoma of 0.0257%.20 To date, no epidemiological correlation between the timing of HCMV infection and the subsequent risk for development of gliomas has been reported. In reviewing the epidemiology of tumor viruses, many factors other than simple association influence the development of cancers associated with these viruses. As examples, the seroprevalence of human papillomavirus and Epstein Barr virus exceeds the incidence of cervical cancer or Burkitt's lymphoma, and the development of liver cancer is associated with a combination of carcinogenic exposure and either hepatitis B virus or hepatitis C virus infection.17 Therefore, a proposed role for HCMV in gliomagenesis is most likely to be associated with a yet undefined event.
Two unique hypotheses are postulated: one presented at the meeting and one recently published. On the basis of studies showing that HCMV enters the cell via PDGFRα21 and that specific PDGFR haplotypes are associated with a greater incidence of glioblastoma,22 A.B. Heimberger suggested that there may be PDGFR receptor haplotype differences that confer differential susceptibility to HCMV infection of glioma cells where the receptor is overexpressed or present on a glioma cell of origin. Alternatively, a recently published proposal suggested that host genes that would affect binding affinity to an HCMV-encoded Fc γ receptor (FcγR) could serve as a risk factor for gliomagenesis. HCMV FcγR is involved in the ability to evade immune detection by interfering with antibody-mediated cellular toxicity. Thus, individuals possessing FcγR receptors with different binding affinities may have variable capability for clearing virally infected cells.23
Role of HCMV in Gliomas
Oncomodulation
The most accepted concept discussed at the meeting is that there is sufficient evidence to support the hypothesis that HCMV could modulate the malignant phenotype in glioblastomas.
The concept of HCMV and oncomodulation was first proposed by Cinatl et al. in 1996, who provided evidence that, although HCMV could modulate the malignant properties of cells, it was not directly involved in transformation.24 Earlier studies have described the transforming capability of HCMV in rodent and human cells, but its DNA was not retained in these cells, and the presence of HCMV antigens, although initially demonstrated, decreased with subsequent passage.25 At present, HCMV is not considered to be an oncogenic virus. Features attributed to known oncogenic viruses (Table 1) have not been identified in HCMV-infected gliomas, such as sustained expression of oncoproteins or genomic integration. Genetic mapping of gliomas as a means of establishing tumor phenotype has not shown HCMV gene products, but these platforms did not include HCMV genes.
Table 1.
Basic mechanisms of established tumor viruses
| Virus | Oncogenic Mechanism |
|---|---|
| Human Papilloma Virus | Oncoprotein, Integration into host genome |
| Hepatitis B Virus | Oncoprotein, Integration into host genome |
| Epstein Barr Virus | Oncoprotein, Translocation signature |
| Human Herpesvirus-8 (Kaposi's Sarcoma) | Oncoprotein |
In 2000, Hanahan and Weinberg described 6 essential alterations in cell physiology that are the hallmarks of cancer, including (1) sustaining proliferative signaling, (2) evading growth suppressors, (3) activating invasion and metastasis, (4) enabling replicative immortality, (5) inducing angiogenesis, and (6) resisting cell death.26 These criteria were recently updated in March 2011 to include (7) deregulating cellular energetics, (8) avoiding immune detection, (9) genome instability, and (10) mutation- and/or tumor-promoting inflammation.27 Cobbs, Alwine, and Kalejta presented studies showing an overlap of HCMV biology with the essential alterations of cell physiology that are hallmarks of cancer (Table 2).10,11,28–60 On the basis of these publications and the findings that we outline below, we consider these altered physiologies with respect to glioblastoma and how HCMV biology can enable the characteristics of cancer and function as an oncomodulator.
Table 2.
Overlap of Human Cytomegalovirus (HCMV) biology with altered cellular physiologies classified as hallmarks of cancer
| Cancer Hallmark | HCMV Activity | HCMV Protein Involved |
|---|---|---|
| Sustaining Proliferative Signaling | Induces cell cycle progression to S phase | IE2 |
| Induces expression of E2F genes | pp71 | |
| Phosphorylates Rb | UL97 | |
| Evading Growth Suppressors | Activates EGFR | HCMV infection |
| Dysregulates Cyclin E expression | IE1 | |
| Inhibits p53 degradation | mtrII | |
| Decreases levels of p21 | ||
| Induces expression of p53 | ||
| Binds to p53 | ||
| Activating Invasion and Metastasis | Activation of RhoA dependent motility of U373 cells | US28 |
| Activates smooth muscle cells | ||
| Enabling Replicative Immortality | Activation of telomerase | IE1 |
| Inducing Angiogenesis | Induction of VEGF expression | US28 |
| Induction of IL-8 | IE1 | |
| Resisting Cell Death | Inhibits apoptosis | IE1 |
| Activates PI3-K/Akt pathway | IE2 vMIA, vICA | |
| Deregulating Cellular Energetics | Increases flux through glycolytic pathway, | HCMV infection |
| acetyl CoA, flux of carbon, nucleotide biosynthesis | ||
| Avoiding Immune Destruction | Production of homologs to immunosuppressive cytokines | HCMV IL-10 |
| Inhibits expression of MCH I | US2 | |
| Intracellular retention of NKG2D | UL16 | |
| Induces expression of TGF-β1 | IE2 | |
| Genome Instability and Mutation | Chromosome damage | Unidentified protein |
| Inhibits DNA damage repair | HCMV infection | |
| Increases mutation frequency | pp65 and pp71 | |
| Induces chromosome aberrations in cell lines | IE1 | |
| UL76 | ||
| Tumor Promoting Inflammation | Induces production of RANTES, fraktalkine, MCP-1 | HCMV infection |
| NF-κB activation & IL-6 production | US28 |
Sustaining Proliferative Signaling, Evading Growth Suppressors, and Enabling Replicative Immortality
The PDGFRα polypeptide is a strong candidate as the portal of access of HCMV into malignant glioma cells or their cells of origin. The HCMV envelope protein gB has been shown to directly interact with and to phosphorylate this receptor.21 Furthermore, viral entry into the cell was shown to activate the PI(3)K pathway with induction of Akt, with none of these events being detected after either blockade of the receptor or deletion of the gene encoding it. Although focal gene amplification and expression of PDGFRα is highest in the proneural phenotype, amplification of PDGFRα is seen in all phenotypes of glioblastoma.61
To evaluate the effects of persistent IE1 expression in glioblastomas, stable expression of this viral protein in human glioblastoma cell lines (U87, U251, LN229, UL118) was evaluated.14 Such expression demonstrated differential effects on cellular proliferation, such that in some cases, it was increased and, in others, unaffected or decreased. MAPK and AKT signaling was considered as a possible mechanism to explain this finding and was found to show a sustained increase. Another important finding was the increase in phosphorylation of the cell-cycle regulator Rb in all the malignant glioma cell lines studied. The inactivation of Rb by HCMV in nonglioblastoma cell lines has been described elsewhere in the virology literature, whereby 2 different viral proteins both degrade and phosphorylate the tumor suppressor.36,38 Numerous studies have documented HCMV interference with the cell cycle, showing p53 and Rb being targeted, as reviewed in Michaelis et al.43 Although not extensively studied in glioblastomas, this represents an important area for further study.
In 2009, Straat et al. reported constitutive telomerase reverse transcriptase (hTERT) expression and telomerase activation as a result of HCMV infection in multiple malignant glioma cell lines.11 To examine a potential mechanism for the activation of telomerase subsequent to HCMV infection, the authors evaluated the behavior of one of its regulators, the transcription factor specificity protein 1 (Sp1). ChIP analysis in fibroblast cells (MRC 5) indicated that Sp1 and IE1 were bound to the hTERT promoter after HCMV infection. When human glioblastoma tissue samples were examined using immunohistochemistry, a direct correlation between immediate early antigen (IEA) and hTERT expression was seen in all 10 patient samples.
Activating Invasion and Metastasis
HCMV infection of U87 cells was found to enhance focal adhesion kinase activity, in addition to significantly increasing cell migration relative to that in immortalized human astrocytes.13 This suggests that HCMV can be associated with a more aggressive phenotype of glioblastoma.
Inducing Angiogenesis
US28 expression has been identified in glioblastomas, primarily in vascular endothelial cells.10 It is a constitutively active HCMV-encoded G protein-coupled receptor expressed with early-stage kinetics during infection. It has gene sequence homology to human chemokine receptors CCR1and CX3R and is capable of binding chemokines CCL2, CCL5, and CX3CL1, among others.62 Stable expression of US28 in NIH 3T3 cells resulted in a significant increase in IL-6 and VEGF production,10 and implantation of these cells in nude mice resulted in tumor formation.63 The intracellular effects of US28 are multiple and include upregulation of cyclin D163 and NF-κB.10 The NF-κB activity increased production of IL-6, which activated the signal transducer and activator of transcription 3 (STAT3) in both an autocrine and paracrine manner.10 STAT3 has previously been described as a key molecular hub of tumorigenesis64,65 and immune suppression, especially in gliomas.66 Its induction in the neural progenitor cells of mice has been shown to induce high-grade gliomas, along with increased VEGF expression and angiogenesis.67
Genomic Instability and Interference with DNA Damage Response
Human neural progenitor cells are fully permissive to HCMV infection, which results in premature and abnormal differentiation.68 As a consequence of infection, altered attachment, migration, loss of multipotency, and down-regulation of MIR21, OLIG 1, and SOX2 have been described.68 In the context of persistent infection of vulnerable stem cells, HCMV-mediated genomic injury could promote oncogenesis, because HCMV has been shown to induce specific chromosome damage.69 Purified virions from 3 different strains of HCMV were found to reproducibly induce breaks in chromosome 1 at 1q42 and 1q21 in 2 cell types (foreskin fibroblasts and human embryonic lung cells). Although this event does not occur at high frequency, it does occur reliably (E. A. Fortunato, personal communication). The loss of at least 1 copy of the chromosome 1q42 band has been reported in a small percentage of patients with glioma.70
To date, HCMV interference with the DNA damage response has not been studied in malignant gliomas, although such an interaction has been documented in the virology literature.33,35,71,72 Although this interaction is to facilitate viral replication, an occurrence in parallel with an appropriate genetic mutation could favor development of a glioblastoma.
Tumor-Promoting Inflammation and Avoidance of Immune Detection
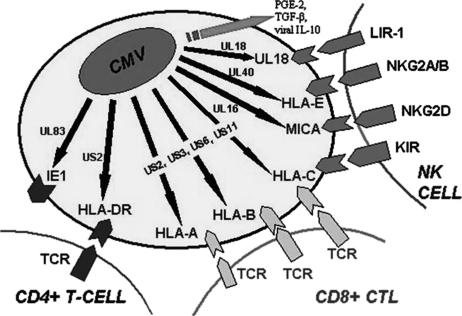
A feature common to both HCMV virulence and glioblastoma malignancy is the ability to evade immune detection. HCMV uses several mechanisms to evade the host-cell immune response and promote immune suppression (Fig. 2).73–77 HCMV has also been shown to promote a chronic inflammatory state associated with increased expression of ROS, RNS, and COX-2.78–82
Fig. 2.
Selected immune subversive human cytomegalovirus (HCMV) proteins blocking CTL and NK cell recognition and antigen presentation pathways. HCMV gene nomenclature designates genes as UL for unique long and US for unique short to reflect regions of the genome from which the gene originates. UL83 (pp65) inhibits presentation of the immunodominant CMV protein immediate early 1 (IE1); US2 mediates degradation of HLA class I and II a chains; US3 causes retention of class I molecules within the ER; US6 inhibits TAP-mediated peptide transportation into the ER; US11 (like US2) causes destruction of class I a chains; UL16 inhibits NK cell recognition via the activating receptor NKG2D by binding to its ligands (ULBPs); UL18 activates LIR-1, an inhibitory receptor found on NK cells, lymphocytes, and most other immune cells; and UL40 activates the inhibitory NKG2A/B receptor by upregulating HLA-E expression. HCMV produces a functional IL-10 homolog (UL111A) and induces expression of cellular PGE-2 and TGF-β, which further inhibit NK cell response.
In most healthy individuals, HCMV remains latent throughout the lifetime of the host. Bone marrow CD34+ progenitor cells have been identified as one site of HCMV latency, and the latent viral genome is carried through the myeloid lineage as these cells differentiate.83 Terminal differentiation of immature myeloid cells into mature macrophages or dendritic cells in the context of inflammation and immunosuppression has been shown to reactivate the virus.16,84–86 Recent evidence suggests that glioblastoma tumor-associated macrophages and microglia are infected with HCMV.9 This population of cells is a major component of the tumor microenvironment.
HCMV can induce a unique M1/M2 polarization signature that promotes viral dissemination and persistence, a process involving the induction of IL-6 and TNF-α.87,88 These cytokines would be expected to contribute to an oncogenic microenvironment, because chronic expression of TNF-α and IL-6 is directly linked to oncogenic transformation in inflammation-induced animal models of cancer.89 Autocrine mechanisms may also exist, considering that TNF-α has been implicated in the reactivation of HCMV in immunosuppressed transplant recipients through enhancement of HCMV IE promoter activity.90 Whereas these cytokines are prototypical of the M1 type proinflammatory cascade, HCMV also simultaneously induces the immunosuppressive M2 type macrophage responses. Recently, it was reported that glioma cancer stem cells (gCSCs) harvested from human glioblastomas produce HCMV IL-10.9 This viral homolog of the human IL-10 immunosuppressive cytokine induces the M2 phenotype that has been described in glioblastoma-associated macrophages and microglia.91 As such, a feed-forward mechanism is proposed in which the HCMV-induced M2 macrophages/microglia produce increased VEGF and TGF-β and stimulate gCSC migration. This study also identified specific glioblastoma cellular subpopulations harboring HCMV, the gCSCs, and cells of monocyte lineage. In monocytes harboring HCMV, IE1 expression was induced after exposure to HCMV IL-10, which further potentiates the feed-forward mechanism.
HCMV as a Therapeutic Target
In Vivo Mouse Model
A recently developed preclinical model of murine CMV (MCMV) was presented by Kwon and Chiocca that could be used to test potential anti-HCMV therapeutics. In this model, transgenic (Mut3) mice were engineered to develop spontaneous gliomas that were then perinatally infected with MCMV. Mice that developed gliomas in the MCMV cohort exhibited more aggressive tumors and showed a marked decrease in median survival time, compared with the uninfected control cohort. This suggests that MCMV infection may accelerate glioma progression.
Clinical Trials: Valganciclovir and Tumor Vaccination
Results are pending from a phase I/II double-blind randomized clinical trial of valganciclovir administered to patients with gliomas as a postsurgical add-on therapy performed at the Karolinska Institute. Valganciclovir is a nucleoside analog and targets HCMV replication through the disruption of DNA synthesis. If a survival advantage is identified, it will be valuable to know whether it is the result of reduced activity of HCMV or of the activity of the drug in disrupting DNA synthesis in actively dividing tumor cells or a combined effect. Because the pathology of HCMV in gliomas does not recapitulate what is seen in active, lytic infections, it is not clear what the trial will demonstrate. However, the reporting of pathological and clinical correlates subsequent to valganciclovir treatment will provide important clues to viral contribution to disease, mechanisms of therapy, and potential interactions between antivirals and other chemotherapeutics.
A second therapeutic strategy targeting HCMV antigens expressed in glioblastomas was presented by Dr. Mitchell from Duke University. A phase I/II autologous dendritic cell vaccine pulsed with HCMV peptides in patients with newly diagnosed glioblastoma multiforme showed a median survival time of 21 months. A follow-up phase II clinical trial of patients with newly diagnosed glioblastoma multiforme is targeted to start by the end of 2011 and will consist of vaccinating patients with multiple HCMV peptides sequentially, along with temozolomide treatment in a manner similar to that used in the epidermal growth factor variant III peptide clinical trials.92,93 The feasibility of this type of approach is supported by a phase I clinical trial that investigated the use of vaccination with autologous dendritic cells pulsed with autologous tumor lysate. In this trial, a patient developed a robust HCMV-specific CD8+ T-cell response to the pp65 HCMV immunodominant epitope that began immediately after one injection of autologous tumor lysate-pulsed dendritic cells.5 Because HCMV proteins have not been found to be expressed outside the confines of tumor tissue, responses are not expected to target uninvolved surrounding brain tissue.
Although a specific role for HCMV in gliomas remains to be defined, there was agreement that it could serve as a novel target for a variety of therapeutic strategies.
Key Future Investigative Directions
Epidemiology and Risk Factors
No epidemiological study to date has been undertaken to ascertain why such a small percentage of the population with latent HCMV develops gliomas. It is unknown whether there are additional risk factors that predispose patients with glioma to the development of their disease, such as genetic polymorphisms that render susceptibility to the oncomodulatory effects of HCMV. Studies addressing possible genetic factors, such as the PDGFR haplotype, FcγR, or environmental factors need to be conducted to identify risk factors and to further elucidate the mechanisms involved in the role of HCMV in glioma pathology. Efforts are under way to develop a HCMV vaccine to prevent congenital birth defects, and ultimately, this cohort could be followed up longitudinally to ascertain the risk of glioma development.
Identification of Therapeutic Targets
Further elaboration of how HCMV contributes to glioma malignancy could identify novel therapeutic targets. Although a portal for cellular entry has been identified,21 how HCMV gains access to the central nervous system is unknown. It is possible that the virus is trafficked and introduced to the tumor milieu via circulating monocytes and that the immunosuppressive glioma microenvironment stimulates reactivation of disease, perpetuating a feed-forward mechanism, but this does not address the initiating event. A blockade of this mechanism could potentially reduce the aggressive nature of glioblastomas. Furthermore, there may be only specific HCMV strains that can initiate gliomagenesis. Irradiation of live HCMV renders the virus noninfectious. Does the same occur in the context of glioma? Does HCMV infect neural, glial, or glioma progenitor cells at an early stage of gliomagenesis? Would this allow an opportunity for immune clearance or immediately trigger immune suppression during early stages of gliomagenesis?
As data from the aforementioned clinical trials become available, it could serve as a platform in the formulation of hypotheses to address these questions.
Conclusions
Sufficient evidence has emerged to suggest that HCMV could modulate the malignant phenotype in glioblastomas, and elements of its biology overlap those considered to be hallmarks of cancer. Recent evidence supports the continued development of therapeutic HCMV vaccine to reduce glioblastoma's malignancy. Studies of the mechanisms used by HCMV should include a major initiative to understand the contributions of HCMV to gliomagenesis.
Conflict of interest statement. None declared.
Appendix
HCMV and gliomas symposium participants: James C. Alwine, Ph.D. (Cancer Biology and the Abramson Family Cancer Research Institute, School of Medicine, the University of Pennsylvania, Philadelphia, PA), William J. Britt, M.D. (Pediatrics, the University of Alabama at Birmingham, Birmingham, AL), Kevin Cassady (Pediatrics, the University of Alabama at Birmingham, Birmingham, AL), Susan M. Chang, M.D. (Neurological Surgery, the University of California at San Francisco, San Francisco, CA), E. Antonio Chiocca, M.D., Ph.D (Dardinger Neuro-Oncology Center, Neurological Surgery, James Cancer Hospital and the Ohio State University Medical Center, Columbus, OH), Charles S. Cobbs, M.D. (Neurological Surgery, the University of California at San Francisco, San Francisco, CA; California Pacific Medical Center Research Institute, San Francisco, CA), Kristine Dziurzynski, M.D. (Neurosurgery, The University of Texas M. D. Anderson Cancer Center, Houston, TX), Elizabeth A. Fortunato, Ph.D. (Biological Sciences, the University of Idaho Moscow, ID), Amy B. Heimberger, M.D. (Neurosurgery, The University of Texas M. D. Anderson Cancer Center, Houston, TX), Robert F. Kalejta, Ph.D. (Institute for Molecular Virology and McArdle Laboratory for Cancer Research, the University of Wisconsin-Madison), Timothy F. Kowalik, Ph.D. (Microbiology and Physiological Systems, the University of Massachusetts Medical Center, Worcester, MA), Chang-Hyuk Kwon, Ph.D. (Dardinger Neuro-Oncology Center, Neurological Surgery, James Cancer Hospital and the Ohio State University Medical Center, Columbus, OH), Stuart R. McGregor Dallas, Ph.D. (Molecular Biology, Lewis Thomas Laboratory, Princeton University, Princeton, NJ), Lisa Matlaf, Ph.D. (California Pacific Medical Center Research Institute, San Francisco, CA), Duane Mitchell, M.D., Ph.D. (Neurosurgery, Duke University Medical Center, Durham, NC), Rick Price, M.D. (Dardinger Neuro-Oncology Center, Neurological Surgery, James Cancer Hospital and the Ohio State University Medical Center, Columbus, OH), Martine J. Smit, Ph.D. (Department Medicinal Chemistry, Faculty of Sciences, VU University Amsterdam, The Netherlands), Cecilia Söderberg-Naucler, M.D., Ph.D. (Medicine, Solna, Karolinska Institutet), and Liliana Soroceanu, Ph.D. (California Pacific Medical Center Research Institute, San Francisco, CA).
HCMV and Gliomas Workshop Meeting Attendees: James C. Alwine, Ph.D. (Cancer Biology and the Abramson Family Cancer Research Institute, School of Medicine, the University of Pennsylvania, Philadelphia, PA), William J. Britt, M.D. (Pediatrics, the University of Alabama at Birmingham, Birmingham, AL), Kevin Cassady (Pediatrics, the University of Alabama at Birmingham, Birmingham, AL), Susan M. Chang, M.D. (Neurological Surgery, the University of California at San Francisco, San Francisco, CA), E. Antonio Chiocca, M.D. Ph.D (Dardinger Neuro-Oncology Center, Neurological Surgery, James Cancer Hospital and the Ohio State University Medical Center, Columbus, OH), Charles S. Cobbs, M.D. (Neurological Surgery, the University of California at San Francisco, San Francisco, CA; California Pacific Medical Center Research Institute, San Francisco, CA), Phillip J. Daschner, Mc.S. (NIH/NCI), Kristine Dziurzynski, M.D. (Neurosurgery, The University of Texas M. D. Anderson Cancer Center, Houston, TX), Elizabeth A. Fortunato, Ph.D. (Biological Sciences, the University of Idaho Moscow, ID), Jane W. Fountain, Ph.D. (NIH/NINDS), Amy B. Heimberger, M.D. (Neurosurgery, The University of Texas M. D. Anderson Cancer Center, Houston, TX), Robert F. Kalejta, Ph.D. (Institute for Molecular Virology and McArdle Laboratory for Cancer Research, the University of Wisconsin-Madison), Timothy F. Kowalik, Ph.D. (Microbiology and Physiological Systems, the University of Massachusetts Medical Center, Worcester, MA), Chang-Hyuk Kwon, Ph.D. (Dardinger Neuro-Oncology Center, Neurological Surgery, James Cancer Hospital and the Ohio State University Medical Center, Columbus, OH), Stuart R. McGregor Dallas, Ph.D. (Molecular Biology, Lewis Thomas Laboratory, Princeton University, Princeton, NJ), Lisa Matlaf, Ph.D. (California Pacific Medical Center Research Institute, San Francisco, CA), Duane Mitchell, M.D., Ph.D. (Neurosurgery, Duke University Medical Center, Durham, NC), Rick Price, M.D. (Dardinger Neuro-Oncology Center, Neurological Surgery, James Cancer Hospital and the Ohio State University Medical Center, Columbus, OH), David Sandak (ABC2), Martine J. Smit, Ph.D. (Department Medicinal Chemistry, Faculty of Sciences, VU University Amsterdam, The Netherlands), Cecilia Söderberg-Naucler, M.D., Ph.D. Medicine, Solna, Karolinska Institutet, Liliana Soroceanu, Ph.D. (California Pacific Medical Center Research Institute, San Francisco, CA), William C. Timmer, Ph.D. (NIH/NCI), Carrie Treadwell (NBTS) and Max Wallace (ABC2).
Funding
This work was supported by the Accelerate Brain Cancer Cure and National Brain Tumor Society foundations.
References
- 1.Cobbs C, Harkins L, Samanta M, et al. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 2002;62:3347–3350. [PubMed] [Google Scholar]
- 2.Lau SK, Chen YY, Chen WG, et al. Lack of association of cytomegalovirus with human brain tumors. Mod Pathol. 2005;18:838–843. doi: 10.1038/modpathol.3800352. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell DA, Xie W, Schmittling R, et al. Sensitive detection of human cytomegalovirus in tumors and peripheral blood of patients diagnosed with glioblastoma. Neuro Oncol. 2008;10:10–18. doi: 10.1215/15228517-2007-035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poltermann S, Schlehofer B, Steindorf K, Schnitzler P, Geletneky K, Schlehofer JR. Lack of association of herpesviruses with brain tumors. J Neurovirol. 2006;12:90–99. doi: 10.1080/13550280600654573. [DOI] [PubMed] [Google Scholar]
- 5.Prins RM, Cloughesy TF, Liau LM. Cytomegalovirus immunity after vaccination with autologous glioblastoma lysate. N Engl J Med. 2008;359:539–541. doi: 10.1056/NEJMc0804818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabatier J, Uro-Coste E, Pommepuy I, et al. Detection of human cytomegalovirus genome and gene products in central nervous system tumours. Br J Cancer. 2005;92:747–750. doi: 10.1038/sj.bjc.6602339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheurer ME, Bondy ML, Aldape KD, Albrecht T, El-Zein R. Detection of human cytomegalovirus in different histological types of gliomas. Acta Neuropathol. 2008;116:79–86. doi: 10.1007/s00401-008-0359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucas KG, Bao L, Bruggeman R, Dunham K, Specht C. The detection of CMV pp65 and IE1 in glioblastoma multiforme. J Neurooncol. 2010;103:231–238. doi: 10.1007/s11060-010-0383-6. [DOI] [PubMed] [Google Scholar]
- 9.Dziurzynski K, Wei J, Qiao W, et al. Glioma-associated cytomegalovirus mediates subversion of the monocyte lineage to a tumor propagating phenotype. Clin Cancer Res. 2011;17:4642–4649. doi: 10.1158/1078-0432.CCR-11-0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slinger E, Maussang D, Schreiber A, et al. Sci Signal. 2010;3:ra58. doi: 10.1126/scisignal.2001180. [DOI] [PubMed] [Google Scholar]
- 11.Straat K, Liu C, Rahbar A, et al. Activation of telomerase by human cytomegalovirus. J Natl Cancer Inst. 2009;101:488–497. doi: 10.1093/jnci/djp031. [DOI] [PubMed] [Google Scholar]
- 12.Renzette N, Bhattacharjee B, Jensen JD, Gibson L, Kowalik TF. Extensive genome-wide variability of human cytomegalovirus in congenitally infected infants. PLoS Pathog. 2011;7:e1001344. doi: 10.1371/journal.ppat.1001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cobbs CS, Soroceanu L, Denham S, et al. Human cytomegalovirus induces cellular tyrosine kinase signaling and promotes glioma cell invasiveness. J Neurooncol. 2007;85:271–280. doi: 10.1007/s11060-007-9423-2. [DOI] [PubMed] [Google Scholar]
- 14.Cobbs CS, Soroceanu L, Denham S, Zhang W, Kraus MH. Modulation of oncogenic phenotype in human glioma cells by cytomegalovirus IE1-mediated mitogenicity. Cancer Res. 2008;68:724–730. doi: 10.1158/0008-5472.CAN-07-2291. [DOI] [PubMed] [Google Scholar]
- 15.Mocarski ES, Jr, Shenk T, Pass RF. Cytomegaloviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Vol 2. Philadelphia: Lippincott, Williams & Wilkins; 2007. pp. 2702–2772. [Google Scholar]
- 16.Reeves M, Sinclair J. Aspects of Latency and Reactivation. In: Shenk TE, Stinski MF, editors. Human Cytomegalovirus. Vol 325. Berlin Heidelberg: Springer-Verlag; 2008. pp. 297–313. [Google Scholar]
- 17.Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer. 2010;10:878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng J, Ke Q, Jin Z, et al. Cytomegalovirus infection causes an increase of arterial blood pressure. PLoS Pathog. 2009;5:e1000427. doi: 10.1371/journal.ppat.1000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Congenital Cytomegalovirus Foundation; 2011. [Google Scholar]
- 20.National CI. National Cancer Institute; 2011. [Google Scholar]
- 21.Soroceanu L, Akhavan A, Cobbs CS. Platelet-derived growth factor-alpha receptor activation is required for human cytomegalovirus infection. Nature. 2008;455:391–395. doi: 10.1038/nature07209. [DOI] [PubMed] [Google Scholar]
- 22.Toepoel M, Joosten PHLJ, Knobbe CB, et al. Haplotype-specific expression of the human PDGFRA gene correlates with the risk of glioblastomas. Int J Cancer. 2008;123:322–329. doi: 10.1002/ijc.23432. [DOI] [PubMed] [Google Scholar]
- 23.Pandey JP. Genetic and Viral Etiology of Glioblastoma–a Unifying Hypothesis. Cancer Epidemiol Biomarkers Prev. 2011;20:1061–1063. doi: 10.1158/1055-9965.EPI-11-0247. [DOI] [PubMed] [Google Scholar]
- 24.Cinatl J, Jr, Cinatl J, Vogel JU, Rabenau H, Kornhuber B, Doerr HW. Modulatory effects of human cytomegalovirus infection on malignant properties of cancer cells. Intervirology. 1996;39:259–269. doi: 10.1159/000150527. [DOI] [PubMed] [Google Scholar]
- 25.Doniger J, Muralidhar S, Rosenthal LJ. Human cytomegalovirus and human herpesvirus 6 genes that transform and transactivate. Clin Microbiol Rev. 1999;12:367–382. doi: 10.1128/cmr.12.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanahan D, Weinberg R. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 27.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Hwang J, Winkler L, Kalejta RF. Ubiquitin-independent proteasomal degradation during oncogenic viral infections. Biochim Biophys Acta. 2011;1816:147–157. doi: 10.1016/j.bbcan.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.AbuBakar S, Au WW, Legator MS, Albrecht T. Induction of chromosome aberrations and mitotic arrest by cytomegalovirus in human cells. Environ Mol Mutagen. 1988;12:409–420. doi: 10.1002/em.2860120409. [DOI] [PubMed] [Google Scholar]
- 30.Castillo JP, Yurochko AD, Kowalik TF. Role of human cytomegalovirus immediate-early proteins in cell growth control. J Virol. 2000;74:8028–8037. doi: 10.1128/jvi.74.17.8028-8037.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan G, Nogalski MT, Yurochko AD. Activation of EGFR on monocytes is required for human cytomegalovirus entry and mediates cellular motility. Proc Natl Acad Sci USA. 2009;106:22369–22374. doi: 10.1073/pnas.0908787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z, Knutson E, Kurosky A, Albrecht T. Degradation of p21cip1 in cells productively infected with human cytomegalovirus. J Virol. 2001;75:3613–3625. doi: 10.1128/JVI.75.8.3613-3625.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.E X, Pickering MT, Debatis M, et al. An E2F1-mediated DNA damage response contributes to the replication of human cytomegalovirus. PLoS Pathog. 2011;7:e1001342. doi: 10.1371/journal.ppat.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fortunato EA, Spector DH. p53 and RPA are sequestered in viral replication centers in the nuclei of cells infected with human cytomegalovirus. J Virol. 1998;72:2033–2039. doi: 10.1128/jvi.72.3.2033-2039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaspar M, Shenk T. Human cytomegalovirus inhibits a DNA damage response by mislocalizing checkpoint proteins. Proc Natl Acad Sci USA. 2006;103:2821–2826. doi: 10.1073/pnas.0511148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hume AJ, Finkel JS, Kamil JP, Coen DM, Culbertson MR, Kalejta RF. Phosphorylation of retinoblastoma protein by viral protein with cyclin-dependent kinase function. Science. 2008;320:797–799. doi: 10.1126/science.1152095. [DOI] [PubMed] [Google Scholar]
- 37.Kalejta RF, Shenk T. Manipulation of the cell cycle by human cytomegalovirus. Front Biosci. 2002;7:d295–d306. doi: 10.2741/kalejta. [DOI] [PubMed] [Google Scholar]
- 38.Kalejta RF, Shenk T. Proteasome-dependent, ubiquitin-independent degradation of the Rb family of tumor suppressors by the human cytomegalovirus pp71 protein. Proc Natl Acad Sci USA. 2003;100:3263–3268. doi: 10.1073/pnas.0538058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kotenko SV, Saccani S, Izotova LS, Mirochnitchenko OV, Pestka S. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10) Proc Natl Acad Sci USA. 2000;97:1695–1700. doi: 10.1073/pnas.97.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luleci G, Sakizli M, Gunalp A. Selective chromosomal damage caused by human cytomegalovirus. Acta Virol. 1980;24:341–345. [PubMed] [Google Scholar]
- 41.McElroy AK, Dwarakanath RS, Spector DH. Dysregulation of cyclin E gene expression in human cytomegalovirus-infected cells requires viral early gene expression and is associated with changes in the Rb-related protein p130. J Virol. 2000;74:4192–4206. doi: 10.1128/jvi.74.9.4192-4206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melnychuk RM, Streblow DN, Smith PP, Hirsch AJ, Pancheva D, Nelson JA. Human cytomegalovirus-encoded G protein-coupled receptor US28 mediates smooth muscle cell migration through Galpha12. J Virol. 2004;78:8382–8391. doi: 10.1128/JVI.78.15.8382-8391.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michaelis M, Baumgarten P, Mittelbronn M, Driever PH, Doerr HW, Cinatl J., Jr Oncomodulation by human cytomegalovirus: novel clinical findings open new roads. Med Microbiol Immunol. 2011;200:1–5. doi: 10.1007/s00430-010-0177-7. [DOI] [PubMed] [Google Scholar]
- 44.Michaelis M, Doerr HW, Cinatl J. The story of human cytomegalovirus and cancer: increasing evidence and open questions. Neoplasia. 2009;11:1–9. doi: 10.1593/neo.81178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog. 2006;2:e132. doi: 10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muralidhar S, Doniger J, Mendelson E, et al. Human cytomegalovirus mtrII oncoprotein binds to p53 and down-regulates p53-activated transcription. J Virol. 1996;70:8691–8700. doi: 10.1128/jvi.70.12.8691-8700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murayama T, Mukaida N, Sadanari H, et al. The immediate early gene 1 product of human cytomegalovirus is sufficient for up-regulation of interleukin-8 gene expression. Biochem Biophys Res Commun. 2000;279:298–304. doi: 10.1006/bbrc.2000.3923. [DOI] [PubMed] [Google Scholar]
- 48.Murphy EA, Streblow DN, Nelson JA, Stinski MF. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of permissive cells. J Virol. 2000;74:7108–7118. doi: 10.1128/jvi.74.15.7108-7118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rolle A, Mousavi-Jazi M, Eriksson M, et al. Effects of human cytomegalovirus infection on ligands for the activating NKG2D receptor of NK cells: up-regulation of UL16-binding protein (ULBP)1 and ULBP2 is counteracted by the viral UL16 protein. J Immunol. 2003;171:902–908. doi: 10.4049/jimmunol.171.2.902. [DOI] [PubMed] [Google Scholar]
- 50.Shen Y, Zhu H, Shenk T. Human cytomagalovirus IE1 and IE2 proteins are mutagenic and mediate “hit-and-run” oncogenic transformation in cooperation with the adenovirus E1A proteins. Proc Natl Acad Sci USA. 1997;94:3341–3345. doi: 10.1073/pnas.94.7.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Siew VK, Duh CY, Wang SK. Human cytomegalovirus UL76 induces chromosome aberrations. J Biomed Sci. 2009;16:107. doi: 10.1186/1423-0127-16-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soderberg-Naucler C. Does cytomegalovirus play a causative role in the development of various inflammatory diseases and cancer? J Intern Med. 2006;259:219–246. doi: 10.1111/j.1365-2796.2006.01618.x. [DOI] [PubMed] [Google Scholar]
- 53.Song YJ, Stinski MF. Effect of the human cytomegalovirus IE86 protein on expression of E2F-responsive genes: a DNA microarray analysis. Proc Natl Acad Sci USA. 2002;99:2836–2841. doi: 10.1073/pnas.052010099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spencer JV, Lockridge KM, Barry PA, et al. Potent immunosuppressive activities of cytomegalovirus-encoded interleukin-10. J Virol. 2002;76:1285–1292. doi: 10.1128/JVI.76.3.1285-1292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Straat K, de Klark R, Gredmark-Russ S, Eriksson P, Soderberg-Naucler C. Infection with human cytomegalovirus alters the MMP-9/TIMP-1 balance in human macrophages. J Virol. 2009;83:830–835. doi: 10.1128/JVI.01363-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vomaske J, Varnum S, Melnychuk R, et al. HCMV pUS28 initiates pro-migratory signaling via activation of Pyk2 kinase. Herpesviridae. 2010;1:2. doi: 10.1186/2042-4280-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wiertz EJ, Tortorella D, Bogyo M, et al. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 58.Yoo YD, Chiou CJ, Choi KS, et al. The IE2 regulatory protein of human cytomegalovirus induces expression of the human transforming growth factor beta1 gene through an Egr-1 binding site. J Virol. 1996;70:7062–7070. doi: 10.1128/jvi.70.10.7062-7070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu Y, Alwine JC. Human cytomegalovirus major immediate-early proteins and simian virus 40 large T antigen can inhibit apoptosis through activation of the phosphatidylinositide 3′-OH kinase pathway and the cellular kinase Akt. J Virol. 2002;76:3731–3738. doi: 10.1128/JVI.76.8.3731-3738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu H, Shen Y, Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Casarosa P, Bakker RA, Verzijl D, et al. Constitutive signaling of the human cytomegalovirus-encoded chemokine receptor US28. J Biol Chem. 2001;276:1133–1137. doi: 10.1074/jbc.M008965200. [DOI] [PubMed] [Google Scholar]
- 63.Maussang D, Verzijl D, van Walsum M, et al. Human cytomegalovirus-encoded chemokine receptor US28 promotes tumorigenesis. Proc Natl Acad Sci USA. 2006;103:13068–13073. doi: 10.1073/pnas.0604433103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu H, Jove R. The STATs of cancer–new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 65.Brantley EC, Benveniste EN. Signal transducer and activator of transcription-3: a molecular hub for signaling pathways in gliomas. Mol Cancer Res. 2008;6:675–684. doi: 10.1158/1541-7786.MCR-07-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abou-Ghazal M, Yang DS, Qiao W, et al. The incidence, correlation with tumor-infiltrating inflammation, and prognosis of phosphorylated STAT3 expression in human gliomas. Clin Cancer Res. 2008;14:8228–8235. doi: 10.1158/1078-0432.CCR-08-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Doucette T. 15th Annual Meeting of the Society of Neuro-Oncology; Montreal, Quebec, Canada: Oxford Journals; 2010. p. iv130. [Google Scholar]
- 68.Luo MH, Hannemann H, Kulkarni AS, Schwartz PH, O'Dowd JM, Fortunato EA. Human cytomegalovirus infection causes premature and abnormal differentiation of human neural progenitor cells. J Virol. 2010;84:3528–3541. doi: 10.1128/JVI.02161-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fortunato EA, Dell'Aquila ML, Spector DH. Specific chromosome 1 breaks induced by human cytomegalovirus. Proc Natl Acad Sci USA. 2000;97:853–858. doi: 10.1073/pnas.97.2.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li YS, Ramsay DA, Fan YS, Armstrong RF, Del Maestro RF. Cytogenetic evidence that a tumor suppressor gene in the long arm of chromosome 1 contributes to glioma growth. Cancer Genet Cytogenet. 1995;84:46–50. doi: 10.1016/0165-4608(95)00065-8. [DOI] [PubMed] [Google Scholar]
- 71.Castillo JP, Frame FM, Rogoff HA, Pickering MT, Yurochko AD, Kowalik TF. Human cytomegalovirus IE1-72 activates ataxia telangiectasia mutated kinase and a p53/p21-mediated growth arrest response. J Virol. 2005;79:11467–11475. doi: 10.1128/JVI.79.17.11467-11475.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luo MH, Rosenke K, Czornak K, Fortunato EA. Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection. J Virol. 2007;81:1934–1950. doi: 10.1128/JVI.01670-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hengel H, Brune W, Koszinowski UH. Immune evasion by cytomegalovirus–survival strategies of a highly adapted opportunist. Trends Microbiol. 1998;6:190–197. doi: 10.1016/s0966-842x(98)01255-4. [DOI] [PubMed] [Google Scholar]
- 74.Loenen WA, Bruggeman CA, Wiertz EJ. Immune evasion by human cytomegalovirus: lessons in immunology and cell biology. Semin Immunol. 2001;13:41–49. doi: 10.1006/smim.2001.0294. [DOI] [PubMed] [Google Scholar]
- 75.Scholz M, Doerr HW, Cinatl J. Human cytomegalovirus retinitis: pathogenicity, immune evasion and persistence. Trends Microbiol. 2003;11:171–178. doi: 10.1016/s0966-842x(03)00066-0. [DOI] [PubMed] [Google Scholar]
- 76.Wiertz E, Hill A, Tortorella D, Ploegh H. Cytomegaloviruses use multiple mechanisms to elude the host immune response. Immunol Lett. 1997;57:213–216. doi: 10.1016/s0165-2478(97)00073-4. [DOI] [PubMed] [Google Scholar]
- 77.Michelson S. Human cytomegalovirus escape from immune detection. Intervirology. 1999;42:301–307. doi: 10.1159/000053964. [DOI] [PubMed] [Google Scholar]
- 78.Suzuki S, Kameoka M, Nakaya T, et al. Superoxide generation by monocytes following infection with human cytomegalovirus. Immunopharmacology. 1997;37:185–190. doi: 10.1016/s0162-3109(97)00047-7. [DOI] [PubMed] [Google Scholar]
- 79.Hsu WM, Chen SS, Peng CH, et al. Elevated nitric oxide level in aqueous humor of AIDS patients with cytomegalovirus retinitis. Ophthalmologica. 2003;217:298–301. doi: 10.1159/000070639. [DOI] [PubMed] [Google Scholar]
- 80.Harkins L, Volk AL, Samanta M, et al. Specific localisation of human cytomegalovirus nucleic acids and proteins in human colorectal cancer. Lancet. 2002;360:1557–1563. doi: 10.1016/S0140-6736(02)11524-8. [DOI] [PubMed] [Google Scholar]
- 81.Maussang D, Langemeijer E, Fitzsimons CP, et al. The human cytomegalovirus-encoded chemokine receptor US28 promotes angiogenesis and tumor formation via cyclooxygenase-2. Cancer Res. 2009;69:2861–2869. doi: 10.1158/0008-5472.CAN-08-2487. [DOI] [PubMed] [Google Scholar]
- 82.Zhu H, Cong JP, Yu D, Bresnahan WA, Shenk TE. From the Cover: Inhibition of cyclooxygenase 2 blocks human cytomegalovirus replication. Proc Natl Acad Sci USA. 2002;99:3932–3937. doi: 10.1073/pnas.052713799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mendelson M, Monard S, Sissons P, Sinclair J. Detection of endogenous human cytomegalovirus in CD34+ bone marrow progenitors. J Gen Virol. 1996;77(Pt 12):3099–3102. doi: 10.1099/0022-1317-77-12-3099. [DOI] [PubMed] [Google Scholar]
- 84.Taylor-Wiedeman J, Sissons P, Sinclair J. Induction of endogenous human cytomegalovirus gene expression after differentiation of monocytes from healthy carriers. J Virol. 1994;68:1597–1604. doi: 10.1128/jvi.68.3.1597-1604.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sinclair J, Sissons P. Latency and reactivation of human cytomegalovirus. J Gen Virol. 2006;87:1763–1779. doi: 10.1099/vir.0.81891-0. [DOI] [PubMed] [Google Scholar]
- 86.Soderberg-Naucler C, Fish KN, Nelson JA. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell. 1997;91:119–126. doi: 10.1016/s0092-8674(01)80014-3. [DOI] [PubMed] [Google Scholar]
- 87.Chan G, Bivins-Smith ER, Smith MS, Smith PM, Yurochko AD. Transcriptome analysis reveals human cytomegalovirus reprograms monocyte differentiation toward an M1 macrophage. J Immunol. 2008;181:698–711. doi: 10.4049/jimmunol.181.1.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chan G, Bivins-Smith ER, Smith MS, Yurochko AD. NF-kappaB and phosphatidylinositol 3-kinase activity mediates the HCMV-induced atypical M1/M2 polarization of monocytes. Virus Res. 2009;144:329–333. doi: 10.1016/j.virusres.2009.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reinke P, Prosch S, Kern F, Volk HD. Mechanisms of human cytomegalovirus (HCMV) (re)activation and its impact on organ transplant patients. Transpl Infect Dis. 1999;1:157–164. doi: 10.1034/j.1399-3062.1999.010304.x. [DOI] [PubMed] [Google Scholar]
- 91.Wu A, Wei J, Kong LY, et al. Glioma cancer stem cells induce immunosuppressive macrophages/microglia. Neuro Oncol. 2010;12:1113–1125. doi: 10.1093/neuonc/noq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sampson JH, Aldape KD, Archer GE, et al. Greater chemotherapy-induced lymphopenia enhances tumor-specific immune responses that eliminate EGFRvIII-expressing tumor cells in patients with glioblastoma. Neuro Oncol. 2011;13:324–333. doi: 10.1093/neuonc/noq157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sampson JH, Heimberger AB, Archer GE, et al. Immunologic escape after prolonged progression-free survival with epidermal growth factor receptor variant III peptide vaccination in patients with newly diagnosed glioblastoma. J Clin Oncol. 2010;28:4722–4729. doi: 10.1200/JCO.2010.28.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]