During murine B cell development, PI3 kinase inhibits Ig gene rearrangement by suppressing FoxO1, which mediates Ikaros mRNA splicing; Ikaros is needed for Ig gene recombination.
Abstract
Somatic rearrangement of immunoglobulin (Ig) genes is a key step during B cell development. Using pro–B cells lacking the phosphatase Pten (phosphatase and tensin homolog), which negatively regulates phosphoinositide-3-kinase (PI3K) signaling, we show that PI3K signaling inhibits Ig gene rearrangement by suppressing the expression of the transcription factor Ikaros. Further analysis revealed that the transcription factor FoxO1 is crucial for Ikaros expression and that PI3K-mediated down-regulation of FoxO1 suppresses Ikaros expression. Interestingly, FoxO1 did not influence Ikaros transcription; instead, FoxO1 is essential for proper Ikaros mRNA splicing, as FoxO1-deficient cells contain aberrantly processed Ikaros transcripts. Moreover, FoxO1-induced Ikaros expression was sufficient only for proximal VH to DJH gene rearrangement. Simultaneous expression of the transcription factor Pax5 was needed for the activation of distal VH genes; however, Pax5 did not induce any Ig gene rearrangement in the absence of Ikaros. Together, our results suggest that ordered Ig gene rearrangement is regulated by distinct activities of Ikaros, which mediates proximal VH to DJH gene rearrangement downstream of FoxO1 and cooperates with Pax5 to activate the rearrangement of distal VH genes.
The development of B cells is a highly regulated process that includes the generation of B cell antigen receptors (BCRs; Rajewsky, 1996). The enormous variability observed in BCRs largely results from recombination of Ig heavy chain (HC; IgH) and light chain (LC; IgL) gene loci (Schlissel, 2003). During B cell development, the first recombination events take place at the HC gene locus leading to expression of a μ-class HC (μHC) in pre–B cells. Together with the surrogate LC (SLC), composed of λ5 and VpreB, µHC associates with the signal transduction subunits Ig-α and Ig-β to form a pre-BCR. As a result of auto–cross-linking, the pre-BCR shows a distinctive capacity for autonomous signaling required for normal pre–B cell development (Ubelhart et al., 2010). The pre–B cell stage is characterized by several pre-BCR–induced rounds of cell division and subsequent recombination at the IgL gene locus, which leads to surface expression of a BCR and progression to the early immature B cell stage (Herzog et al., 2009).
Ig gene recombination requires the action of recombination-activating gene products Rag-1 and -2 (Schatz, 2004). To achieve correct and sequential recombination, Rag expression must be tightly regulated. The activity of phosphoinositide-3-kinase (PI3K; Schatz, 2004; Manning and Cantley, 2007), which is largely orchestrated by cytokines, as well as pre-BCR and BCR signaling in B-lymphocytes, has been described to play an important role in negatively regulating Ig gene rearrangement by inhibiting Rag transcription (Amin and Schlissel, 2008; Dengler et al., 2008). PI3K generates phosphatidylinositol-3,4,5-trisphosphate, a lipid second messenger that facilitates membrane recruitment and activation of numerous proteins, of which protein kinase B (PKB, also known as Akt; Okkenhaug and Vanhaesebroeck, 2003) plays a central role in PI3K-mediated negative regulation of Ig gene recombination. For instance, SLP-65 (SH2 domain-containing lymphocyte protein of 65 kD, also called BLNK or BASH), a central adaptor protein acting downstream of pre-BCR and BCR, was shown to promote IgL gene recombination by down-regulating PKB activity (Herzog et al., 2008). Moreover, these studies demonstrated the importance of FoxO transcription factors in the process of Ig gene recombination. FoxO proteins are the mammalian counterparts of Caenorhabditis elegans decay-accelerating factor 16 and share an evolutionarily conserved DNA-binding domain (Coffer and Burgering, 2004). They are negatively regulated by PKB-mediated phosphorylation, which results in their export from the nucleus and proteasomal degradation (Biggs et al., 1999; Kops and Burgering, 1999; Takaishi et al., 1999). The activity of PKB is negatively regulated by phosphatase and tensin homolog (Pten; Maehama and Dixon, 1998), whose main substrate is phosphatidylinositol-3,4,5-trisphosphate generated by PI3K. The role of Pten in B cells has been studied by analyzing floxed Pten mice crossed to mice expressing Cre-recombinase from the B cell–specific CD19 promoter. The resulting animals displayed hyperproliferation of B cells and defects in class switch recombination (Suzuki et al., 2003; Omori et al., 2006).
Ig gene recombination requires the action of the Kruppel-like zinc finger transcription factor Ikaros (Cobb and Smale, 2005), which is involved in activating Rag expression and the accessibility of the IgH gene locus (Reynaud et al., 2008). Hence, disruption of Ikaros leads to an early blockade in lymphocyte development and complete absence of the earliest B cell progenitors in mice (Georgopoulos et al., 1994; Wang et al., 1996; Merkenschlager, 2010). Another crucial factor for IgH gene recombination is the B-lineage commitment factor Pax5 (Cobaleda et al., 2007), whose absence leads to a halt at the pro–B cell stage with cells retaining a broad potential to develop into lymphoid and myeloid lineages (Urbánek et al., 1994). Pax5 acts as a transcriptional regulator and has been demonstrated to activate the transcription of genes involved in pre-BCR and BCR signaling such as CD19, Ig-α, and SLP-65 (Nutt et al., 1997; Schebesta et al., 2002). In addition to transcriptional regulation of signaling proteins, it is also involved in VH-DJH gene recombination. Interestingly, Pax5 appears to be mainly required for the recombination of distal VH gene segments. Proximal VH segments are efficiently recombined in the absence of Pax5 (Fuxa et al., 2004; Johnson et al., 2004). This suggests that other factors initiate proximal VH-DJH recombination before Pax5 action (Yancopoulos et al., 1984; Marshall et al., 1996; ten Boekel et al., 1997; Jhunjhunwala et al., 2009; Hewitt et al., 2010). It has been shown that the transcription factors Pax5, early B cell factor 1 (EBF1), FoxO1, and Ikaros play major roles during Ig gene rearrangement and are thus essential for early B cell development (Hagman et al., 1993; Rajewsky, 1996; Fuxa et al., 2004; Amin and Schlissel, 2008; Reynaud et al., 2008). However, it remained ill defined whether they play redundant roles or whether they are specifically involved in particular steps of early B cell development. Here, we show that the function of FoxO1 in Ig gene recombination is largely mediated by Ikaros, whose transcripts show an unusual splicing pattern in the absence of FoxO1. We show that proper splicing of Ikaros transcripts and Ig gene recombination can be achieved by Pten expression, nuclear FoxO1, or inhibition of PI3K activity.
RESULTS
Pten mediates μHC recombination in pro–B cells
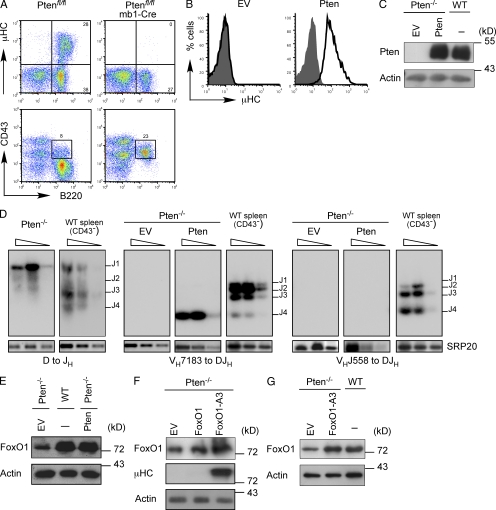
To study the role of FoxO1 in B cell development, we first generated conditional FoxO1-deficient mice by crossing mb1-Cre mice (Hobeika et al., 2006) to FoxO1fl/fl mice (Tothova et al., 2007). As described before (Dengler et al., 2008), the resulting animals displayed a severe block in B cell development (unpublished data). We were unable to establish IL-7–dependent in vitro cultures of FoxO1-deficient B cell precursors from FoxO1fl/fl/mb1-Cre mice. Because FoxO1 activity is largely dependent on Pten, we reasoned that Pten-deficient mice (Suzuki et al., 2003) might be a suitable source for B cell progenitors that would allow us to study FoxO1 function. Thus, we crossed mb1-Cre mice to Ptenfl/fl mice to generate conditional Pten-deficient mice (Ptenfl/fl/mb1-Cre), in which Pten is inactivated at an early stage of B cell development. We analyzed BM cells from Ptenfl/fl/mb1-Cre mice by flow cytometry and found that B cell development was blocked at the pro–B cell stage (Fig. 1 A and not depicted). Comparison of BM cells from conditional Pten-deficient mice with those of conditional FoxO1-deficient mice revealed a nearly identical block in B cell development (unpublished data). We isolated BM cells from different Ptenfl/fl/mb1-Cre mice and cultured them in medium complemented with IL-7. From these slowly growing and fragile cultures we obtained two independent cell lines (#10 and #71) that had deleted Pten (Pten−/−). The results presented in this study were obtained using #71 and were reproducibly repeated with #10. Flow cytometric analysis revealed that these Pten−/− cells lacked μHC expression and were trapped in the pro–B cell stage of development (Fig. 1 B and not depicted). To test whether reconstitution of Pten expression induces μHC expression, we retrovirally transduced the Pten−/− pro–B cells with either an empty vector (EV) or a Pten expression vector and analyzed μHC expression by flow cytometry. We found that the amount of retrovirally expressed Pten was comparable to endogenous Pten in WT pro–B cells and that only Pten-reconstituted cells were positive for μHC (Fig. 1, B and C).
Figure 1.
Pten-deficient pro–B cells do not express µHC. (A) Flow cytometric analysis of BM cells from Ptenfl/fl mice and Ptenfl/fl/mb1-Cre mice (n > 3 per genotype), stained for the indicated surface markers. Numbers in quadrants indicate percentages of cells. (B) Flow cytometric analysis of µHC expression in Pten-deficient pro–B cells reconstituted with either an empty expression vector (EV) or with a Pten expression vector (black lines) as compared with untransduced cells (filled gray). (C) Immunoblot analysis of Pten expression in Pten-deficient pro–B cells transduced with an empty expression vector (EV) or with a Pten expression vector as compared with WT cells. Immunoblot of actin served as a loading control. (D) Southern blot analysis of PCR fragments amplified with specific primers for VHDQ52, VH7183, VHJ558, and JH4 from genomic DNA of Pten-deficient cells transduced with an empty expression vector (EV) or with a Pten expression vector. Wedges above lines indicate 2× serial dilutions. PCR for the splicing factor SRP20 was used as control. Purified CD43− splenic B cells from WT mice served as a control. (E) Immunoblot analysis of FoxO1 expression in Pten-deficient cells transduced with either an empty expression vector (EV) or a Pten expression vector as compared with WT cells. Immunoblot of actin served as a loading control. (F) Immunoblot analysis of FoxO1 and µHC expression in Pten-deficient cells transduced with either an empty expression vector (EV), a FoxO1 expression vector, or an expression vector encoding activated FoxO1 (FoxO1-A3). Immunoblot of actin served as a loading control. (G) Immunoblot analysis of Pten-deficient cells transduced with EV or FoxO1-A3 and analyzed for FoxO1 expression as compared with WT cells. Actin was used as a loading control. Data are representative of at least two to three independent experiments.
To test whether the inability of the Pten−/− pro–B cells to express μHC was a result of defective IgH gene recombination, we analyzed two independent Pten-deficient cell lines for the recombination status of the IgH genes. This analysis showed that Pten−/− cells had completed D to JH1 gene rearrangement, whereas VH to DJH gene recombination was not detectable neither for proximal VH7183 nor distal VHJ558 genes (Fig. 1 D).
Upon reconstitution with Pten, we detected VH to DJH rearrangement in Pten−/− cells; however, it was restricted to the proximal VH7183 gene family, as we did not observe any recombination of the distal J558 family of VH segments after reconstitution with Pten (Fig. 1 D). Interestingly, only VH7183 to DJH4 gene rearrangements were detected, which might suggest that secondary DJH recombination had occurred, thereby deleting the DJH1 recombination observed in the parental Pten−/− cells (Fig. 1 D). Sequencing of the respective band revealed that the detected VH7183 to DJH4 recombination was clonal. This suggests that the rearrangement of proximal VH elements was inefficient and that a cell with a productive VH to DJH recombination has a strong selective advantage induced by pre-BCR expression (unpublished data).
Next, we analyzed FoxO1 expression in Pten−/− pro–B cells and found that they had reduced FoxO1 expression as compared with WT cells and that FoxO1 amounts were increased upon reconstitution of Pten expression (Fig. 1 E). To test whether induction of FoxO1 is involved in Ig gene rearrangement, we expressed WT FoxO1 or a constitutively nuclear form of FoxO1 (FoxO1-A3; Brunet et al., 1999) in Pten−/− cells and analyzed μHC expression. Retroviral transduction of WT FoxO1 resulted in only a minor increase of total FoxO1 protein amounts and no induction of μHC expression. In contrast, transduction of FoxO1-A3 increased FoxO1 protein amounts and induced μHC expression in Pten−/− pro–B cells (Fig. 1 F). Importantly, expression levels of FoxO1-A3 were comparable to FoxO1 protein amounts in WT cells (Fig. 1 G). Together, the data suggest that activation of FoxO1 is the main Pten-dependent step for IgH gene rearrangement.
Ikaros restores IgH gene recombination in Pten−/− cells
Our data are in agreement with available results, suggesting an important role of FoxO1 during the process of Ig gene recombination. However, it remained unclear whether the ubiquitously expressed FoxO1 exerts its actions directly or via additional proteins that promote IgH gene rearrangement. Therefore, we screened for lymphocyte-specific factors that might act downstream of FoxO1.
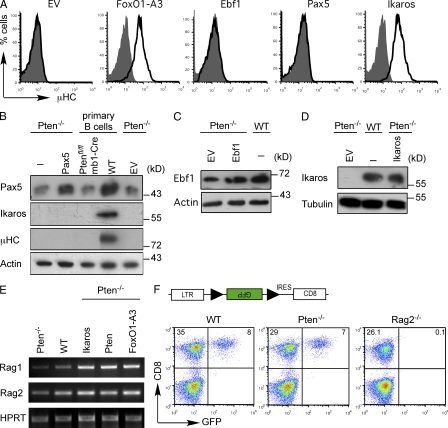
Pax5 is an essential transcription factor for B cell development and IgH gene recombination (Hagman et al., 1993; Rajewsky, 1996; Fuxa et al., 2004; Amin and Schlissel, 2008; Reynaud et al., 2008). It has been shown that Pax5 binds to regulatory elements in the IgH gene locus to promote distal VH to DJH gene recombination by chromatin remodeling (Nutt et al., 1997; Hesslein et al., 2003; Fuxa et al., 2004; Johnson et al., 2004). Indeed, immunoblot analysis showed that Pax5 expression was reduced in Pten−/− pro–B cells as compared with WT cells (Fig. 2 B). Thus, we tested whether expression of Pax5 in Pten−/− cells may lead to restoration of μHC expression. However, retroviral transduction of Pten−/− pro–B cells with a Pax5 expression vector substantially increased Pax5 protein levels but did not induce any μHC expression (Fig. 2, A and B). Together, these data suggest that although low levels of Pax5 are sufficient for normal CD19 expression in Pten−/− cells (unpublished data), Pax5 is not sufficient to induce VHDJH recombination in the absence of Pten even after retroviral transduction to increase Pax5 expression. Similar to Pax5, EBF1 was not able to induce μHC expression in Pten−/− pro–B cells (Fig. 2, A and C).
Figure 2.
FoxO1 and Ikaros restore µHC expression in Pten-deficient pro–B cells. (A) Flow cytometric analysis of µHC expression in Pten-deficient pro–B cells transduced with either an empty expression vector (EV) or expression vectors for FoxO1-A3, Ebf1, Pax5, or Ikaros (black lines) as compared with untransduced cells (filled gray). (B) Immunoblot analysis of Pax5, Ikaros, and µHC expression in freshly isolated CD19+ cells from Ptenfl/fl/mb1-Cre, and WT mice, and Pten-deficient pro–B cells transduced with either an empty expression vector (EV) or with an expression vector encoding Pax5. Immunoblotting for actin served as a loading control. (C) Immunoblot analysis of Pten-deficient cells transduced with either EV or EBF1 and WT cells for expression of EBF1. Actin served as a loading control. (D) Immunoblot analysis for Ikaros expression in Pten-deficient cells transduced with EV or Ikaros and in WT cells. Immunoblotting for tubulin served as a loading control. (E) RT-PCR analysis of Rag1 and Rag2 mRNA in Pten-deficient cells, WT cells, and Pten-deficient cells transduced with expression vectors encoding Ikaros, Pten, or FoxO1-A3. HPRT served as a loading control. (F) GFP expression after recombination of an artificial reporter system in WT cells, Pten-deficient cells, and in a recombination-defective cell line (Rag2−/−). As shown on the top, Rag-1/2 activity leads to the recombination of the two RSS (black triangles) flanking the inverted GFP cDNA, resulting in GFP expression. Data are representative of at least three independent experiments.
The Kruppel-like zinc finger transcription factor Ikaros has been implicated in IgH gene recombination (Reynaud et al., 2008) and was shown to bind to regulatory elements in the Ig HC locus (Reynaud et al., 2008; Sellars et al., 2009). Furthermore, we found that Pten−/− pro–B cells lacked endogenous Ikaros protein (Fig. 2 B). Therefore, we analyzed whether Ikaros expression could restore the defect in Ig gene recombination observed in Pten−/− pro–B cells. Indeed, we found that retroviral expression of Ikaros to levels comparable to WT cells led to μHC expression in Pten−/− cells (Fig. 2, A and D).
Because Ikaros was shown to induce the expression of Rag1/2 (Reynaud et al., 2008; Sellars et al., 2009), we tested whether Pten−/− pro–B cells lacked Rag1/2 expression and were therefore unable to show Ig gene recombination after Pax5 expression. Our experiments show that Rag1/2 expression is slightly reduced in Pten−/− pro–B cells as compared with WT pro–B cells and that introducing Ikaros, Pten, or FoxO1-A3 induces Rag1/2 expression (Fig. 2 E). However, the Pten−/− pro–B cells were capable of recombining an artificial recombination substrate to a similar extent as WT cells (Fig. 2 F). It seems therefore unlikely that the reduced Rag1/2 expression levels account for the inability of Pax5 to induce any IgH gene recombination in Pten−/− pro–B cells.
Pax5 requires Ikaros for activating distal VH segments
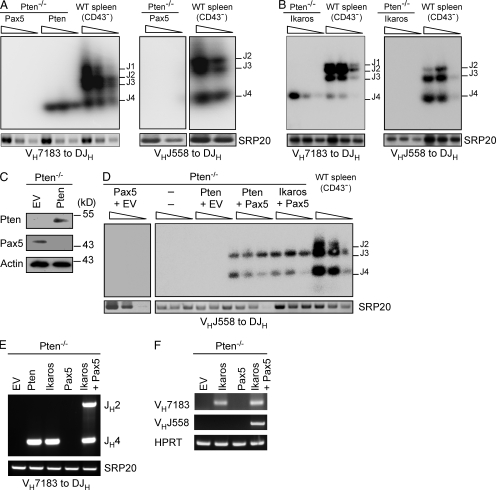
To investigate why Pax5 was unable to induce μHC expression in Pten−/− cells, we analyzed the recombination status of the IgH genes. This analysis showed that retrovirally expressed Pax5 failed to induce any VH to DJH rearrangement in Pten−/− pro–B cells (Fig. 3 A). In contrast, similar to Pten, expression of Ikaros resulted in efficient recombination of only proximal (VH7183) but not distal (VHJ558) gene segments (Fig. 3, A and B). Pax5 has been described to be important for the recombination of distal VH segments (Hesslein et al., 2003; Fuxa et al., 2004; Johnson et al., 2004). The inability of Pten-reconstituted Pten−/− pro–B cells to induce any distal VHDJH recombination might be explained by the reduced Pax5 expression in Pten−/− cells. Therefore, we tested Pax5 expression upon Pten reconstitution and found that Pax5 levels were even further reduced (Fig. 3 C). We then tested whether coexpression of Pax5 with either Pten or Ikaros would result in distal VH to DJH rearrangements. Indeed, Pten-deficient cells recombined distal VHJ558 segments when Pax5 was coexpressed with either Pten or Ikaros (Fig. 3, D and E). In addition, different recombinations of proximal VH7183 segments were detected as determined by usage of different JH segments, indicating that the increased efficiency is not limited to distal VH gene segments (Fig. 3 E).
Figure 3.
Pax5 requires Ikaros for the recombination of distal VH gene segments. (A) Southern blot analysis of PCR fragments amplified from genomic DNA of Pten-deficient cells reconstituted with expression vectors encoding Pax5 or Pten, with specific primers for VH7183, VHJ558, and JH4. Wedges above lines indicate 2× serial dilutions. PCR for the splicing factor SRP20 was used as a loading control. Purified CD43− splenic B cells from WT mice served as a control. (B) Southern blot analysis of PCR fragments amplified from genomic DNA of Pten-deficient cells transduced with an Ikaros expression vector, with specific primers for VH7183 and JH4, and VHJ558 and JH4. Purified CD43− splenic B cells from WT mice served as a control. (C) Immunoblot analysis of Pten-deficient cells transduced either with EV or Pten and analyzed for Pten and Pax5 expression. Actin was used as a loading control. (D) Southern blot analysis of PCR fragments amplified from genomic DNA of Pten-deficient cells transduced with the indicated combinations of expression vectors, with specific primers for VHJ558 and JH4. Wedges above lines indicate 2× serial dilutions. PCR for the splicing factor SRP20 was used as loading control. Purified CD43− splenic B cells from WT mice served as a control. (E) PCR analysis of genomic DNA of Pten-deficient cells transduced with the indicated expression vectors. Primers were specific for VH7183 and JH4. The splicing factor SRP20 was used as a control. (F) RT-PCR analysis of Igh germline transcripts in Pten-deficient cells transduced with the indicated combinations of expression vectors with specific primers for VH7183 and VHJ558. HPRT served as loading control. Data are representative of at least two to three independent experiments.
Because gene activation is a prerequisite for Ig gene rearrangement, we analyzed germline transcripts of proximal and distal VH genes as direct signs of gene activation and accessibility of the respective segments for the recombination machinery. These experiments clearly show that Ikaros is sufficient to activate proximal VH7183, but not distal VHJ558 transcripts. For induction of VHJ558 germline transcripts Pax5 is an obligate factor (Fig. 3 F). These results suggest that, although required, Pax5 is not sufficient for the activation of distal VH gene recombination and is strictly dependent on Ikaros for this function.
Pten/FoxO1 are essential for Ikaros expression
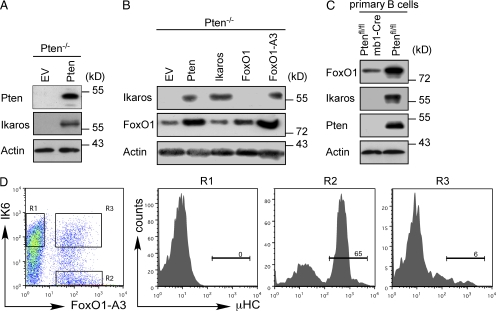
The facts that Ikaros was not expressed in Pten−/− cells and that we observed similar effects on IgH gene rearrangement when we expressed Pten, FoxO1-A3, or Ikaros prompted us to test whether Pten may induce Ikaros expression by activation of FoxO1. Indeed, immunoblot analysis showed that introduction of Pten up-regulates Ikaros expression in Pten−/− pro–B cells (Fig. 4 A). Moreover, the constitutively nuclear form of FoxO1 (FoxO1-A3) resulted in a strong induction of Ikaros expression, whereas WT FoxO1 was unable to induce Ikaros expression in the absence of Pten (Fig. 4 B, lanes 4 and 5). To exclude that Ikaros deficiency can only be observed after cultivating Pten−/− cells in vitro, we tested Ikaros expression in freshly isolated BM B cells from Ptenfl/fl/mb1-Cre mice. Immunoblot analysis showed that BM B cells from these mice lacked Ikaros expression and were similar to the in vitro–cultured pro–B cells (Fig. 4 C). These data, and the fact that Ikaros expression in Pten-deficient cells induced µHC expression without altering FoxO1 protein amounts (Fig. 4 B, lane 3), suggest that Ikaros acts downstream of FoxO1 in promoting IgH gene recombination. Induction of Ikaros by FoxO1 implies that expression of a dominant-negative version of Ikaros should reverse the effects of FoxO1-A3 on IgH gene recombination. Thus, we coexpressed FoxO1-A3 together with IK6, a dominant-negative splice variant of Ikaros (Klein et al., 2006; Iacobucci et al., 2008; Reynaud et al., 2008). Interestingly, IK6 efficiently abolished µHC expression mediated by FoxO1-A3 (Fig. 4 D). These data suggest that induction of Ikaros activity is key for FoxO1-mediated IgH gene rearrangement.
Figure 4.
Ikaros expression and function is essential for μHC expression. (A) Immunoblot analysis for Pten and Ikaros expression in Pten-deficient cells transduced with an EV or with a Pten expression vector. Immunoblotting for actin served as loading control. (B) Immunoblot analysis of Ikaros and FoxO1 expression in Pten-deficient cells transduced with the indicated expression vectors. Immunoblotting for actin served as loading control. (C) Immunoblot analysis of FoxO1, Ikaros, and Pten expression in freshly isolated CD19+ BM cells from Pten-sufficient and Pten-deficient mice. Immunoblotting for actin served as loading control. (D) Flow cytometric analysis of Pten-deficient cells transduced with expression vectors encoding FoxO1-A3 and dominant-negative Ikaros (IK6). Single- (R1 and R2) and double-positive cells (R3) were gated and are displayed as histograms (right) showing µHC expression. Numbers indicate the percentages of cells expressing μHC. Data are representative of at least two to three independent experiments.
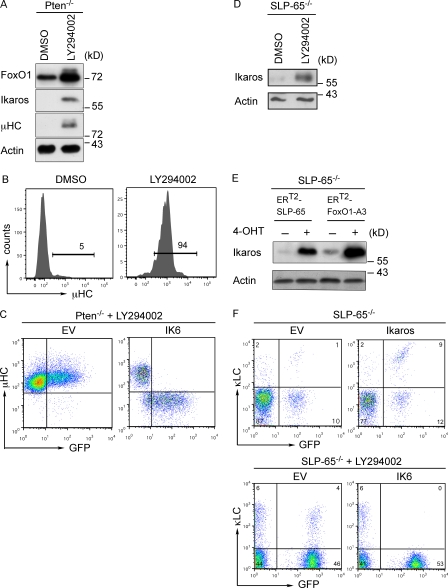
PI3K signaling represses Ikaros expression
Because increased FoxO1 expression was required for induction of Ikaros and because the FoxO transcription factors are negatively regulated by PKB-mediated phosphorylation, we tested whether inhibition of PI3K signaling would induce Ikaros and result in μHC expression in Pten−/− cells. We therefore treated Pten−/− cells with the PI3K inhibitor LY294002 and performed immunoblot and FACS analysis to test for μHC expression. The results show a strong increase of FoxO1 and induction of Ikaros expression upon treatment of Pten-deficient cells with LY294002 (Fig. 5 A). Moreover, μHC was readily observed after 48 h of treatment with the PI3K inhibitor LY294002 (Fig. 5, A and B). The expression of μHC was also restored by inhibiting PKB/Akt (unpublished data). To test whether the induction of μHC expression by LY294002 treatment was mediated by Ikaros, we compared μHC expression in LY294002-treated cells that were transduced either with an empty control vector or with dominant negative IK6. Our results show that the positive effect of LY294002 treatment on μHC expression was reversed when dominant-negative IK6 was expressed in Pten−/− pro–B cells (Fig. 5 C). Collectively, these results identify Ikaros as a crucial element downstream of Pten and FoxO1 in PI3K-mediated negative regulation of IgH gene rearrangement.
Figure 5.
Inhibition of PI3K leads to expression of Ikaros and to μHC generation. (A) Immunoblot analysis of FoxO1, Ikaros, and μHC expression in Pten-deficient cells treated with either DMSO or the PI3K inhibitor LY294002. Immunoblotting for actin served as loading control. (B) Flow cytometric analysis of µHC expression in Pten-deficient cells treated with either DMSO or the PI3K inhibitor LY294002. (C) Flow cytometric analysis for µHC and GFP expression of Pten-deficient cells transduced with an EV or an expression vector encoding dominant-negative Ikaros (IK6) and treated with LY294002. (D) Immunoblot analysis for Ikaros expression in SLP-65–deficient pre–B cells treated with DMSO or the PI3K inhibitor LY294002. Immunoblotting for actin served as loading control. (E) Immunoblot analysis for Ikaros expression in SLP-65–deficient pre–B cells transduced with 4-hydroxytamoxifen (4-OHT)–inducible forms of either SLP-65 (ERT2-SLP65) or FoxO1-A3 (ERT2-FoxO1-A3). Upon sorting of positively transduced cells, the cells were treated with the solvent ethanol (−) or 4-OHT (+) to activate ERT2-SLP65 and ERT2-FoxO1-A3. Immunoblotting for actin served as loading control. (F) Top, FACS plots for κLC expression in SLP-65–deficient pre–B cells transduced with an EV or with an Ikaros expression vector. Cells were cultured in the presence of IL-7. Bottom, dot plots for κLC expression in SLP-65–deficient pre–B cells transduced with an EV or with an expression vector for DN-Ikaros (IK6); both were treated with the PI3K inhibitor LY294002. Numbers in quadrants indicate percentages of cells in the respective region. Data are representative of at least two to three independent experiments.
To investigate whether Ikaros also acts downstream of FoxO1 in IgL gene recombination, we used our SLP-65 reconstitution system for the induction of κLC expression. Our previous results suggested that SLP-65 interferes with PI3K signaling to activate FoxO proteins and induce IgL gene recombination (Herzog et al., 2008). This predicts that induction of SLP-65 should lead to induction of Ikaros expression. Indeed, we found that induction of Ikaros expression in SLP-65–deficient pre–B cells can be achieved by LY294002 treatment, by activation of SLP-65, or by activation of FoxO1-A3 (Fig. 5, D and E). To show that Ikaros is involved in LC expression in SLP-65–deficient pre–B cells, we expressed Ikaros in these cells and found that this was sufficient to induce κLC expression. Moreover, combining LY294002 treatment with the inhibition of Ikaros activity by IK6 completely blocked κLC expression in SLP-65–deficient pre–B cells (Fig. 5 F). This suggests that Ikaros acts downstream of SLP-65 and FoxO1 for the induction of IgL gene rearrangement.
Aberrant Ikaros splicing in Pten−/− pro–B cells
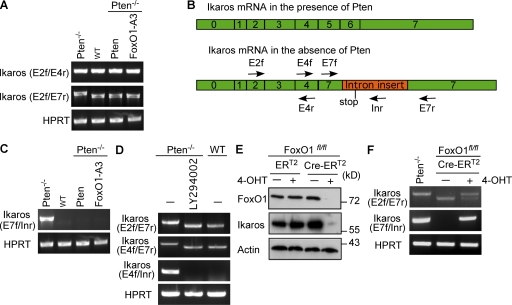
To investigate the underlying mechanism of FoxO1-mediated activation of Ikaros, we analyzed Ikaros transcripts in Pten−/− pro–B cells before and after expression of Pten or Foxo1-3A. RT-PCR analysis for the Ikaros 5′-region showed normal amounts of Ikaros transcripts in Pten−/− pro–B cells. RT-PCR analysis for full-length Ikaros, however, showed that Pten−/− pro–B cells lacked normal full-length transcripts and revealed the existence of aberrant Ikaros transcripts in these cells (Fig. 6 A and not depicted). Sequence analyses showed that the aberrant transcript lacked exons 5 and 6 and contained a 378-nucleotide insertion in exon 7, which was obviously derived from the preceding intron and introduced a stop codon at the beginning of the transactivation domain encoded by exon 7 (Fig. 6 B and Fig. S1). The underlying mechanism for generating this aberrant Ikaros transcript is unclear because there are no conventional splice sites and no obvious homology to known nonconventional splicing pathways (Yoshida, 2007). To confirm the presence of the aberrant Ikaros transcript in Pten−/− pro–B cells, we designed specific primers for this transcript and used them for RT-PCR analyses (Fig. 6 B). This analysis confirmed that the aberrant Ikaros transcript is expressed in Pten−/− pro–B cells and that it was completely abolished after expression of Pten or FoxO1-A3 in these cells (Fig. 6 C). To test whether aberrant Ikaros splicing was induced by PI3K signaling, we treated Pten−/− pro–B cells with LY294002 and found that normal Ikaros splicing is restored after inhibition of PI3K (Fig. 6 D). Thus, our results show that PI3K regulates Ikaros expression at the level of RNA processing by activation of a yet unknown splicing pathway. To confirm that this novel abnormal splicing of Ikaros is regulated by FoxO1 and can also be observed in other cells, we used BM-derived pro–B cells from FoxO1fl/fl mice and introduced a tamoxifen-inducible Cre-recombinase by retroviral transduction. Cre activation led to deletion of the floxed FoxO1 gene and abolished FoxO1 expression resulting in a severe reduction of Ikaros protein amounts (Fig. 6 E and not depicted), which confirms the essential role of FoxO1 for Ikaros expression. Furthermore, RT-PCR analyses showed that, similar to Pten−/− pro–B cells, deletion of FoxO1 leads to abnormal Ikaros transcripts (Fig. 6 F). These data show that aberrant Ikaros transcripts are not restricted to the Pten−/− pro–B cells and that FoxO1 is required for normal Ikaros splicing and expression. By down-regulating the amounts of FoxO1, PI3K activates aberrant splicing of Ikaros, thereby inhibiting the generation of normal transcripts and Ikaros protein expression.
Figure 6.
Aberrant Ikaros splicing in the absence of Pten. (A) RT-PCR analysis of Ikaros transcripts in Pten-deficient cells, WT cells, and Pten-deficient cells transduced with expression vectors for Pten or FoxO1-A3, using gene-specific primers. The forward primer is located in exon 2 (E2f), the reverse primers either in exon 4 (E4r) or in exon 7 (E7r) of the Ikaros transcript. HPRT was used as a loading control. (B) Schematic view of Ikaros transcripts and pre-mRNA splicing in the presence and absence of Pten. Arrows specify the location of the indicated primers. Forward primers were located in exons 2 (E2f), 4 (E4f), and 7 (E7f). Reverse primers were located in exons 4 (E4r), 7 (E7r), and the inserted sequence from intron 6 (Inr). (C) RT-PCR analysis of Ikaros transcripts in cells from A. The forward primer is located in exon 7 (E7f) and the reverse primer is located in the inserted intron fragment (Inr). HPRT was used as a control. (D) RT-PCR analysis of Ikaros transcripts in Pten-deficient cells and Pten-deficient cells treated with LY294002 as compared with WT cells. Forward primers were either located in exon 2 (E2f) or exon 4 (E4f), the reverse primers were located either in exon 7 (E7r) or in the inserted intron fragment (Inr). HPRT was used as a loading control. (E) Immunoblot analysis of FoxO1 and Ikaros expression in BM-derived cells from FoxO1fl/fl mice transduced with a tamoxifen-inducible Cre-recombinase (Cre-ERT2) before and after deletion of FoxO1 by 4-OHT treatment. Treatment with the solvent EtOH and cells transduced with an expression vector encoding ERT2 were used as controls. Immunoblotting for actin served as loading control. (F) RT-PCR analysis of Ikaros transcripts in Pten-deficient cells and Cre-ERT2–transduced FoxO1fl/fl cells with or without 4-OHT treatment. Forward primers were either located in exon 2 (E2f) or in exon 7 (E7f), the reverse primers were either located in exon 7 (E7r) or in the inserted intron fragment (Inr). HPRT was used as a loading control. Data are representative of two to three independent experiments.
DISCUSSION
This study establishes the transcription factor Ikaros as a novel component of the PI3K-regulated signaling machinery in developing B cells. Furthermore, our study shows that the activation of Ikaros is a central event during the induction of Ig gene rearrangement by FoxO1. In fact, the dominant-negative Ikaros version IK6 blocked FoxO1-mediated Ig gene recombination in our experiments. Interestingly, Ikaros and FoxO1 were both shown to directly bind and activate Rag1/2 genes (Liu et al., 2006; Amin and Schlissel, 2008; Dengler et al., 2008; Reynaud et al., 2008; Sellars et al., 2009). However, our results show that residual Rag1/2 expression is present in Ikaros-deficient cells, which is sufficient for recombining an artificial VDJ recombination substrate. Moreover, the induction of Ikaros expression required an increase in FoxO1 activity, which was achieved by either reconstituting Pten expression or by introducing a constitutively nuclear FoxO1 form (FoxO1-A3). The basal FoxO1 expression, which is observed in Pten-deficient pro–B cells, seems to be important for the survival of the cells and might be responsible for the Rag expression that was detected in these cells. Surprisingly, the observed Rag expression together with Pax5 was not sufficient to induce any VH-DJH recombination in our cells unless Ikaros activity was available either by retroviral transduction of Ikaros expression constructs or by activation of endogenous Ikaros expression through Pten. However, the ability of Pten−/− cells to recombine an artificial substrate demonstrated that the recombination machinery is expressed and, in principle, functional in Pten−/− pro–B cells. This suggests that VH gene segments cannot be recombined when only Pax5 is expressed in these cells.
Interestingly, retroviral expression of Pten or Ikaros in Pten-deficient pro–B cells did not completely restore VDJ recombination either, as only usage of proximal VH7183 segments could be detected. Furthermore, the clonal nature of cells carrying these rearrangements suggests that the recombination process is rather inefficient. One reason for this might be the reduction of Pax5 expression in these cells. In addition, although the recombination machinery is expressed in these cells, the Rag proteins might be inefficiently recruited to VH gene segments.
Although Pax5 has been described to be the central factor for the activation and recombination of distal VH gene segments (Hesslein et al., 2003; Fuxa et al., 2004; Johnson et al., 2004), our results clearly show that Pax5 requires the cooperation of Ikaros for this function. Thus, Pax5 alone is essential but not sufficient for the recombination of distal VH gene segments. In addition to its role in the recombination of distal VH segments, Pax5 seems to also be involved in the recombination of proximal VH segments, as our data show additional proximal VH to DJH recombinations when Pax5 was coexpressed with Ikaros.
Assuming that Ikaros increases the general accessibility of VH gene segments (Reynaud et al., 2008), it is unclear how it cooperates with Pax5 to increase the accessibility of distal VH gene segments. It is possible that Ikaros induces chromatin modifications on both proximal and distal VH gene segments and that these modifications are sufficient for proximal VH gene usage but not for distal VH segments. Such modifications might then facilitate the recruitment of Pax5, which introduces additional modifications that lead to changes in chromatin architecture and subsequent rearrangement of distal VH genes (Jhunjhunwala et al., 2009; Hewitt et al., 2010). Together, the emerging scheme suggests that the recombination of different VH gene segments is regulated by networks of transcription factors that operate in hierarchical (FoxO1/Ikaros) and cooperative (Pax5/Ikaros) manners. This divergent regulation of Pax5 and Ikaros may provide a mechanism for the fine-tuned regulation of VH gene segment activation, which might explain the observed differential usage of VH gene segments during B cell development. For instance, fetal liver B cells display a clear bias toward proximal VH segments, such as VH81X, a member of the VH7183 family (Yancopoulos et al., 1984; Marshall et al., 1996). Interestingly, also during early B cell development in the BM, VH usage is biased toward proximal gene segments, whereas they are underrepresented in peripheral B cells (Yancopoulos et al., 1984; Huetz et al., 1993; Decker et al., 1995; Marshall et al., 1996; ten Boekel et al., 1997).
After successful IgH gene rearrangement and pre-BCR expression, FoxO expression is induced by SLP-65–mediated down-regulation of PKB (Herzog et al., 2008) at the end of the pre–B cell stage. At this stage, FoxO proteins induce IgL gene recombination and, as in the case for IgH gene rearrangement, our data suggest that Ikaros acts downstream of FoxO1 to allow IgL gene rearrangement. The role of Ikaros in IgL gene recombination is supported by several observations. First, Ikaros has been shown to bind to the κLC gene locus (Liu et al., 2006). Second, the Ikaros family member Aiolos was shown to directly down-regulate the expression of the SLC component λ5, thereby effecting pre-BCR down-regulation, which is required for the induction of IgL gene recombination (Thompson et al., 2007). Third, Ikaros expression is induced by pre-BCR signaling and is required for the cell cycle arrest at the end of the pre–B cell stage (Trageser et al., 2009), where Ikaros has been reported to repress c-Myc and to induce p27 expression. These are important steps for the activation of IgL recombination because c-Myc activity interferes with κLC gene recombination, whereas induction of p27 results in cell cycle arrest and stabilization of Rag2 protein (Lee and Desiderio, 1999; Duy et al., 2010; Ma et al., 2010). The ability of Ikaros to interfere with cell cycle progression is in full agreement with the tumor suppressor role of Ikaros (Wang et al., 1996; Mullighan et al., 2008; Trageser et al., 2009). Indeed, deletions within IKZF1, the gene encoding Ikaros, seem to be a characteristic feature of BCR/ABL1-positive acute lymphoblastic leukemia (ALL) (Wang et al., 1996; Mullighan et al., 2008; Trageser et al., 2009). Moreover, our previous results suggested that BCR/ABL1 induces aberrant Ikaros splicing and the generation of the dominant-negative IK6 form (Wang et al., 1996; Mullighan et al., 2008; Trageser et al., 2009). This inactivation of Ikaros function is important for BCR/ABL1-mediated transformation, as Ikaros induces cell cycle arrest of BCR/ABL1-transformed ALL cells (Trageser et al., 2009).
Interestingly, the aberrant nonconventional splicing identified in the present study has not been described previously and does not seem to encode any Ikaros protein isoforms, as antibodies directed against the N terminus of Ikaros failed to detect any Ikaros isoforms in Pten−/− pro–B cells. Although we cannot exclude the presence of such isoforms, they are unlikely to act in a dominant-negative manner because expression of WT Ikaros was sufficient to induce efficient IgH gene recombination. An urgent question is how intron sequences can be inserted into exon 7 of Ikaros to disrupt the open reading frame. The fact that we observed this insertion in independent cell lines, in primary cells, and under different conditions suggests that this is a physiological PI3K-activated process, which differs from conventional alternative splicing. It is conceivable that this PI3K-activated nonconventional splicing pathway may regulate the expression of further genes, thereby providing a reversible mechanism for switching off genes at the level of RNA processing by inserting intron sequences into mature transcripts to disrupt their open reading frame. The emerging scheme suggests that PI3K signaling induces proliferation and blocks differentiation (as Ig gene rearrangement) by interfering with conventional splicing of tumor suppressors such as Ikaros and that FoxO1 exerts its tumor suppressor function partially by restoring the conventional splicing machinery to allow the expression of downstream targets.
MATERIALS AND METHODS
Mice.
Ptenfl/fl mice (provided by T. Mak, Campbell Family Institute for Breast Cancer Research at Princess Margaret Hospital, Toronto, ON, Canada) and FoxO1fl/fl mice (provided by R.A. DePinho, Belfer Institute for Applied Cancer Science, Dana-Farber Cancer Institute, Boston, MA) were bred to mb1-Cre (CD79a-Cre) transgenic mice to achieve B cell–specific Cre-mediated recombination in early pro–B cells. Mice used for peripheral B cell analysis and BM cultures were 6–8 wk old.
Mice were bred and housed in the animal facility of the Max-Planck-Institute of Immunobiology and Epigenetics. All animal experiments were done in compliance with the guidelines of the German law and were approved by the Animal Care and Use Committees of the Max-Planck-Institute of Immunobiology and Epigenetics and the local government.
Cell culture and biochemistry.
Cells were cultured in Iscove’s medium (Biochrom) containing 10% heat-inactivated FCS (Vitromex), 2 mM l-glutamine, 100 U/ml penicillin, 100 U/ml streptomycin (Invitrogen), and 5 × 10−5 M 2-mercaptoethanol. For IL-7–dependent growth, the supernatant of J558L cells stably transfected with a vector encoding mouse IL-7 was supplemented in excess. Pten-deficient cell lines were established by culturing BM cells of the respective mice for 14 d in IL-7–supplemented medium. For inhibition of PI3K, cells were treated with 30 µM LY294002 (Merck Biosciences) or as control with DMSO (Fluka Analytical) for the indicated times.
Plasmids and retroviral transduction.
To generate Pten-IRES-GFP, a full-length cDNA encoding Pten was subcloned into the retroviral vector backbone pMIG. The plasmid for expression of WT FoxO1 was generated by PCR amplification of the cDNA encoding full-length FoxO1, which was subsequently ligated into the pMIG retroviral vector. To generate mutant FoxO1-A3, three point mutations were inserted by PCR (T24A, S254A, and S319A). This mutant lacks the three phosphorylation sites of Akt and is therefore not underlying the regulation by PI3K/Akt signaling. Full-length FoxO1-A3 was subcloned into the pMIG vector. The plasmid for expression of Ikaros was kindly provided by I. Ferreirós Vidal and M. Merkenschlager (Imperial College London, London, England, UK). The dominant-negative mutant of Ikaros was generated by PCR amplification of a cDNA fragment lacking exons 2–6, which encode the DNA binding site (Mullighan et al., 2008), and was subcloned into the bicistronic pMIG vector. Subsequently, the GFP fragment was replaced by the CD8 sequence. pMITom-Pax5 was provided by R. Grosschedl and E. Mandel (Max-Planck-Institute for Immunobiology and Epigenetics).
Viral supernatants were generated using the Phoenix retroviral producer cell line as described in the manufacturer’s instructions. In brief, Phoenix cells were cultured for 48 h in Iscove’s culture medium + 10% FCS. Cells were plated at a density of 106 cells per ml to generate supernatants after transfection using GeneJuice (EMD). Retroviral supernatants were harvested after 36 and 60 h. For the subsequent transduction, Pten-deficient cells were mixed with supernatants and centrifuged at 300 g at 37°C for 3 h. Transduced cells were returned to culture and propagated in medium containing IL-7 for 4–5 d and were analyzed by flow cytometry.
Flow cytometry.
Cell suspensions were stained by standard procedures for flow cytometry analysis. Intracellular FACS staining was done using Fix and perm cell permeabilization kit (ADG), and then measured by FACSCalibur or LSRII flow cytometers (BD) using Cy5–anti-μHC (SouthernBiotech), FITC–anti-CD43 (SouthernBiotech), PE–anti-B220 (SouthernBiotech), Cy5–anti-CD25 (BD), PE–anti-cKIT (BD), and Alexa Fluor 647–anti-CD19 (BD).
Immunoblotting.
For immunoblot analysis, 1.5 × 106 cells/ml were collected and lysed in 50 µl of modified RIPA buffer (50 mM Tris HCl, pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, pH 8, 1 mM sodium orthovanadate, 1 mM NaF, and protease inhibitor cocktail [Sigma-Aldrich]). Lysates were subjected to 10% SDS-PAGE and transferred to PVDF membranes (Millipore). Membranes were blocked with 5% milk powder in PBT (PBS and 0.1% Tween-20) for 2 h at room temperature with constant agitation. Primary antibodies were diluted in PBT supplemented with 4% BSA fraction V (Enzo Life Sciences); secondary antibodies were diluted in blocking solution. Immunoreactive proteins were detected using a chemiluminescence detection system (ECL; GE Healthcare). Antibodies used were anti-Pten (Cell Signaling Technologies), anti-Ikaros (Active Motif Biotech), anti-FoxO1 (Cell Signaling Technologies), anti-Pax5 (Abcam), anti-μHC (SouthernBiotech), and anti-Actin (I-19; Santa Cruz Biotechnology, Inc.).
PCR analysis of IgH gene rearrangements.
DNA recombination at the IgH locus was assessed as previously described (Bertolino et al., 2005). Serial dilutions of genomic DNA were analyzed by PCR with primers specific for the VHJ558, VH7183, DQ52 gene segments and a primer 3′ of JH4 (Ehlich et al., 1994; Jumaa and Nielsen, 1997), and SRP20 was used as loading control (Jumaa and Nielsen, 1997). Subsequently, PCR products were separated by gel electrophoresis on a 1% agarose gel and were analyzed by Southern blotting with a 500-bp probe downstream of JH4.
PCR analysis of Ikaros transcripts.
PCR analysis of Ikaros transcripts were performed with specific forward primers located in exon 2 (E2), exon 4 (E4), or exon 7 (E7f). The reverse primers were located in exon 4 (E4), exon 7 (E7), or in the inserted intron fragment (Inr). Primer sequences were E2f, 5′-TCGAGGCATGGCCAGTAATGT-3′; E4f, 5′-CATCTTTGCAACTATGCCTGC-3′; E7f, 5′-AGACAAGTGCCTGTCAGACAT-3′; E4r, 5′-GTCCTCAGGTGGCCGGTGAGG-3′; E7r, 5′-TGTACACCTTCAGCTGCTCG-3′; and Inr, 5′-TAGCCCTCACTGTCCTTGG-3′.
Online supplemental material.
Fig. S1 shows a schematic overview of the Ikaros transcript in the absence of Pten and its sequence. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20110216/DC1.
Acknowledgments
We thank Peter J. Nielsen and Michael Reth for critical discussion and reading of the manuscript, Tak W. Mak (University Health Network, Toronto) for providing Ptenfl/fl mice, and Ronald A. DePinho for the FoxO1fl/fl mice. We also thank Andreas Wuerch and Sebastian Hobitz for cell sorting.
Our work is supported by the Deutsche Forschungsgemeinschaft (SFB 620 and SFB 746).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- BCR
- B cell antigen receptor
- EBF1
- early B cell factor 1
- EV
- empty vector
- HC
- heavy chain
- IgH
- Ig HC
- IgL
- Ig LC
- LC
- light chain
- PI3K
- phosphoinositide-3-kinase
- PKB
- protein kinase B
- Pten
- phosphatase and tensin homolog
- SLC
- surrogate LC
References
- Amin R.H., Schlissel M.S. 2008. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat. Immunol. 9:613–622 10.1038/ni.1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino E., Reddy K., Medina K.L., Parganas E., Ihle J., Singh H. 2005. Regulation of interleukin 7-dependent immunoglobulin heavy-chain variable gene rearrangements by transcription factor STAT5. Nat. Immunol. 6:836–843 10.1038/ni1226 [DOI] [PubMed] [Google Scholar]
- Biggs W.H., III, Meisenhelder J., Hunter T., Cavenee W.K., Arden K.C. 1999. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc. Natl. Acad. Sci. USA. 96:7421–7426 10.1073/pnas.96.13.7421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A., Bonni A., Zigmond M.J., Lin M.Z., Juo P., Hu L.S., Anderson M.J., Arden K.C., Blenis J., Greenberg M.E. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 96:857–868 10.1016/S0092-8674(00)80595-4 [DOI] [PubMed] [Google Scholar]
- Cobaleda C., Schebesta A., Delogu A., Busslinger M. 2007. Pax5: the guardian of B cell identity and function. Nat. Immunol. 8:463–470 10.1038/ni1454 [DOI] [PubMed] [Google Scholar]
- Cobb B.S., Smale S.T. 2005. Ikaros-family proteins: in search of molecular functions during lymphocyte development. Curr. Top. Microbiol. Immunol. 290:29–47 10.1007/3-540-26363-2_3 [DOI] [PubMed] [Google Scholar]
- Coffer P.J., Burgering B.M. 2004. Forkhead-box transcription factors and their role in the immune system. Nat. Rev. Immunol. 4:889–899 10.1038/nri1488 [DOI] [PubMed] [Google Scholar]
- Decker D.J., Kline G.H., Hayden T.A., Zaharevitz S.N., Klinman N.R. 1995. Heavy chain V gene-specific elimination of B cells during the pre-B cell to B cell transition. J. Immunol. 154:4924–4935 [PubMed] [Google Scholar]
- Dengler H.S., Baracho G.V., Omori S.A., Bruckner S., Arden K.C., Castrillon D.H., DePinho R.A., Rickert R.C. 2008. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat. Immunol. 9:1388–1398 10.1038/ni.1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duy C., Yu J.J., Nahar R., Swaminathan S., Kweon S.M., Polo J.M., Valls E., Klemm L., Shojaee S., Cerchietti L., et al. 2010. BCL6 is critical for the development of a diverse primary B cell repertoire. J. Exp. Med. 207:1209–1221 10.1084/jem.20091299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlich A., Martin V., Müller W., Rajewsky K. 1994. Analysis of the B-cell progenitor compartment at the level of single cells. Curr. Biol. 4:573–583 10.1016/S0960-9822(00)00129-9 [DOI] [PubMed] [Google Scholar]
- Fuxa M., Skok J., Souabni A., Salvagiotto G., Roldan E., Busslinger M. 2004. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 18:411–422 10.1101/gad.291504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos K., Bigby M., Wang J.H., Molnar A., Wu P., Winandy S., Sharpe A. 1994. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 79:143–156 10.1016/0092-8674(94)90407-3 [DOI] [PubMed] [Google Scholar]
- Hagman J., Belanger C., Travis A., Turck C.W., Grosschedl R. 1993. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 7:760–773 10.1101/gad.7.5.760 [DOI] [PubMed] [Google Scholar]
- Herzog S., Hug E., Meixlsperger S., Paik J.H., DePinho R.A., Reth M., Jumaa H. 2008. SLP-65 regulates immunoglobulin light chain gene recombination through the PI(3)K-PKB-Foxo pathway. Nat. Immunol. 9:623–631 10.1038/ni.1616 [DOI] [PubMed] [Google Scholar]
- Herzog S., Reth M., Jumaa H. 2009. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat. Rev. Immunol. 9:195–205 10.1038/nri2491 [DOI] [PubMed] [Google Scholar]
- Hesslein D.G., Pflugh D.L., Chowdhury D., Bothwell A.L., Sen R., Schatz D.G. 2003. Pax5 is required for recombination of transcribed, acetylated, 5′ IgH V gene segments. Genes Dev. 17:37–42 10.1101/gad.1031403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt S.L., Chaumeil J., Skok J.A. 2010. Chromosome dynamics and the regulation of V(D)J recombination. Immunol. Rev. 237:43–54 10.1111/j.1600-065X.2010.00931.x [DOI] [PubMed] [Google Scholar]
- Hobeika E., Thiemann S., Storch B., Jumaa H., Nielsen P.J., Pelanda R., Reth M. 2006. Testing gene function early in the B cell lineage in mb1-cre mice. Proc. Natl. Acad. Sci. USA. 103:13789–13794 10.1073/pnas.0605944103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huetz F., Carlsson L., Tornberg U.C., Holmberg D. 1993. V-region directed selection in differentiating B lymphocytes. EMBO J. 12:1819–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobucci I., Lonetti A., Messa F., Cilloni D., Arruga F., Ottaviani E., Paolini S., Papayannidis C., Piccaluga P.P., Giannoulia P., et al. 2008. Expression of spliced oncogenic Ikaros isoforms in Philadelphia-positive acute lymphoblastic leukemia patients treated with tyrosine kinase inhibitors: implications for a new mechanism of resistance. Blood. 112:3847–3855 10.1182/blood-2007-09-112631 [DOI] [PubMed] [Google Scholar]
- Jhunjhunwala S., van Zelm M.C., Peak M.M., Murre C. 2009. Chromatin architecture and the generation of antigen receptor diversity. Cell. 138:435–448 10.1016/j.cell.2009.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K., Pflugh D.L., Yu D., Hesslein D.G., Lin K.I., Bothwell A.L., Thomas-Tikhonenko A., Schatz D.G., Calame K. 2004. B cell-specific loss of histone 3 lysine 9 methylation in the V(H) locus depends on Pax5. Nat. Immunol. 5:853–861 10.1038/ni1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumaa H., Nielsen P.J. 1997. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 16:5077–5085 10.1093/emboj/16.16.5077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F., Feldhahn N., Herzog S., Sprangers M., Mooster J.L., Jumaa H., Müschen M. 2006. BCR-ABL1 induces aberrant splicing of IKAROS and lineage infidelity in pre-B lymphoblastic leukemia cells. Oncogene. 25:1118–1124 10.1038/sj.onc.1209133 [DOI] [PubMed] [Google Scholar]
- Kops G.J., Burgering B.M. 1999. Forkhead transcription factors: new insights into protein kinase B (c-akt) signaling. J. Mol. Med. 77:656–665 10.1007/s001099900050 [DOI] [PubMed] [Google Scholar]
- Lee J., Desiderio S. 1999. Cyclin A/CDK2 regulates V(D)J recombination by coordinating RAG-2 accumulation and DNA repair. Immunity. 11:771–781 10.1016/S1074-7613(00)80151-X [DOI] [PubMed] [Google Scholar]
- Liu Z., Widlak P., Zou Y., Xiao F., Oh M., Li S., Chang M.Y., Shay J.W., Garrard W.T. 2006. A recombination silencer that specifies heterochromatin positioning and ikaros association in the immunoglobulin kappa locus. Immunity. 24:405–415 10.1016/j.immuni.2006.02.001 [DOI] [PubMed] [Google Scholar]
- Ma S., Pathak S., Mandal M., Trinh L., Clark M.R., Lu R. 2010. Ikaros and Aiolos inhibit pre-B-cell proliferation by directly suppressing c-Myc expression. Mol. Cell. Biol. 30:4149–4158 10.1128/MCB.00224-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T., Dixon J.E. 1998. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 273:13375–13378 10.1074/jbc.273.22.13375 [DOI] [PubMed] [Google Scholar]
- Manning B.D., Cantley L.C. 2007. AKT/PKB signaling: navigating downstream. Cell. 129:1261–1274 10.1016/j.cell.2007.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall A.J., Wu G.E., Paige G.J. 1996. Frequency of VH81x usage during B cell development: initial decline in usage is independent of Ig heavy chain cell surface expression. J. Immunol. 156:2077–2084 [PubMed] [Google Scholar]
- Merkenschlager M. 2010. Ikaros in immune receptor signaling, lymphocyte differentiation, and function. FEBS Lett. 584:4910–4914 10.1016/j.febslet.2010.09.042 [DOI] [PubMed] [Google Scholar]
- Mullighan C.G., Miller C.B., Radtke I., Phillips L.A., Dalton J., Ma J., White D., Hughes T.P., Le Beau M.M., Pui C.H., et al. 2008. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 453:110–114 10.1038/nature06866 [DOI] [PubMed] [Google Scholar]
- Nutt S.L., Urbánek P., Rolink A., Busslinger M. 1997. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 11:476–491 10.1101/gad.11.4.476 [DOI] [PubMed] [Google Scholar]
- Okkenhaug K., Vanhaesebroeck B. 2003. PI3K in lymphocyte development, differentiation and activation. Nat. Rev. Immunol. 3:317–330 10.1038/nri1056 [DOI] [PubMed] [Google Scholar]
- Omori S.A., Cato M.H., Anzelon-Mills A., Puri K.D., Shapiro-Shelef M., Calame K., Rickert R.C. 2006. Regulation of class-switch recombination and plasma cell differentiation by phosphatidylinositol 3-kinase signaling. Immunity. 25:545–557 10.1016/j.immuni.2006.08.015 [DOI] [PubMed] [Google Scholar]
- Rajewsky K. 1996. Clonal selection and learning in the antibody system. Nature. 381:751–758 10.1038/381751a0 [DOI] [PubMed] [Google Scholar]
- Reynaud D., Demarco I.A., Reddy K.L., Schjerven H., Bertolino E., Chen Z., Smale S.T., Winandy S., Singh H. 2008. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat. Immunol. 9:927–936 10.1038/ni.1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz D.G. 2004. V(D)J recombination. Immunol. Rev. 200:5–11 10.1111/j.0105-2896.2004.00173.x [DOI] [PubMed] [Google Scholar]
- Schebesta M., Pfeffer P.L., Busslinger M. 2002. Control of pre-BCR signaling by Pax5-dependent activation of the BLNK gene. Immunity. 17:473–485 10.1016/S1074-7613(02)00418-1 [DOI] [PubMed] [Google Scholar]
- Schlissel M.S. 2003. Regulating antigen-receptor gene assembly. Nat. Rev. Immunol. 3:890–899 10.1038/nri1225 [DOI] [PubMed] [Google Scholar]
- Sellars M., Reina-San-Martin B., Kastner P., Chan S. 2009. Ikaros controls isotype selection during immunoglobulin class switch recombination. J. Exp. Med. 206:1073–1087 10.1084/jem.20082311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Kaisho T., Ohishi M., Tsukio-Yamaguchi M., Tsubata T., Koni P.A., Sasaki T., Mak T.W., Nakano T. 2003. Critical roles of Pten in B cell homeostasis and immunoglobulin class switch recombination. J. Exp. Med. 197:657–667 10.1084/jem.20021101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaishi H., Konishi H., Matsuzaki H., Ono Y., Shirai Y., Saito N., Kitamura T., Ogawa W., Kasuga M., Kikkawa U., Nishizuka Y. 1999. Regulation of nuclear translocation of forkhead transcription factor AFX by protein kinase B. Proc. Natl. Acad. Sci. USA. 96:11836–11841 10.1073/pnas.96.21.11836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Boekel E., Melchers F., Rolink A.G. 1997. Changes in the V(H) gene repertoire of developing precursor B lymphocytes in mouse bone marrow mediated by the pre-B cell receptor. Immunity. 7:357–368 10.1016/S1074-7613(00)80357-X [DOI] [PubMed] [Google Scholar]
- Thompson E.C., Cobb B.S., Sabbattini P., Meixlsperger S., Parelho V., Liberg D., Taylor B., Dillon N., Georgopoulos K., Jumaa H., et al. 2007. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity. 26:335–344 10.1016/j.immuni.2007.02.010 [DOI] [PubMed] [Google Scholar]
- Tothova Z., Kollipara R., Huntly B.J., Lee B.H., Castrillon D.H., Cullen D.E., McDowell E.P., Lazo-Kallanian S., Williams I.R., Sears C., et al. 2007. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 128:325–339 10.1016/j.cell.2007.01.003 [DOI] [PubMed] [Google Scholar]
- Trageser D., Iacobucci I., Nahar R., Duy C., von Levetzow G., Klemm L., Park E., Schuh W., Gruber T., Herzog S., et al. 2009. Pre-B cell receptor–mediated cell cycle arrest in Philadelphia chromosome-positive acute lymphoblastic leukemia requires IKAROS function. J. Exp. Med. 206:1739–1753 10.1084/jem.20090004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubelhart R., Bach M.P., Eschbach C., Wossning T., Reth M., Jumaa H. 2010. N-linked glycosylation selectively regulates autonomous precursor BCR function. Nat. Immunol. 11:759–765 10.1038/ni.1903 [DOI] [PubMed] [Google Scholar]
- Urbánek P., Wang Z.Q., Fetka I., Wagner E.F., Busslinger M. 1994. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 79:901–912 10.1016/0092-8674(94)90079-5 [DOI] [PubMed] [Google Scholar]
- Wang J.H., Nichogiannopoulou A., Wu L., Sun L., Sharpe A.H., Bigby M., Georgopoulos K. 1996. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 5:537–549 10.1016/S1074-7613(00)80269-1 [DOI] [PubMed] [Google Scholar]
- Yancopoulos G.D., Desiderio S.V., Paskind M., Kearney J.F., Baltimore D., Alt F.W. 1984. Preferential utilization of the most JH-proximal VH gene segments in pre-B-cell lines. Nature. 311:727–733 10.1038/311727a0 [DOI] [PubMed] [Google Scholar]
- Yoshida H. 2007. Unconventional splicing of XBP-1 mRNA in the unfolded protein response. Antioxid. Redox Signal. 9:2323–2333 10.1089/ars.2007.1800 [DOI] [PubMed] [Google Scholar]