2′-O-methylation of guanosine 18 is a naturally occurring tRNA modification that can suppress immune TLR7 responses.
Abstract
Naturally occurring nucleotide modifications within RNA have been proposed to be structural determinants for innate immune recognition. We tested this hypothesis in the context of native nonself-RNAs. Isolated, fully modified native bacterial transfer RNAs (tRNAs) induced significant secretion of IFN-α from human peripheral blood mononuclear cells in a manner dependent on TLR7 and plasmacytoid dendritic cells. As a notable exception, tRNATyr from Escherichia coli was not immunostimulatory, as were all tested eukaryotic tRNAs. However, the unmodified, 5′-unphosphorylated in vitro transcript of tRNATyr induced IFN-α, thus revealing posttranscriptional modifications as a factor suppressing immunostimulation. Using a molecular surgery approach based on catalytic DNA, a panel of tRNATyr variants featuring differential modification patterns was examined. Out of seven modifications present in this tRNA, 2′-O-methylated Gm18 was identified as necessary and sufficient to suppress immunostimulation. Transplantation of this modification into the scaffold of yeast tRNAPhe also resulted in blocked immunostimulation. Moreover, an RNA preparation of an E. coli trmH mutant that lacks Gm18 2′-O-methyltransferase activity was significantly more stimulatory than the wild-type sample. The experiments identify the single methyl group on the 2′-oxygen of Gm18 as a natural modification in native tRNA that, beyond its primary structural role, has acquired a secondary function as an antagonist of TLR7.
Microbial nucleic acid recognition has been identified as a common theme in innate immunity involving a variety of receptors (Toll-like receptors [TLRs] 3, 7, 8, and 9, RIG-I, Mda5, Nalp3, AIM2, and IFI16; Kumar et al., 2011). Generally, three principles have been elucidated that allow for differentiation of foreign and self–nucleic acids: spatial restrictions, sequence composition, and nucleotide modifications.
RNA is abundantly modified with >100 different known chemical modifications that are introduced posttranscriptionally (Czerwoniec et al., 2009; Grosjean, 2009; Cantara et al., 2011). Because the extent of modifications depends on the RNA species as well as its evolutionary origin, and because eukaryotic RNA in general is more abundantly modified, nucleotide modifications might conceivably allow discrimination of self and nonself (Koski et al., 2004; Karikó et al., 2005). Indeed, random incorporation of naturally occurring m5C, m6A, m5U, s2U, or pseudouridine into in vitro transcripts (IVTs) of messenger RNAs abrogated stimulation of TLRs (Koski et al., 2004). Similarly, multiple studies showed that incorporation of 2′-O-Me nucleotides into small interfering RNA (siRNA) suppressed unwanted immunostimulation (Robbins et al., 2007; Sioud et al., 2007; Eberle et al., 2008). As a point in case, 2′-O-Me is a regularly occurring modification in natural RNA that occurs in higher frequencies in eukaryotic RNA than in bacterial or mitochondrial RNA (Karikó and Weissman, 2007; Jühling et al., 2009).
However, the aforementioned concepts have not yet been tested with natural, full-length RNA species of defined sequence. Because access to pure native RNA species is limited, all previous studies, when specifically analyzing single modifications, made use either of short synthetic oligoribonucleotides or of IVTs. Moreover, investigations are biased toward self-RNA, and most interpretations have not been cross-validated with nonself-RNA. A point in case is again given by 2′-O-Me: although a suppressive effect on secretion of IFN-α from PBMCs is documented in RNAs of artificial or eukaryotic sequence, effects in foreign RNAs are not understood. This is particularly relevant because 2′-O-Me occurs in bacteria as well (Persson et al., 1997; Hori et al., 2002; Czerwoniec et al., 2009; Cantara et al., 2011). Conceivably, the ensemble of other bacterial modifications as well as positioning and structural context of such modifications is probably of additional importance.
Therefore, we investigated native bacterial RNA species to analyze nucleotide modifications in their natural context. We used purified transfer RNA (tRNA) species of bacterial origin, and the resulting TLR7-mediated stimulation, as measured by ELISA-based detection of IFN-α secretion from PBMCs, was compared with purified eukaryotic tRNA species. We report the development of a method to produce tRNA hybrids that allows dissecting single modification and analyzing their immunostimulatory potential.
Using such a molecular surgical approach, we unequivocally identify 2′-O-Me G18 (Gm18) as a highly efficient suppressive modification in bacterial native tRNATyr. This single methyl group is necessary and sufficient for immunosuppression in a classical tRNA macromolecule, as could be shown by transplantation experiments into otherwise highly stimulatory IVTs. Analysis of eukaryotic tRNAs reveals that Gm18 is not the singular discriminator of self- versus nonself-tRNA. However, increased stimulation by total tRNA preparations from Escherichia coli mutants lacking the Gm18 methyltransferase (MTase) trmH suggests a potential role of this enzyme as a virulence factor.
RESULTS AND DISCUSSION
As opposed to previous approaches identifying nucleotide modifications with effects on immunostimulation in self-RNA, we set out to identify such moieties in nonself-RNA. Although previous studies have relied on ill-defined preparations of total RNA, we based our investigations on the fact that native bacterial tRNAs contain a large number of modifications at precise and well-known positions.
Native E. coli tRNATyr lacks immunostimulatory activity in PBMCs
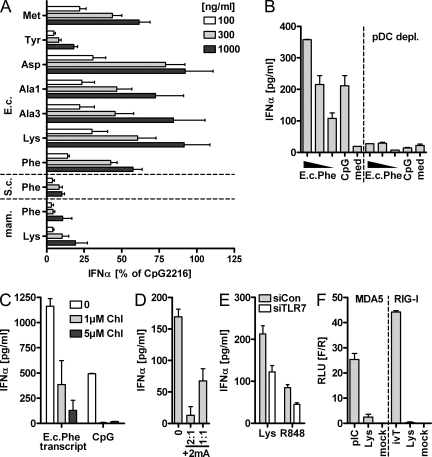
We first tested a panel of native modified bacterial tRNAs. Importantly, tRNA accounts for around 10% of the bacterial RNA population and thus is the second most abundant RNA species. Considering that the majority of the ribosomal RNA population is complexed in ribosomal particles and thus likely to be shielded from TLR recognition, tRNA is probably a relevant RNA species for immune recognition. PBMCs were tested for secretion of IFN-α upon stimulation with RNA. To account for donor variance, experimental results were normalized to stimulation with the TLR9 ligand CpG2216. Various tRNA species from E. coli in general showed strong immunostimulation (Fig. 1 A). In contrast, tRNAPhe from Saccharomyces cerevisiae, a heavily modified eukaryotic tRNA, induced only minor amounts of IFN-α when compared side by side with E. coli tRNAPhe. Similarly, mammalian tRNAPhe and tRNALys3 were devoid of immunostimulatory activity. Among the strongly stimulatory tRNA species from E. coli, i.e., Met, Ala1, Ala3, Phe, Lys, and Asp (sequences in Jühling et al. [2009]), only moderate differences could be evidenced. In surprising contrast to these bacterial tRNAs, native E. coli tRNATyr was not stimulatory.
Figure 1.
Native E. coli tRNATyr lacks immunostimulation in PBMCs. (A) PBMCs were stimulated with E. coli (E.c.) tRNAs, tRNAPhe from eukaryotic S. cerevisiae (S.c.), mammalian (mam.) tRNAPhe, or mammalian tRNALys3 and analyzed for secretion of IFN-α. Data are normalized to 1 µM CpG2216 (n = 3–10; mean + SEM). (B) PBMCs or PBMCs depleted of pDCs were stimulated with E. coli tRNAPhe and analyzed as in A (mean of triplicate values + SD). (C) PBMCs were preincubated with 1 or 5 µM chloroquine (Chl) and stimulated with 1 µg/ml IVT of E. coli tRNAPhe. (D) PBMCs were stimulated with 300 ng/ml E. coli tRNALys in the absence or presence of a 1:1 or 1:2 ratio of the TLR7-blocking RNA oligonucleotide 2mA. (C and D) One of two experiments is shown (mean of duplicate values + SD). (E) PBMCS were treated with siRNA against TLR7 and subsequently stimulated with 1 µg/ml E. coli tRNALys or 1 µg/ml R848 and tested for IFN-α secretion (one of two experiments is shown; mean + SD). (F) Huh7.5 cells harboring an IRF3 reporter gene were transfected with either RIG-I or MDA5. Cells were stimulated with 1 µg/ml E. coli tRNALys; 0.2 µg/ml of 400-bp ds IVT RNA or poly (dI:dC) was used as control, and reporter gene activity was measured (n = 2; mean + SD). RLU, raw light units.
We verified that the observed IFN-α response was caused by stimulation of TLR7 in plasmacytoid DCs (pDCs). Thus, depletion of pDCs from PBMCs abolished tRNA-mediated IFN-α secretion (Fig. 1 B). The observed tRNA activity was abolished either by blocking endosomal maturation with chloroquine or by inhibition of TLR7 with an antagonistic RNA oligonucleotide (2mA; Fig. 1, C and D). Furthermore, partial knockdown of TLR7 in PBMCs by siRNA clearly decreased tRNA-mediated induction of IFN-α in proportion to RNA interference efficiency by a known TLR7 ligand (Fig. 1 E). Contributions by other RNA-sensing entities were largely excluded. No activation of RIG-I but minimal stimulation of MDA5 could be observed in Huh7.5 cells expressing the respective receptors (Fig. 1 F), thus ruling out a major contribution of intracellular MDA5 recognition. Moreover, siRNA knockdown of IPS1, the downstream adaptor of MDA5 and RIG-I, did not reduce tRNA stimulation (not depicted).
Lack of activity of E. coli tRNATyr is caused by naturally occurring nucleotide modifications
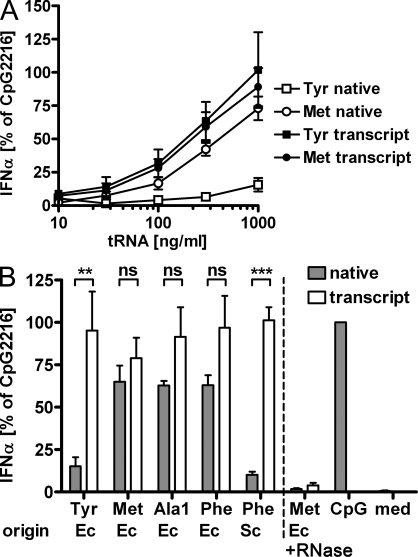
We compared the lack of immunostimulatory activity of E. coli tRNATyr with the immune response to an unmodified version, obtained by in vitro transcription. Of special importance, IVTs were generated from a precursor with a self-excising hammerhead (hh) leader sequence, resulting in a 5′-OH end, effectively excluding the presence of 5′-triphosphates, whose known stimulatory properties might activate RIG-I (Hornung et al., 2006). Processed tRNA IVTs were detected and purified by PAGE. The IVTs of tRNATyr and tRNAMet were directly compared with the native tRNA preparations (Fig. 2 A). Of note, the IVT of tRNATyr strongly induced IFN-α, thus arguing for suppressive activity within the native tRNA. Testing yet further IVTs of various tRNAs (E. coli Phe, E. coli Ala1, and S. cerevisiae Phe) showed that in general all IVTs are stimulatory, and among the tested bacterial tRNAs, only tRNATyr showed a significant difference between native and IVT tRNA (Fig. 2 B). Comparison of native and IVT E. coli tRNATyr clearly indicates that the silencing effect must be caused by one or more of the seven posttranscriptional nucleotide modifications present in the native tRNA, namely s4U8, Gm18, Q34, ms2i6A37, Ψ39, T54, and Ψ55 (Fig. 3 A).
Figure 2.
IVT tRNAs are stimulatory. (A) TLR7 response of native E. coli tRNAs (open symbols) and IVTs (closed symbols) of tRNATyr (squares) and tRNAMet (circles) normalized to CpG2216 (n = 4–6; mean + SEM). (B) Comparison at 1 µg/ml of native tRNAs (closed bars) and the corresponding IVTs (open bars) of E. coli (Ec) tRNAs for Tyr, Met, Ala1, and Phe and S. cerevisiae (Sc) tRNAPhe (n = 4–7; mean + SEM; IVT tRNAPhe Sc, n = 2). The control consist of an RNase A digest of native tRNAMet and its IVT. med, medium control. **, P < 0.01; ***, P < 0.001.
Figure 3.
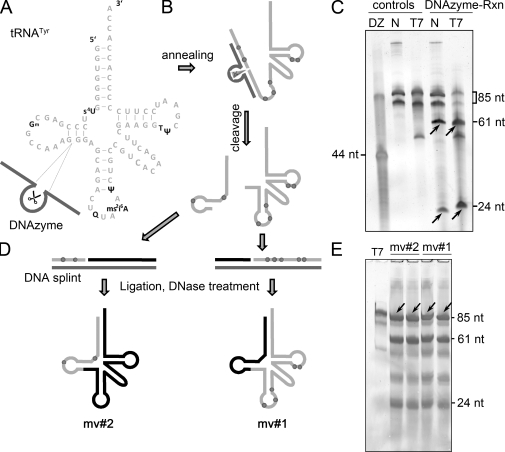
Molecular surgery approach based on catalytic DNA. (A and B) A DNAzyme was engineered to cut between residues 24 and 25 of tRNATyr (85 nt), generating fragments of 61 nt and 24 nt. (C) Fragments of cleaved native tRNATyr1 and its IVT were purified by PAGE (indicated by arrows). (D) Isolated unmodified (black lines) and modified fragments (gray lines with dots indicating modifications) were annealed on splint cDNA to yield modivariants mv#1 and mv#2 after ligation and digestion of cDNA. (E) PAGE purification of modivariants (indicated by arrows).
Molecular surgery identifies 2′-O-Me at G18 to suppress IFN-α induction by E. coli tRNATyr
To deconvolute the contribution of single modifications or groups thereof, we synthesized modivariants, i.e., differentially modified tRNATyr species, in a two-step procedure of molecular surgery as depicted in Fig. 3. By judicious choice of the targeting sequences (Hengesbach et al., 2008), the RNA cleavage activity of an 8-17 DNAzyme was directed between nucleotides 24 and 25 to generate defined tRNA fragments carrying modifications from native tRNATyr or their unmodified counterparts from the corresponding IVT (Fig. 3, B and C). Repeated thermocycling was used to maximize cleavage, and the resulting fragments were isolated from reactions on a preparative scale (i.e., 800 pmol). After phosphorylation on their 5′ end and removal of cyclic 3′-phosphates by treatment with T4-PNK, reassembly of permutated fragments was achieved by hybridization onto a full-length cDNA. This resulted in the formation of nicked DNA–RNA hybrids, which are ligation-competent complexes for T4-DNA ligase (Fig. 3, D and E; Kurschat et al., 2005). After ligation, the DNA splint was removed by enzymatic digestion, and the full-length modivariant tRNAs mv#1 and mv#2 were purified by PAGE in final quantities of ∼2 µg.
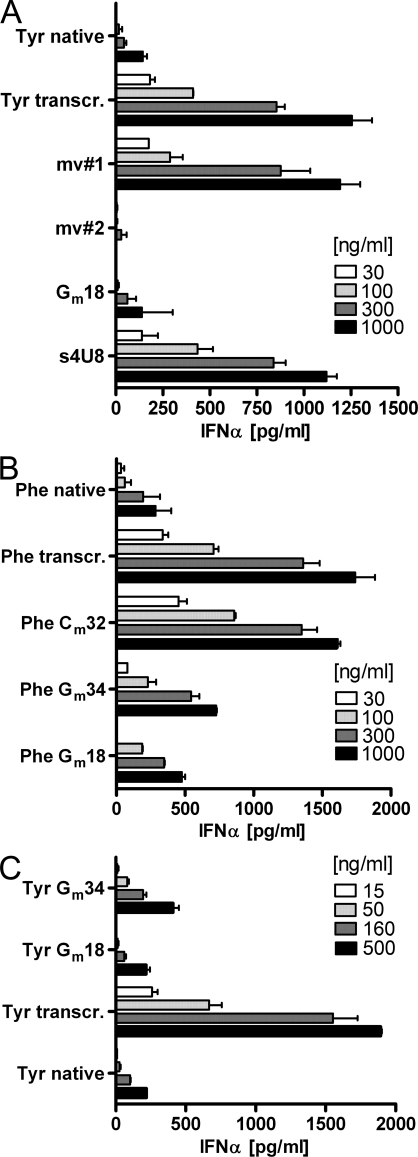
Analysis of these modivariants in the IFN-α secretion assay revealed that the modifications suppressing immunostimulation are situated in the 5′ half of the tRNA (Fig. 4 A): mv#1, which contains the five modifications Q34, ms2i6A37, Ψ39, T54, and Ψ55 of the 3′ fragment, was nearly as strongly stimulatory as the IVT, whereas mv#2, containing s4U8 and Gm18, was nearly as silent as the native tRNATyr. Next, modivariants Gm18 and s4U8 were generated from synthetic fragments including only Gm18 or s4U8 in the 5′ end of the tRNA. We observed that s4U8 did not show any effect (Fig. 4 A), Gm18 alone was sufficient to completely ablate any immunostimulatory properties of the 85-nt full-length bacterial tRNATyr, thus emerging as the singular effective modification in this tRNA.
Figure 4.
Unique effect of Gm18 in suppression of immunostimulation. (A) PBMC stimulation with native E. coli tRNATyr, its IVT, and modivariants mv#1 and mv#2, as well as modivariants Gm18 and s4U8 (n = 4; mean + SD). (B) Stimulation of PBMCs with native S. cerevisiae tRNAPhe, its IVT, and variants bearing Gm18 or single, naturally occurring 2′-O-Me residues at different positions (Cm32 and Gm34; n = 2; mean + SD of triplicates). (C) Stimulation of PBMCs with variants of E. coli tRNATyr with positionally altered Gm (n = 3; mean + SD).
Different modification events lead to immunosilencing of eukaryotic tRNA
Because Gm is common in eukaryotic tRNAs as well, the question arises of whether this modification represents a fundamental principle in the recognition of tRNA by TLR7. Native tRNAPhe from yeast, which showed no immunostimulation (Fig. 1), lacks Gm18 (Jühling et al., 2009) but bears other modifications that obviously act in a suppressive manner. Using a similar molecular surgery approach as for tRNATyr, we set out to identify these. However, tRNAPhe is more heavily modified than tRNATyr, and the accompanying increase in thermostability protected native tRNA from DNAzyme cleavage, whereas the IVT was an efficient substrate (not depicted). We therefore resorted to fragments resulting from chemical cleavage of the tRNA by hydrochloric acid, which causes a depurination of the wybutosine base 37, followed by acid cleavage (Sprinzl et al., 1976). From the resulting fragments, several modivariants were synthesized in small amounts and tested, which pointed to the anticodon as possible location of silencing modifications with a potential similar to Gm18 (Fig. S1). According to these observations and the results from tRNATyr, we constructed modivariants carrying single 2′-O-Me modifications found in native tRNAPhe (Gm34 and Cm32; Fig. 4 B). Cm32 was without effect, but Gm34 efficiently suppressed TLR7 recognition of tRNAPhe. Because certain modivariants carrying modifications in the 3′ part of the tRNA were not synthetically accessible, the possibility that further modifications influence TLR7 recognition could not be ruled out. Indeed, fully modified tRNAPhe was yet less stimulatory than its Gm34 modivariant (Fig. 4). Of note, mammalian tRNALys3 does not contain any Gm at all and must thus rely on yet a different modification.
Gm contains a methylation of the ribose 2′-OH, which has been described as a factor lowering TLR-mediated immunostimulation that results in IFN secretion (Karikó et al., 2005; Karikó and Weissman, 2007; Robbins et al., 2007; Sioud et al., 2007; Eberle et al., 2008). However, the previously discovered effect of Um nucleotides (Robbins et al., 2007) was much less pronounced; indeed, to silence a 21mer (single-stranded) siRNA, all uridines in this RNA were 2′-O-Me. Tluk et al. (2009) have then described a silencing effect of 2′-O-Me of all four major nucleotides and at various positions within short self-RNA or related sequences. The sequences of the latter were derived by insertion of an immunostimulatory 5′-UUGU-3′ tetranucleotide sequence. Although such modifications did indeed silence the TLR7-mediated IFN-α response, their provenance was not native, i.e., neither was the position of the natural modification maintained, nor was the investigation extended to a full-length natural RNA. Previously published results included a density of at least one 2′-O-Me residue per ∼15–20 nt (Karikó et al., 2005). In contrast, we have here identified a silencing modification that acts on a significantly larger, full-length RNA that is of physiological significance and unrelated to self-RNA.
Molecular transplantation experiments demonstrate that bacterial and eukaryotic tRNA scaffolds are interchangeable
The IVT of tRNAPhe from S. cerevisiae was as strongly stimulatory as all other tRNA IVTs (Fig. 1), suggesting that there are no cryptic details inherent in eukaryotic tRNA sequences that might govern TLR7 recognition on a fundamental level. To clearly demonstrate that bacterial and eukaryotic tRNA scaffolds are interchangeable, transplantation experiments were performed in which both identified effective Gm modifications (Gm18 from prokaryotic and Gm34 from eukaryotic tRNA) were assayed in a nonnative tRNA context. Transplantation by molecular surgery of Gm18 into the scaffold of the tRNAPhe IVT efficiently silenced the strong response provoked by the IVT (Phe Gm18 in Fig. 4 B). This shows that the TLR7 response to eukaryotic self-tRNA may be silenced by Gm18 also in a tRNA context in which it does not naturally occur. Similarly, in the analogous transplantation of the eukaryotic modification into a bacterial tRNA context, the Gm34 modification also silenced the tRNATyr response, albeit with somewhat reduced efficiency (Tyr Gm34 in Fig. 4 C).
Gm18 reduces immunostimulatory activity of total bacterial tRNA preparations
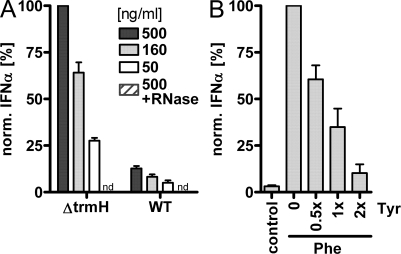
In E. coli, the gene product of trmH has been identified to act as 2′-O-MTase, which is crucial for tRNA Gm18 modification (Persson et al., 1997). trmH deficiency results in loss of Gm18. As Gm18 occurs in 13 out of 45 tRNAs in E. coli, loss of Gm18 might increase the immunostimulatory properties of whole tRNA preparations. Surprisingly, total tRNA from WT E. coli was only weakly stimulatory, although the majority of tRNAs did not possess the Gm18 modification. We speculated that Gm18 might not only lack immunostimulation but even suppress otherwise activating tRNA. This notion is strongly supported by the finding that total tRNA from a trmH mutant (ΔtrmH::kan; Persson et al., 1997) elicited a significantly stronger IFN-α response (Fig. 5 A). This result implies an antagonistic property of Gm18-containing tRNAs, which appear to silence the TLR7-mediated response to the remainder of the total tRNA population. We therefore decided to evaluate the antagonistic properties of Gm18-containing tRNATyr in a titration experiment with stimulatory, G18-unmethylated tRNA. Increasing amounts of E. coli tRNATyr were added to stimulatory E. coli tRNAPhe (Fig. 5 B). Indeed, E. coli tRNATyr decreased IFN-α secretion induced by tRNAPhe in an inhibitory manner.
Figure 5.
Increased immunostimulation by tRNA lacking Gm18. (A) PBMCs were stimulated with whole tRNA preparations from either WT E. coli or a deletion mutant of the Gm18 MTase trmH (ΔtrmH). RNA preparations (500 ng/ml) were also digested with RNase A. To account for donor variance, the IFN-α response at 500 ng/ml RNA from ΔtrmH was used for normalization. nd, not detectable (n = 4–6; mean + SEM; RNase A digests, n = 2). (B) PBMCs were stimulated with a fixed concentration (830 ng/ml) of tRNAPhe, and tRNATyr was added in a ratio as indicated (n = 3 donors with each donor stimulated by tRNAPhe alone set to 100% to account for donor variance; mean + SEM).
At this point, we can state that self-tRNA is guarded against TLR7 activation (which was the most important receptor triggered by tRNA in our study setting) not by a singular principle, but by a variety of at least three different posttranscriptional modifications, of which we have characterized Gm18 and Gm34 in detail, and another one is contained in mammalian tRNALys3. Systematic investigation of all of the >40 tRNA species in humans must be postponed to future research. We have already shown (Fig. 4) that Cm32 does not affect recognition of the tRNAPhe scaffold by TLR7, indicating that not all 2′-O-methylations are effective in this matter. Naturally occurring 2′-O-Me residues in tRNAs include positions 4, 44, or 54, which are thus prime candidates for further investigations (Jühling et al., 2009; Cantara et al., 2011).
Gm18 in bacterial tRNA is likely to have evolved from selection pressure unrelated to the mammalian innate immune system. Indeed, the Gm18 modification is found in bacterial, archaeal, and eukaryotic tRNAs alike, where it is part of a class of modifications in the structural core of the tRNA deemed to be responsible for structural and metabolic stabilization (Motorin and Helm, 2010). Lack of a growth phenotype, i.e., normal growth in modification-deficient mutants, as observed for trmH mutants in E. coli (Persson et al., 1997), is a common feature of such modifications, which are thought to contribute to a network of cooperative structural effects. Although a structural effect still awaits biophysical confirmation (Kumagai et al., 1980; Björk, 1995; Motorin and Helm, 2010), a widespread occurrence supports the notion that an effect of Gm18 on TLR7 recognition is a secondary function acquired late in evolution.
Most interestingly, a potential physiological importance of Gm18 for the immunological recognition of bacterial RNA could be demonstrated by using an E. coli mutant that lacked the 2′-O-MTase trmH. Whole tRNA preparations from mutant bacteria showed increased immunostimulation as compared with WT bacteria. This result verifies that Gm18 in a physiological setting is important to shape bacterial immunogenicity for innate immune cells. Our results show that among natural tRNA modifications, Gm18 plays an outstanding role in modulating the tRNA’s immunostimulatory properties. The fact that trmH mutants are more immunostimulatory although only a minority of tRNAs (Persson et al., 1997) bear a Gm18 modification (Fig. 5 A) and decreased immunostimulation of tRNA upon addition of Gm18-modified species (Fig. 5 B) indicates a dominant inhibitory effect, which de facto categorizes tRNATyr as a TLR7 antagonist.
Assuming a secondary role of bacterial Gm18 in avoiding a mammalian immune response, a correlation between the Gm content in bacterial tRNA from various species and the adaptation of the respective prokaryote to mammals might reveal evolutionary immunological pressure that selected for this modification. Certainly, differences in Gm18 frequencies between different bacteria can be observed, e.g., Haemophilus influenzae is lacking trmH (Fleischmann et al., 1995), but extensive systematic analysis of Gm content in bacterial tRNA is needed to address this point. It remains unknown whether bacteria can regulate their Gm18 modification pattern to cope with immune recognition by a mammalian host. Interestingly, many of the stimulatory E. coli tRNAs present in total tRNA, including, e.g., tRNAPhe and tRNALys, possess the relevant target residue G18 but are no substrates for TrmH and remain unmethylated. This is not untypical for tRNA MTases, which normally require a limited set of structural and sequence elements for substrate recognition (Motorin and Helm, 2011). Although the precise substrate requirements for TrmH are yet unknown, they are obviously present in tRNATyr but not in tRNAPhe and tRNALys. In turn, this points to the substrate specificity of TrmH as a potentially important contribution to the overall immunostimulation of total tRNA from E. coli in particular and from any given potential pathogen in general. Future research must elucidate whether broader substrate specificity and a correspondingly higher fraction of Gm18-containing tRNAs leads to a further decrease in TLR7 stimulation and, possibly, increased virulence of the respective bacterium. Also, future tests must include less canonical tRNA structures such as initiator tRNA, human mitochondrial tRNAs, or mutated tRNAs with deliberate mutations of tertiary structure to evaluate whether our findings apply to substrates without the classical tRNA structure context.
MATERIALS AND METHODS
tRNA preparation
Native tRNATyr1, tRNAPhe, tRNAAla1, tRNAAla2, tRNAMet, tRNAAsp, and tRNALys1 from E. coli had been purified as described previously (Holmes et al., 1975) and were repurified by PAGE after long-term storage. S. cerevisiae tRNAPhe and certain batches of tRNATyr1 were obtained from Sigma-Aldrich. Mammalian tRNAPhe and tRNALys3 were isolated as described previously (Keith and Dirheimer, 1978; Bénas et al., 2000). Fragments of yeast tRNAPhe originated from chemical cleavage via depurination of the wybutosine nucleotide at position 37, followed by strand scission under acidic conditions. These fragments had been isolated earlier (Sprinzl et al., 1976).
Total tRNA lacking Gm18 was prepared from strain CF4409, an E. coli K-12 derivate in which spoU/trmH 2′-O-MTase (Persson et al., 1997) had been deleted (ΔtrmH::kan). The parental strain CF897 was used as a control (Xiao et al., 1991). All strains were provided by M. Cashel (National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD). Bacteria were grown at 37°C for 6 h in Luria-Bertani medium supplemented with 50 µg/ml kanamycin. Bacteria were pelleted, frozen at −80°C overnight, and then lysed in 40 mg/ml lysozyme for 20 min. Total RNA was isolated using TRIZOL reagent (Invitrogen) according to the manufacturer’s instructions. RNA was separated by denaturing PAGE (10% acrylamide and 50% urea). The bands containing the tRNA were cut and shaken in 0.3 M sodium acetate overnight. tRNA was precipitated overnight and resuspended in RNase-free water. The concentration was controlled via UV absorbance.
tRNA synthesis by transcription
Unmodified full-length tRNAs were obtained by in vitro transcription with T7 RNA polymerase as described previously (Fechter et al., 1998) using a PCR-amplified template coding for the T7 promoter, an hh ribozyme, and the tRNA sequence. Synthetic oligonucleotides (IBA) used for transcription of E. coli tRNAs were as follows: forward primer (for all tRNAs), 5′-CGCGCGAAGCTTAATACGACTCACTATA-3′; template Tyr, 5′-TGGTGGTGGGGGAAGGATTCGAACCTTCGAAGTCTGTGACGGCAGATTTACAGTCTGCTCCCTTTGGCCGCTCGGGAACCCCACCGACGGTACCGGGTACCGTTTCGTCCTCACGGACTCATCAGGGTGGGGTTTCTCCCTATAGTGAGTCGTATT-3′; reverse primer Tyr, 5′-TGGTGGTGGGGG-3′; template Ala1, 5′-TGGTGGAGCTATGCGGGATCGAACCGCAGACCTCCTGCGTGCAAAGCAGGCGCTCTCCCAGCTGAGCTATAGCCCCGACGGTACCGGGTACCGTTTCGTCCTCACGGACTCATCAGGGGGCTATATCTCCCTATAGTGAGTCGTATT-3′; reverse primer Ala1, 5′-TGGTGGAGCTATGC-3′; template Phe, 5′-TGGTGCCCGGACTCGGAATCGAACCAAGGACACGGGGATTTTCAATCCCCTGCTCTACCGACTGAGCTATCCGGGCGACGGTACCGGGTACCGTTTCGTCCTCACGGACTCATCAGGCCCGGATATCTCCCTATAGTGAGTCGTATT-3′; reverse primer Phe, 5′-TGGTGCCCGGAC-3′; template Met, 5′-TGGTGGCTACGACGGGATTCGAACCTGTGACCCCATCATTATGAGTGATGTGCTCTAACCAACTGAGCTACGTAGCCGACGGTACCGGGTACCGTTTCGTCCTCACGGACTCATCAGGGCTACGTATCTCCCTATAGTGAGTCGTATT-3′; and reverse primer Met, 5′-TGGTGGCTACGAC-3′.
Upon transcription, tRNA precursor IVTs are generated, which self-process into full-length tRNAs by autocatalytic cleavage of the hh leader sequence creating a hydroxyl group at the 5′ end of the tRNA. RNAs were purified by denaturing PAGE, band excision, and elution of the processed tRNA. S. cerevisiae tRNAPhe was transcribed from a linearized plasmid containing the tRNAPhe gene (p67YF0; Sampson and Uhlenbeck, 1988), and 5 µg tRNA was 5′-dephosphorylated with 0.6 U/µl shrimp alkaline phosphatase in shrimp alkaline phosphatase buffer (10 mM Tris-HCl, pH 7.5, 10 mM MgCl2, and 0.1 mg/ml BSA) in a final volume of 50 µl for 1 h at 37°C and purified by phenol-ether extraction.
Synthesis of modivariants
DNAzyme catalyzed fragmentation of E. coli tRNATyr.
DNAzyme cycling reactions of 250-µl final volume contained 150 mM KCl, 10 mM MgCl2, 50 mM Tris-HCl, pH 7.4, 20 µg of either native or in vitro IVT E. coli tRNATyr, and a 2× molar excess of DNAzyme (5′-AGTCTGCTCTGTCAGCGACACGAATTTGGCCGCTCGGGAACCCC-3′). The mixture was subjected to 20 repetitions of temperature cycling: 30 s at 85°C, a temperature decrease by 0.5°C/s to 37°C, and 3 min at 37°C. DNAzyme was digested by directly adding DNase I (4–6 U/µg DNA substrate; Fermentas) to the reaction mixture and incubating for 1 h at 37°C.
Phosphorylation and dephosphorylation.
For 5′-phosphorylation and 3′-dephosphorlyation, 200 pmol of each fragment was incubated in Buffer A (50 mM Tris-HCl, pH 7.6, 10 mM MgCl2, 5 mM DTT, 0.1 mM spermidine, and 0.1 mM EDTA) supplemented with 2 mM ATP and 0.8 U/µl T4-PNK (Fermentas) in a final volume of 17–24 µl for 1 h at 37°C. The reaction mixture was directly used for subsequent steps.
Splinted ligation.
Ligation of prephosphorylated fragments was performed in a final volume of 50 µl, at final concentrations of 50 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 4 µM of each oligoribonucleotide, and 0.9 equivalents of DNA splint (5′-TGGTGGTGGGGGAAGGATTCGAACCTTCGAAGTCTGTGACGGCAGATTTACAGTCTGCTCCCTTTGGCCGCTCGGGAACCCCACC-3′). After heating for 4 min to 75°C and slow subsequent cooling to room temperature for 10 min, T4-DNA ligase (Fermentas) was added to 4 U/µl, and the mixture was incubated overnight at 16°C. The DNA splint was digested, and the ligation product was PAGE purified as described in tRNA synthesis by transcription. For the construction of modivariants Tyr Gm18, Gm34, and s4U8, synthetic oligonucleotides corresponding to nucleotides 1–24, 25–53, and 54–86 of the natural tRNA sequence and carrying appropriate 2′-O-Me or s4U modifications were used.
Synthesis of S. cerevisiae tRNAPhe modivariants.
Complementing fragments to the chemically derived native ones were generated via in vitro transcription from T7 hh templates as described in tRNA synthesis by transcription using the same forward primer, template 1, 5′-TGGTGCGAATTCTGTGGATCGAACACAGGACCTCCAGATCGACGGTACCGGGTACCGTTTCGTCCTCACGGACTCATCAGGATCTGGACTCTCCCTATAGTGAGTCGTATT-3′; reverse primer, 5′-TGGTGCGAATTCTGTGGATCGAAC-3′; template 2, 5′-CTTCAGTCTGGCGCTCTCCCAACTGAGCTAAATCCGCGACGGTACCGGGTACCGTTTCGTCCTCACGGACTCATCAGGCGGATTTATCTCCCTATAGTGAGTCGTATT-3′; and reverse primer, 5′-CTTCAGTCTGGCGCTCT-3′. Modivariants Phe Gm18, Cm32, Gm34, and CmGm were constructed of synthetic oligonucleotides corresponding to nucleotides 1–36 of the natural tRNA sequence, and carrying appropriate 2′-O-Me modification, nucleotides 37–52 and 53–76. Native fragments were 3′-dephosphorylated and 5′-phosphorylated as described in Phosphorylation and dephosphorylation. Ligation was performed as described in Splinted ligation, but at 37°C and using the DNA splint 5′-TGGTGCGAATTCTGTGGATCGAACACAGGACCTCCAGATCTTCAGTCTGGCGCTCTCCCAACTGAGCTAAATCCGCATTTTGGGTACC-3′.
Stimulation of PBMCs with tRNAs
Human PBMCs were isolated from heparinized blood of healthy donors by standard Ficoll-Hypaque density-gradient centrifugation (Bicoll 1.078 g/ml; Eberle et al., 2008). PBMCs were filtered through a 100-µm cell strainer and resuspended in complete medium prepared of RPMI 1640 supplemented with heat-inactivated (1 h, 56°C) 2% autologous serum. Cells were plated at 4 × 105 cells/well in a 96-well flat-bottom plate. 1 µg tRNA sample was diluted in a volume of 5 µl. The RNA was encapsulated with 2.5 µl of 1 mg/ml DOTAP (N-[1-(2, 3-dioleoyloxy)propyl]-N, N, N-trimethylammonium methylsulfate) by mixing with serum-free medium and incubation for 10 min. PBMCs were stimulated in a humidified 5% CO2 atmosphere at 37°C for 16 h. As internal positive control, PBMCs were stimulated with 1 µM of the TLR9-specific stimulus CpG2216. In inhibition experiments, two different tRNAs were mixed as indicated and added together to the cells. Cell-free supernatant was analyzed for secretion of IFN-α using a sandwich ELISA (Bender MedSystems).
Where indicated, cells were pretreated with 1 or 5 µM chloroquine (Sigma-Aldrich) for 30 min or with RNA oligonucleotide 2mA (5′-GMAMACUUCMAGGGUCMAGCUUGCCG-3′) to block endosomal TLRs or TLR7, respectively (Eberle et al., 2009). PBMCs were also tested when depleted of BDCA-4–positive pDCs through magnetic cell separation (MACS; Miltenyi Biotec) according to the manufacturer’s protocol. Induction of TNF by LPS stimulation was not affected through pDC depletion or any of the inhibitors. Analysis of stimulation of RIG-I or MDA5 was essentially performed as described previously (Eberle et al., 2009) by overexpressing these receptors in otherwise inactive Huh7.5 cells harboring an IRF3 reporter.
Knockdown of TLR7 in PBMCs to verify engagement of TLR7 by tRNAs was performed as follows: in brief, 2.5 pmol siRNA was incubated with Lipofectamine RNAiMax in Optimem in 96-well plates. PBMCs were plated after 20 min of incubation (105 cells/well) and placed at 37°C, 5% CO2. After 24 h, cells were stimulated with the respective tRNA or control.
Statistical analysis
Data were analyzed by the Prism 5 program (GraphPad Software). Significant differences were assessed by the Student’s t test. In all figures, the p-value is indicated by *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Online supplemental material
Fig. S1 shows the preparation of modivariants mv#3 and mv#4 of S. cerevisiae tRNAPhe, similar to the scheme in Fig. 3 except that acid hydrolysis was used instead of a DNAzyme to site-specifically cleave native tRNA, and also shows stimulation data of these modivariants mv#3 and mv#4, as well as of one more construct tRNAPheGmCm, which point to the identification of 2′-O-Me modifications in the anticodon loop of tRNAPhe as being important for silencing of the IFN-α response to this tRNA. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20111044/DC1.
Acknowledgments
We thank René Karayilan for excellent technical support and Michael Cashel for providing us with bacterial strains.
This work has been supported by supported by contract research “Methoden für die Lebenswissenschaften” of the Baden-Württemberg Stiftung (P-LS-Meth/10 to A.H. Dalpke and M. Helm).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- hh
- hammerhead
- IVT
- in vitro transcript
- MTase
- methyltransferase
- pDC
- plasmacytoid DC
- siRNA
- small interfering RNA
- TLR
- Toll-like receptor
- tRNA
- transfer RNA
References
- Bénas P., Bec G., Keith G., Marquet R., Ehresmann C., Ehresmann B., Dumas P. 2000. The crystal structure of HIV reverse-transcription primer tRNA(Lys,3) shows a canonical anticodon loop. RNA. 6:1347–1355 10.1017/S1355838200000911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk G.R. 1995. Genetic dissection of synthesis and function of modified nucleosides in bacterial transfer RNA. Prog. Nucleic Acid Res. Mol. Biol. 50:263–338 10.1016/S0079-6603(08)60817-X [DOI] [PubMed] [Google Scholar]
- Cantara W.A., Crain P.F., Rozenski J., McCloskey J.A., Harris K.A., Zhang X., Vendeix F.A., Fabris D., Agris P.F. 2011. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 39:D195–D201 10.1093/nar/gkq1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerwoniec A., Dunin-Horkawicz S., Purta E., Kaminska K.H., Kasprzak J.M., Bujnicki J.M., Grosjean H., Rother K. 2009. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 37:D118–D121 10.1093/nar/gkn710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle F., Giessler K., Deck C., Heeg K., Peter M., Richert C., Dalpke A.H. 2008. Modifications in small interfering RNA that separate immunostimulation from RNA interference. J. Immunol. 180:3229–3237 [DOI] [PubMed] [Google Scholar]
- Eberle F., Sirin M., Binder M., Dalpke A.H. 2009. Bacterial RNA is recognized by different sets of immunoreceptors. Eur. J. Immunol. 39:2537–2547 10.1002/eji.200838978 [DOI] [PubMed] [Google Scholar]
- Fechter P., Rudinger J., Giegé R., Théobald-Dietrich A. 1998. Ribozyme processed tRNA transcripts with unfriendly internal promoter for T7 RNA polymerase: production and activity. FEBS Lett. 436:99–103 10.1016/S0014-5793(98)01096-5 [DOI] [PubMed] [Google Scholar]
- Fleischmann R.D., Adams M.D., White O., Clayton R.A., Kirkness E.F., Kerlavage A.R., Bult C.J., Tomb J.F., Dougherty B.A., Merrick J.M., et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 269:496–512 10.1126/science.7542800 [DOI] [PubMed] [Google Scholar]
- Grosjean H. 2009. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Landes Bioscience, Austin, TX. 682 pp. [Google Scholar]
- Hengesbach M., Meusburger M., Lyko F., Helm M. 2008. Use of DNAzymes for site-specific analysis of ribonucleotide modifications. RNA. 14:180–187 10.1261/rna.742708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes W.M., Hurd R.E., Reid B.R., Rimerman R.A., Hatfield G.W. 1975. Separation of transfer ribonucleic acid by sepharose chromatography using reverse salt gradients. Proc. Natl. Acad. Sci. USA. 72:1068–1071 10.1073/pnas.72.3.1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H., Suzuki T., Sugawara K., Inoue Y., Shibata T., Kuramitsu S., Yokoyama S., Oshima T., Watanabe K. 2002. Identification and characterization of tRNA (Gm18) methyltransferase from Thermus thermophilus HB8: domain structure and conserved amino acid sequence motifs. Genes Cells. 7:259–272 10.1046/j.1365-2443.2002.00520.x [DOI] [PubMed] [Google Scholar]
- Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.K., Schlee M., et al. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 314:994–997 10.1126/science.1132505 [DOI] [PubMed] [Google Scholar]
- Jühling F., Mörl M., Hartmann R.K., Sprinzl M., Stadler P.F., Pütz J. 2009. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 37:D159–D162 10.1093/nar/gkn772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó K., Weissman D. 2007. Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implication for therapeutic RNA development. Curr. Opin. Drug Discov. Devel. 10:523–532 [PubMed] [Google Scholar]
- Karikó K., Buckstein M., Ni H., Weissman D. 2005. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 23:165–175 10.1016/j.immuni.2005.06.008 [DOI] [PubMed] [Google Scholar]
- Keith G., Dirheimer G. 1978. The primary structure of rabbit, calf and bovine liver tRNAPhe. Biochim. Biophys. Acta. 517:133–149 [DOI] [PubMed] [Google Scholar]
- Koski G.K., Karikó K., Xu S., Weissman D., Cohen P.A., Czerniecki B.J. 2004. Cutting edge: innate immune system discriminates between RNA containing bacterial versus eukaryotic structural features that prime for high-level IL-12 secretion by dendritic cells. J. Immunol. 172:3989–3993 [DOI] [PubMed] [Google Scholar]
- Kumagai I., Watanabe K., Oshima T. 1980. Thermally induced biosynthesis of 2′-O-methylguanosine in tRNA from an extreme thermophile, Thermus thermophilus HB27. Proc. Natl. Acad. Sci. USA. 77:1922–1926 10.1073/pnas.77.4.1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H., Kawai T., Akira S. 2011. Pathogen recognition by the innate immune system. Int. Rev. Immunol. 30:16–34 10.3109/08830185.2010.529976 [DOI] [PubMed] [Google Scholar]
- Kurschat W.C., Müller J., Wombacher R., Helm M. 2005. Optimizing splinted ligation of highly structured small RNAs. RNA. 11:1909–1914 10.1261/rna.2170705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y., Helm M. 2010. tRNA stabilization by modified nucleotides. Biochemistry. 49:4934–4944 10.1021/bi100408z [DOI] [PubMed] [Google Scholar]
- Motorin Y., Helm M. 2011. RNA nucleotide methylation. Wiley Interdiscip Rev RNA. 2:611–631 10.1002/wrna.79 [DOI] [PubMed] [Google Scholar]
- Persson B.C., Jäger G., Gustafsson C. 1997. The spoU gene of Escherichia coli, the fourth gene of the spoT operon, is essential for tRNA (Gm18) 2′-O-methyltransferase activity. Nucleic Acids Res. 25:4093–4097 10.1093/nar/25.20.4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins M., Judge A., Liang L., McClintock K., Yaworski E., MacLachlan I. 2007. 2′-O-methyl-modified RNAs act as TLR7 antagonists. Mol. Ther. 15:1663–1669 10.1038/sj.mt.6300240 [DOI] [PubMed] [Google Scholar]
- Sampson J.R., Uhlenbeck O.C. 1988. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. USA. 85:1033–1037 10.1073/pnas.85.4.1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioud M., Furset G., Cekaite L. 2007. Suppression of immunostimulatory siRNA-driven innate immune activation by 2′-modified RNAs. Biochem. Biophys. Res. Commun. 361:122–126 10.1016/j.bbrc.2007.06.177 [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Wagner T., Lorenz S., Erdmann V.A. 1976. Regions of tRNA important for binding to the ribosomal A and P sites. Biochemistry. 15:3031–3039 10.1021/bi00659a015 [DOI] [PubMed] [Google Scholar]
- Tluk S., Jurk M., Forsbach A., Weeratna R., Samulowitz U., Krieg A.M., Bauer S., Vollmer J. 2009. Sequences derived from self-RNA containing certain natural modifications act as suppressors of RNA-mediated inflammatory immune responses. Int. Immunol. 21:607–619 10.1093/intimm/dxp030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H., Kalman M., Ikehara K., Zemel S., Glaser G., Cashel M. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980–5990 [PubMed] [Google Scholar]